Abstract
The intronic splicing silencer (ISS) of CFTR exon 9 promotes exclusion of this exon from the mature mRNA. This negative influence has important consequences with regards to human pathologic events, as lack of exon 9 correlates well with the occurrence of monosymptomatic and full forms of CF disease. We have previously shown that the ISS element interacts with members of the SR protein family. In this work, we now provide the identification of SF2/ASF and SRp40 as the specific SR proteins binding to this element and map their precise binding sites in IVS9. We have also performed a functional analysis of the ISS element using a variety of unrelated SR-binding sequences and different splicing systems. Our results suggest that SR proteins mediate CFTR exon 9 exclusion by providing a ‘decoy’ sequence in the vicinity of its suboptimal donor site. The results of this study give an insight on intron ‘exonization’ mechanisms and provide useful indications for the development of novel therapeutic strategies aimed at the recovery of exon inclusion.
INTRODUCTION
The regulation of CFTR exon 9 splicing has been extensively studied in recent years because of its clear connection with CF disease (1–6). At present, several splicing controlling regions have been characterized near the 3′ and 5′ boundaries of this exon. These include a polymorphic TG(m)T(n) region near the 3′ss, a suboptimal donor site and a Polypyrimidine-rich Controlling Element just downstream of the 5′ss (PCE) (7–9). Moreover, additional controlling regions have been identified inside the exon itself in the form of CERES elements (10) and as an intronic splicing silencer region (ISS) further away in the IVS9 intron sequence (11) (Figure 1A). During the course of these studies several trans-acting elements have also been identified as binding specifically to these regulatory elements: TDP-43 to the (TG)m region near the 3′ss of the exon, which has been recently shown to recruit hnRNP proteins near the 3′ss (6,12,13), TIA-1 to the PCE that promotes exon inclusion (9), and unidentified members of the SR protein family to the ISS sequence (11) (Figure 1B).
Figure 1.
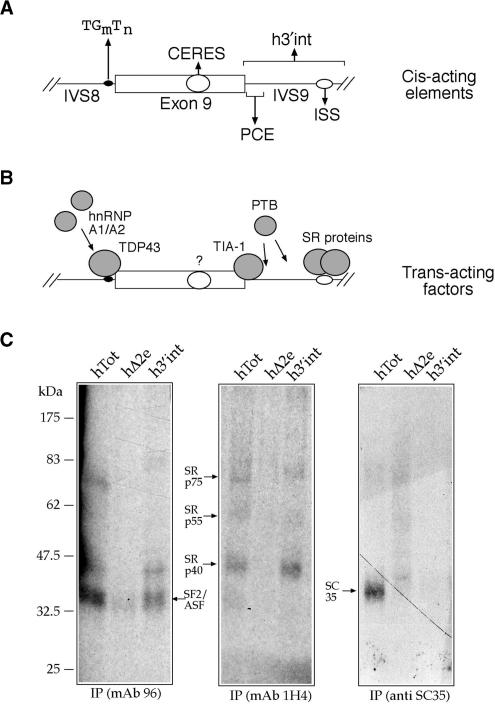
(A) Shows a schematic diagram of the splicing controlling regions of CFTR exon 9, both within the exon (CERES element) and in the flanking IVS8 and IVS9 sequences: TG(m)T(n), PCE and ISS. h3′int defines the IVS9 region that includes both the PCE and ISS controlling elements. (B) Shows the trans-acting factors identified up to now that bind to these elements. (C) Shows an immunoprecipitation analysis of the h3′int region and of the fibronectin EDA ESE element both in its wