Abstract
Heteroplasmy, the presence of more than one type of mtDNA within cells, is common in animals and has been associated with aging and disease in humans. Changes in the frequencies of mtDNA variants between cell and animal generations thus bears on the evolution of mtDNA and the progression of diverse pathologies. We have used densitometry of Southern blots of individual heteroplasmic Drosophila melanogaster to study the effects of age, increased egg production after mating, and temperature on evolution of heteroplasmy within and between generations. The frequency of the longer mtDNA variant consistently increased between early and late cohorts of F1 offspring derived from 18 independent heteroplasmic mothers as they aged. Neither temperature (flies maintained at 25°C and 18°C) nor the holding of flies as virgins for 10 days before mating had significant effects on transmission patterns. However, at the ends of their lives, flies that had laid eggs at 25°C had a greater frequency of the long mtDNA than did their siblings who had laid eggs at 18°C. The evolution of heteroplasmy within a generation was studied in samples of siblings that either were mated or held as virgins, and then scored for mtDNA haplotype frequencies at two different ages (day 2 and day 14). Mated flies showed a significantly greater increase in the frequency of the long mtDNA variant with age than did the unmated flies. This system provides a model for the joint analysis of generational and chronological age in the transmission dynamics of a molecular polymorphism.
In the past decade direct evidence for mtDNA heteroplasmy, the presence of more than one type of mtDNA in cells, has been increasing rapidly from two very different lines of study. Surveys of mtDNA variation in natural populations of animals have revealed that heteroplasmy for mtDNA length variation is quite common. Much of this variation is caused by insertions, deletions, or duplications in and around the control region of the molecule (containing the origin of replication and transcription; reviewed in ref. 1). In humans, examples of mtDNA heteroplasmies have been reported involving both deletions of large coding regions and point mutations in individual genes (2, 3). Although no strong fitness effects have been attributed to mtDNA size variation in the control region (e.g., ref. 4), this type of variation has been useful in documenting the dynamics of cytoplasmic inheritance (5–7). In contrast, human heteroplasmies have been shown to accumulate in different tissues with increasing age, and the presence of specific mtDNA mutations has been linked to particular neuromuscular diseases (2, 8–10). Because the severity of some of these diseases is a function of the proportion of mutant mtDNAs within the cytoplasm, and smaller mtDNAs are thought to outreplicate wild-type molecules (2), information on the fate of variant mtDNA molecules in successive generations of heteroplasmic cells is central to the genetics and evolution of mtDNA as well as the progression of mitochondrial diseases.
Age-related effects of mtDNA mutations have been examined primarily in somatic tissues (9, 11). Although inter-generational studies have played an important role in the identification of mitochondrial diseases (caused by the maternal inheritance of mtDNA), less attention has been focused on the effects of age on mtDNA mutations in the germ line (8, 12, 13). Because mutations in mtDNA may increase with age, age of reproduction may influence the frequency and occurrence of heteroplasmy transmitted to the offspring, which may be related to the health of offspring born early or late in the life of the mother. However, strong selection against mutant mtDNAs, or cells carrying too many mutant mtDNAs, may be important in proper implantation and development of early mammalian embryos (8, 14). This sort of “cleansing” process may have been an important force in the evolution of both aging and organelle genomes (e.g., ref. 15).
Here we use strains of Drosophila melanogaster heteroplasmic for two mtDNA length variants that differ in the A+T-rich control region of the molecule to describe the fate of mtDNA variants within and between generations. By using densitometry of Southern blots of single flies, we document a consistent increase in the frequency of the large mtDNA length variant between early and late cohorts of offspring from single heteroplasmic females, confirming an earlier study with D. mauritiana (6). We extend this analysis to mated and unmated flies of different ages and show that the increase of the long mtDNA is significantly greater in mated flies. This system provides a model for the joint analysis of generational and chronological age in the transmission dynamics of a mtDNA polymorphism.
MATERIALS AND METHODS
Fly Strains.
D. melanogaster were collected in Brownsville, TX (16), obtained from Larry Hale (McMaster University, Hamilton, ONT, Canada), and maintained in population cages. Subsequent Southern blot analysis of individual flies from the laboratory populations revealed flies carrying spontaneously generated heteroplasmy for mtDNA size variation in the A+T-rich control region. To obtain heteroplasmic stocks, isofemale lines were started from the population cages, two of which were used in the current experiments. The 4-19D4 stock contains flies that are heteroplasmic for ≈19.5-kilobase (kb) and ≈20.4-kb mtDNA length variants, and the 5-27H3 stock contains flies heteroplasmic for ≈19.5 kb and ≈20.0 kb. MtDNA length variation in D. melanogaster is caused by the presence of 464-bp (and smaller) repeats in the A+T-rich region of the molecule (17). The long and short mtDNAs in the heteroplasmic lines likely differ by one (5-27H3) and two repeat units (4-19D4).
Southern Blot Analysis.
Southern blots were used because they were determined to be more accurate than a PCR approach. Individual females were ground in a 1.5-ml Eppendorf tube with a pestle in 25 μl of grinding buffer (0.2 M sucrose/50 mM EDTA/0.1 M Tris/0.5% SDS, pH 9.0) and 0.5 μl of diethyl-pyrocarbonate. The subsequent isolation and digestion of DNA followed standard procedures (18, 19). DNA from single flies was digested with HaeIII in 20-μl reactions according to the specifications of the supplier (New England Biolabs; although 0.5 μl of 10 mg/ml RNase was added to each digest). DNA fragments were separated on a 0.7% agarose-Tris acetate, and Southern blots were prepared according to protocols provided with the nylon membrane (Micron Separations, Westboro, MA) and the pressure blotting device (Stratagene).
The probe used to detect the mtDNA length variation was a clone of the 4.8-kb HindIII fragment of purified D. melanogaster mtDNA obtained by using standard methods (19). This probe fragment was gel-purified after HindIII digestion and ≈20 ng then was labeled with dAT32P by using the Prime-It II random priming kit from Stratagene. This probe hybridizes to unique-sequence DNA in the length-variable HaeIII fragment and to the constant-sized 6.2-kb band in all flies (see Fig. 1).
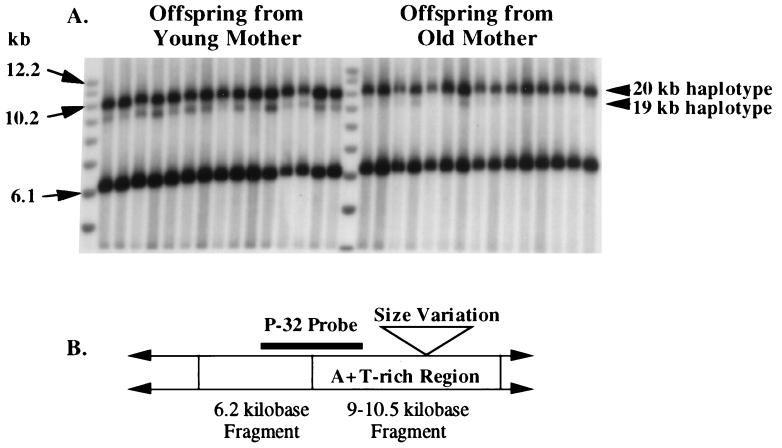
Figure 1.
(A) Southern blot of individuals D. melanogaster F1 offspring from a single heteroplasmic mother. (B) Restriction map of D. melanogaster mtDNA near the A+T rich region. Vertical lines between arrows indicate HaeIII sites used to cleave total genomic DNA. The P-32 probe is a HindIII fragment of D. melanogaster mtDNA.
Autoradiography and Densitometer Tracing.
Washed membranes were exposed to Fuji x-ray film with two intensifying L-Plus screens. Two exposures were obtained for each gel in most experiments. The images were scanned with an imaging densitometer (model GS-670, Bio-Rad) interfaced with a Macintosh computer. By using auto volume integration with a general background subtraction (molecular analyst Software, Version 2.0; Bio-Rad), relative frequencies of the two length variants were estimated from the relative band areas on the scanned image. The averages of the values from the replicate autoradiographs were used as the estimates of the frequencies of mtDNA length variants within a single fly (i.e., a fly’s heteroplasmic “genotype”).
Female Age and the Inheritance of Heteroplasmy.
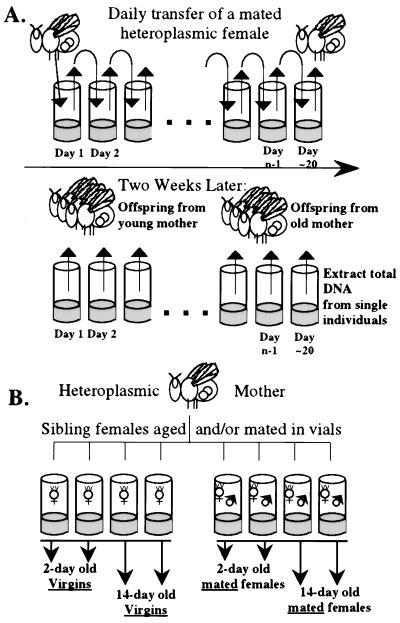
Individual female virgins were collected from heteroplasmic mothers and placed in yeasted vials with a single sibling male at 1 day of age, and each vial was assigned a subline code (such as 4-19D4-D1 or 5-27H3-D). These vials were divided randomly into two sets of isofemale lines to be maintained at different temperatures (18°C and 25°C). All mated pairs were changed to a new vial every other day until the female no longer produced many eggs (about 20 days). This action resulted in samples of 2-day cohorts of F1 offspring from each heteroplasmic female through most of her adult life (for two culture temperatures). After about 20 days, the females (“mothers”) were stored at −80°C until DNA analysis (see Fig. 2A). Each female subline thus is treated as a “replicate” (allowing for some variation in heteroplasmy among sublines caused by cytoplasmic transmission).
Figure 2.
(A) Aging protocol for transmission of mtDNA length variants. This culture protocol was followed at both 25°C and 18°C with the mothers used for the two temperature treatments being siblings of the same heteroplasmic female. The data reported in Table 1 were obtained from the offspring generation, and the data reported in Table 2 were obtained from the mothers after they had laid eggs. (B) Aging and mating protocol. This procedure was replicated across different female sublines.
Some of the virgins collected to start this age- and temperature-specific transmission experiment were held for 10 days at 18°C before they were sib-mated to compare mtDNA transmission between females who were mated immediately and those that hold their eggs before mating. The culture protocol for the “held” flies was the same as described above (see Fig. 2A).
Female Age, Mating, and the Change in Heteroplasmy Within a Generation.
Samples of unmated and mated sibling flies of different ages were examined for heteroplasmy to study the change in frequency of mtDNA haplotypes as heteroplasmic individuals age. Virgin females were collected from four different heteroplasmic females (each within the 4-19D4 line), and each sample was split randomly into two groups. Flies from one group were allowed to age as virgins (one fly per vial; changed every other day). Flies from the other group were sib-mated and allowed to mate and lay eggs as they aged (one pair per vial; changed every other day). On day 2, half of the virgin flies and half of the mated flies were frozen. The remaining flies were allowed to age until day 14 when they were frozen for later DNA analysis (see Fig. 2B). The structure of these data is as follows: female line is a random effect (model II ANOVA), mating status and age status are fixed effects (model I ANOVA), both of which are nested within female line (20). Two-factor nested ANOVA was performed on arcsin-square root transformed frequency data, but the untransformed frequencies are presented in Fig. 3. Mated flies will lay approximately 50–100 eggs per day through age 14 days and continually turn over oocytes from each of their ovarioles. Unmated females will lay a few eggs per day but the production of new cytoplasm by the ovarioles is dramatically reduced (ref. 21, see Discussion).
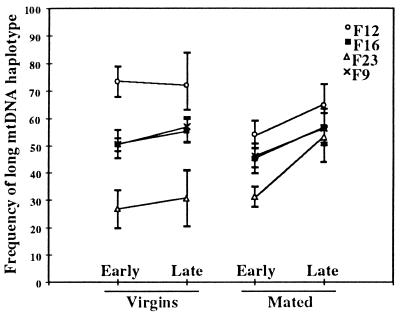
Figure 3.
The effect of mating and age on the proportion of long mtDNA within individual D. melanogaster. See Table 3 for statistical analysis. Means (symbols) and standard deviations (between bars) for samples of offspring from four different heteroplasmic mothers (F9, F12, F16, and F23). Each data point reflects the proportion of long mtDNA in a sample of about seven F1 female offspring from the respective mother. Early = day 2 and late = day 14.
We note an important caveat in these experiments. By using mated vs. unmated flies as a means of manipulating egg turnover we are introducing other uncontrolled factors that may affect the fitness of the experimental females (22, 23). But because there was no death observed between any of our early (day 2) and late (day 14) samples we feel that, from the perspective of mtDNA length variants in the germ line, the mating treatment had its most significant effect on egg (cytoplasm) turnover.
RESULTS
An Age-Specific Transmission Bias of mtDNA Length Variants.
Fig. 1 shows that as a female ages her later cohort of offspring has a greater proportion of long mtDNA than does her early cohort. This same pattern was observed in 16 of 18 heteroplasmic lines of D. melanogaster studied; in 12 of these 16 cases the increase was significant at the 5% level or higher (Table 1; although there is a multiple comparison problem across this table, a Wilcoxon signed-rank test shows that 16 increases of 18 is highly significant, Ts = 4; P < 0.0001). This result holds for the two different stocks of heteroplasmic flies (4-19 and 5-27), which have the same-sized small mtDNA (≈19.5 kb) but different large mtDNAs (≈20.4 and ≈20.0 kb, respectively). Holding the mothers as virgins before mating them does not appear to alter the patterns of transmission dramatically (see bottom section of Table 1). The variance among offspring was not consistently lower in the early cohort as compared with the late cohort (see Table 1). This finding is in contrast to data for D. mauritiana where an increase in the variance in late F1 cohorts has been reported (6).
Table 1.
Mitochondrial genotype frequencies in early and late cohorts of F1 offspring from heteroplasmic D. melanogaster mothers carrying two different length variants of mtDNA
| Line-subline | Temp | Early lay; late lay | Offspring, %L ± var | n | Δ%L, Late-early | Per-day, Δ%L | U | P < |
|---|---|---|---|---|---|---|---|---|
| 4-19D4-D1 | 25°C | 0 | 0.771 ± 0.0038 | 14 | 0.097 | 0.00691 | 183 | 0.001 |
| 15 | 0.868 ± 0.0040 | 15 | ||||||
| D2 | 25°C | 0 | 0.911 ± 0.0036 | 13 | −0.053 | −0.00355 | 116 | 0.1 |
| 15 | 0.858 ± 0.0074 | 13 | ||||||
| G4 | 25°C | 2 | 0.922 ± 0.0018 | 11 | 0.040 | 0.00199 | 129 | 0.005 |
| 22 | 0.962 ± 0.0002 | 14 | ||||||
| H4 | 25°C | 2 | 0.845 ± 0.0031 | 10 | 0.098 | 0.00464 | 207 | 0.001 |
| 23 | 0.944 ± 0.0004 | 14 | ||||||
| B2 | 18°C | 2 | 0.734 ± 0.0048 | 15 | 0.161 | 0.00845 | 146 | 0.001 |
| 21 | 0.895 ± 0.0067 | 10 | ||||||
| D1F | 18°C | 2 | 0.760 ± 0.0114 | 13 | 0.068 | 0.00676 | 121 | 0.1 |
| 12 | 0.828 ± 0.0052 | 14 | ||||||
| D3 | 18°C | 3 | 0.726 ± 0.0048 | 15 | 0.125 | 0.00889 | 201 | 0.001 |
| 17 | 0.850 ± 0.0058 | 15 | ||||||
| D4 | 18°C | 3 | 0.796 ± 0.0037 | 15 | 0.042 | 0.00301 | 116 | 0.11 |
| 17 | 0.838 ± 0.0022 | 12 | ||||||
| G2 | 18°C | 2 | 0.905 ± 0.0072 | 12 | 0.058 | 0.00276 | 152 | 0.001 |
| 23 | 0.963 ± 0.0014 | 15 | ||||||
| H1 | 18°C | 2 | 0.819 ± 0.0025 | 15 | 0.110 | 0.00733 | 222 | 0.001 |
| 17 | 0.929 ± 0.0012 | 15 | ||||||
| 5-27H3-D | 25°C | 0 | 0.488 ± 0.0041 | 15 | 0.105 | 0.00955 | 160 | 0.004 |
| 11 | 0.593 ± 0.0090 | 13 | ||||||
| J | 25°C | 0 | 0.532 ± 0.0010 | 10 | 0.131 | 0.01191 | 88 | 0.001 |
| 11 | 0.663 ± 0.0031 | 9 | ||||||
| O | 25°C | 0 | 0.521 ± 0.0024 | 12 | 0.068 | 0.00618 | 96 | 0.02 |
| 11 | 0.589 ± 0.0038 | 11 | ||||||
| Mothers held as virgins for 10 days before mating | ||||||||
| 4-19D4-B6 | 25°C | 0 | 0.839 ± 0.0086 | 13 | 0.154 | 0.01029 | 195 | 0.001 |
| 15 | 0.993 ± 0.0001 | 13 | ||||||
| D5 | 25°C | 0 | 0.916 ± 0.0037 | 15 | 0.042 | 0.00219 | 142 | 0.07 |
| 19 | 0.958 ± 0.0019 | 14 | ||||||
| B8 | 18°C | 0 | 0.636 ± 0.0056 | 12 | −0.002 | −0.00011 | 88 | 0.7 |
| 15 | 0.635 ± 0.0074 | 14 | ||||||
| G8 | 18°C | 0 | 0.839 ± 0.0058 | 14 | 0.055 | 0.01363 | 121 | 0.1 |
| 4 | 0.893 ± 0.0112 | 13 | ||||||
| H7 | 18°C | 0 | 0.880 ± 0.0035 | 10 | 0.054 | 0.00385 | 98 | 0.01 |
| 14 | 0.934 ± 0.0010 | 12 | ||||||
Line and subline refer to the strain and mother within strain, respectively, of D. melonogaster (see Materials and Methods). Temp. refers to the temperature at which the mothers laid eggs and the eggs developed to emergence of F1 adult. Early and late lay refer to the age of the heteroplasmic mothers (in days) at the start of a 2-day period of egg collection. Heteroplasmic genotypes are expressed as the mean percent of the long (L) mtDNA among individual flies in a sample of offspring ± the variance (n = sample size of F1 off spring). Δ%L is the change in frequency of the L-mtDNA between the early and late cohorts. The per-day Δ%L is the value of the Δ%L divided by the difference in age of the mother between the early and late lay dates. The U value is for Mann–Whitney tests between early and late cohorts.
Temperature does not appear to have a significant effect on this pattern of transmission between generations (mean and SD of the frequency change per day for 18°C and 25°C are 0.0064 ± 0.0024 and 0.0025 ± 0.0045, respectively; not significant). In apparent contradiction to these transmission data, temperature did appear to have an effect on the proportion of mtDNA length variants within a fly during its lifetime (see Table 2). Heteroplasmic mothers who were mated and laid eggs at 25°C for 15–20 days showed significantly greater proportions of the long mtDNA than did samples of their siblings maintained in a similar way, but at 18°C. Note that many of the flies used to obtain the intrageneration data in Table 2 were mothers of the flies used to obtain the transmission data for Table 1; see Materials and Methods).
Table 2.
The effect of temperature on the proportion of long mtDNA within heteroplasmic flies
| Line | 25°C %L | 18°C %L | U | P < |
|---|---|---|---|---|
| 4-19D4B | 0.842 | 0.732 | 12 | 0.035 |
| 0.791 | 0.748 | |||
| 0.762 | 0.659 | |||
| 0.778 | ||||
| Mean | 0.793 | 0.713 | ||
| SD | 0.035 | 0.047 | ||
| 4-19D4C | 0.922 | 0.890 | 5.5 | 0.140 |
| 0.890 | 0.740 | |||
| 0.790 | ||||
| Mean | 0.906 | 0.807 | ||
| SD | 0.023 | 0.076 | ||
| 4-19D4G | 0.815 | 0.795 | 9 | 0.050 |
| 0.857 | 0.750 | |||
| 0.859 | 0.695 | |||
| Mean | 0.844 | 0.747 | ||
| SD | 0.025 | 0.050 | ||
| 4-19D4H | 0.713 | 0.627 | 12 | 0.035 |
| 0.710 | 0.588 | |||
| 0.711 | 0.653 | |||
| 0.604 | ||||
| Mean | 0.711 | 0.618 | ||
| SD | 0.002 | 0.028 |
For each heteroplasmic female identified in the Line column, a sample of sibling females were split randomly into two groups, mated individually, placed at 18°C or 25°C, and allowed to lay eggs for >15 days. Each number indicates the proportion of long mtDNA within a single female fly. The U value is for the Mann–Whitney test between samples of siblings maintained at the two temperatures.
The Effect of Mating and Age on Heteroplasmy Within a Generation.
Heteroplasmic females who were mated and allowed to lay eggs as they aged showed significantly greater increases in the proportions of the long mtDNA after 14 days than did a comparable sample of sibling females who were aged as virgins for 14 days (both groups were maintained at 25°C; Fig. 3). In an ANOVA among 102 genotyped individuals from the four female lines, all effects were highly significant (see Table 3). The female line effect indicates that the four different females had different initial frequencies of the long mtDNAs, which is evident in Fig. 3 (the variation among means is not significant by an F test). The single effects of mating and age are strong individually. The interaction effect is also highly significant, indicating that the difference between young and old samples is different between mated and unmated flies.
Table 3.
Nested ANOVA for frequencies of the long mtDNA haplotype
| Source | df | Sum of squares | Mean square | F value | P < |
|---|---|---|---|---|---|
| Female line | 3 | 1.220 | 0.407 | 91.93 | 0.0001 |
| Mating status within female line | 4 | 0.248 | 0.062 | 14.00 | 0.0001 |
| Age status within female line | 4 | 0.210 | 0.052 | 11.86 | 0.0001 |
| Mating status, age status within female line* | 4 | 0.077 | 0.019 | 4.360 | 0.0030 |
| Residual | 86 | 0.380 | 0.004 |
Frequencies were arcsin square root transformed before analysis.
*Mating status is mated vs. unmated; age status is day 2 vs. day 14.
More than 650 individual D. melanogaster were quantified by densitometry of Southern blots. In no case did we observe bands indicating novel mtDNA length variants, suggesting that mutation was not a major force, relative to replication or segregation of mtDNAs, in the frequency changes we report.
DISCUSSION
Genetics of mtDNA Length Variants.
These data bear on a number issues relating to mitochondrial genetics and evolution. First, why does the long mtDNA variant increase with age both within and between generations? Previous studies in Drosophila (5) and crickets (7) have shown that the smaller of two mtDNA length variants within a germ line increase in frequency between generations. These reports are consistent with the notion that mitochondrial genomes of animals have been shaped by selection for small size, perhaps driven by a “race for replication” (1, 24). The consistent increase in the frequency of the longer mtDNA variant reported here thus seems inconsistent with the race for replication scenario. However, our data are consistent with data reported by Solignac et al. (6) for D. mauritiana in which the frequency of a long mtDNA variant was observed to increase in successive cohorts of flies descended from heteroplasmic females. Because our study included two different heteroplasmic strains (4-19D4 and 5-27H3) whose long mtDNAs differ in size, and both of these strains differ from the heteroplasmy examined in D. mauritiana, the age-dependent increase in the proportion of long mtDNA is well-supported for Drosophila and probably for insects.
At the cytoplasmic level the argument that small plasmid-like genomes should outcompete longer genomes seems sound if the only relevant factor is the total amount of DNA to replicate. It has been suggested that the severity of some mitochondrial diseases are caused by the preferential replication of smaller mtDNAs in cells heteroplasmic for normal and deletion mutant genomes (2). Because the length variation we have studied here is caused by variable number of tandem repeats (VNTRs) in the A+T rich region of mtDNA that contains the origin of replication, there may be a trade-off between the initiation and the completion of replication. Additional repeats may stimulate the binding of polymerase and more than compensate for the additional base pairs of DNA that need to be replicated (1). Suggestive of this trade-off is evidence for the maintenance of heteroplasmy of a normal and a deletion-mutant mtDNA lacking 10.4 kb (63% of the genome) in a pedigree of patients with diabetes and reduced oxidative phosphorylation. The smaller mtDNA in this case was lacking the light strand origin of replication, which may have offset the replicative advantage of the deleted mitochondrial genome (2, 25).
Germ-Line Turnover.
Our observation that the increase in frequency of the long mtDNA as a fly ages is significantly greater in mated than in unmated females suggests that some aspect of egg production is related to this phenomenon. Mated females can lay up to 100 eggs per day, but this number drops off continually as the female ages. In D. melanogaster that are held as virgins after eclosing there is a significant reduction in the rate of egg laying, relative to mated flies (reviewed in ref. 21, pp. 140–149). Although mature oocyte production is arrested in held virgins, stem cell division is not shut down completely (26–28). From these observations we can conclude that holding a fly as a virgin reduces considerably the turnover of germ-line cytoplasm, but does not reduce it to zero.
Our data are consistent with the qualitative information about cytoplasmic turnover in the Drosophila germ line. Flies held as virgins for 14 days did exhibit a slight increase in the proportion of the long mtDNA, but mating significantly increases this rate of increase (see Fig. 3; Table 3). As such, the pattern of mtDNA frequency shifts we have seen may have less to do with chronological age than with cell-generational age determined by the amount of cytoplasmic turnover occurring in a given cell lineage. Although we were not able to distinguish between the mtDNA in somatic vs. germ-line tissues in individual flies, it is likely that the ratio of mtDNA length variants quantified in whole-fly DNA extracts primarily reflect that in oocytes because these tissues are rich in mitochondria and comprise a substantial proportion of the total cytoplasm of a mature female. Moreover, cytoplasmic turnover is likely to be greatest in the production of mature oocytes, relative to other tissues in the fly (which has determinant growth). Thus our data and these observations lead us to the working hypothesis that the rate of age-specific increase in long mtDNAs is a function of the amount of mtDNA turnover in tissues, itself driven by the number of cell generations per unit time, necessitating a replenishment of the cytoplasmic population of mtDNAs. Interestingly, there are a number of different levels at which these frequency shifts could operate: differential replication of mtDNAs, differential inheritance of mitochondria, differential survivorship of oocytes or even larvae and adults.
Mitochondrial Mutations and Aging.
There have been many reports in the literature suggesting a link between mtDNA mutations and the aging process (2, 3, 8–11). Although the evidence for an increase in the proportion of mutant mtDNAs with age is plentiful, compelling evidence for a causal association between the presence of these mutations and true senescence (physiological decay with age) is less common. The lack of any strong phenotype associated with the age-specific changes in heteroplasmy we describe makes it more likely that aging (defined as increasing numbers of cell generations) drives the changes in heteroplasmy, rather than the reverse.
Relatively little is known about the relationship between chronological age and cell-generational age in terms of mtDNA mutations in the germ line. Is there an increased likelihood that late reproduction may increase the probability of transmitting acquired, mutant mtDNAs (see ref. 13)? Or is the population of mtDNAs within a germ line effectively purified before it is transmitted to the next generation (cf. refs. 8 and 15)? Answers to these questions may shed light on the ongoing controversy over the importance of mutation accumulation versus antagonistic pleitropy in the evolution of aging (29–31).
The very distinct patterns of germ-line determination in different organisms (32) also may affect the opportunity for the transmission of accumulated mitochondrial mutations (7). Although there are clear differences between mammals and Drosophila in the biology of oocyte maturation (the lifetime supply of oocytes in mammals are produced around the age of birth, whereas flies continually produce oocytes), in both groups oocytes are arrested in mid-meiosis before fertilization. Because virgin flies hold an initial clutch of eggs if they are not mated, the differences between mammals and flies can be reduced under appropriate experimental conditions. Drosophila thus offers a manipulative system in which the distinction between the causes and effects of aging and its relationship to mitochondrial genetics may be made.
Acknowledgments
We thank Daniel Promislow, Marc Tatar, and an anonymous reviewer for comments on an earlier version. This work was supported by Grants DEB 9120293 and DEB 9707676 from the National Science Foundation to D.M.R.
ABBREVIATION
- kb
kilobase
References
- 1.Rand D M. J Mol Evol. 1993;37:281–295. doi: 10.1007/BF00175505. [DOI] [PubMed] [Google Scholar]
- 2.Wallace D C. Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 3.Wallace D C. Sci Am. 1997;277:40–47. doi: 10.1038/scientificamerican0897-40. [DOI] [PubMed] [Google Scholar]
- 4.Zouros E, Pogson G H, Cook D I, Dadswell M J. Evolution. 1992;46:1466–1476. doi: 10.1111/j.1558-5646.1992.tb01137.x. [DOI] [PubMed] [Google Scholar]
- 5.Solignac M, Genermont J, Monnerot M, Mounolou J. Mol Gen Genet. 1984;197:183–188. [Google Scholar]
- 6.Solignac M, Genermont J, Monnerot M, Mounolou J. Genetics. 1987;117:687–696. doi: 10.1093/genetics/117.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rand D M, Harrison R G. Genetics. 1986;114:955–970. doi: 10.1093/genetics/114.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linnane A W, Zhang C, Baumer A, Nagley P. Mutat Res. 1992;275:195–208. doi: 10.1016/0921-8734(92)90023-i. [DOI] [PubMed] [Google Scholar]
- 9.Kadenbach B, Munscher C, Frank V, Muller-Hocker J, Napiwotzki J. Mutat Res. 1995;338:161–172. doi: 10.1016/0921-8734(95)00021-w. [DOI] [PubMed] [Google Scholar]
- 10.Ozawa T. Exp Gerontol. 1995;30:269–290. doi: 10.1016/0531-5565(94)00057-a. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C, Bills M, Quigley A, Maxwell R J, Linnane A J, Nagley P. Biochem Biophys Res Commun. 1997;230:630–635. doi: 10.1006/bbrc.1996.6020. [DOI] [PubMed] [Google Scholar]
- 12.Casane D, Dennebouy N, de Rochambeau H, Mounolou J C, Monnerot M. Genetics. 1994;138:471–480. doi: 10.1093/genetics/138.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keefe D L, Niven-Fairchild T, Powell S, Buradagunta S. Fertil Steril. 1995;64:577–583. [PubMed] [Google Scholar]
- 14.Cortopassi G, Lui Y. Mutat Res. 1995;338:151–159. doi: 10.1016/0921-8734(95)00020-7. [DOI] [PubMed] [Google Scholar]
- 15.Avise J C. Evolution. 1993;47:1293–1301. doi: 10.1111/j.1558-5646.1993.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 16.Hale L R, Singh R S. Proc Natl Acad Sci USA. 1986;78:8813–8817. doi: 10.1073/pnas.83.22.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis D L, Farr C L, Farquhar A L, Kaguni L S. Mol Biol Evol. 1994;11:523–538. doi: 10.1093/oxfordjournals.molbev.a040132. [DOI] [PubMed] [Google Scholar]
- 18.MacRae A F, Anderson W W. Genetics. 1988;120:485–494. doi: 10.1093/genetics/120.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Sokal R R, Rohlf F J. Biometry. 2nd Ed. San Francisco: Freeman; 1981. [Google Scholar]
- 21.Ashburner M. Drosophila: A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Partridge L, Green A, Fowler K. J Insect Physiol. 1987;33:745–749. [Google Scholar]
- 23.Chapman T, Liddle L F, Kalb J M, Wolfner M F, Partridge L. Nature (London) 1995;373:241–244. doi: 10.1038/373241a0. [DOI] [PubMed] [Google Scholar]
- 24.Attardi G. Int Rev Cytol. 1985;93:93–145. doi: 10.1016/s0074-7696(08)61373-x. [DOI] [PubMed] [Google Scholar]
- 25.Ballinger S W, Shoffner J M, Hedaya E V, Trounce I, Polak M A, Koontz D A, Wallace D C. Nat Genet. 1992;1:11–15. doi: 10.1038/ng0492-11. [DOI] [PubMed] [Google Scholar]
- 26.King R C. Ovarian Development in Drosophila melanogaster. New York: Academic; 1970. [Google Scholar]
- 27.Wyman R. J Embryol Exp Morph. 1979;50:137–144. [PubMed] [Google Scholar]
- 28.Wilson T G. J Insect Physiol. 1985;31:109–117. [Google Scholar]
- 29.Charlesworth B, Hughes K A. Proc Natl Acad Sci USA. 1996;93:6140–6145. doi: 10.1073/pnas.93.12.6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Promislow D E, Tatar M, Khazaeli A A, Curtsinger J W. Genetics. 1996;143:839–848. doi: 10.1093/genetics/143.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatar M, Promislow D E, Khazaeli A A, Curtsinger J W. Genetics. 1996;143:849–858. doi: 10.1093/genetics/143.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buss L W. Proc Natl Acad Sci USA. 1983;80:1387–1391. doi: 10.1073/pnas.80.5.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]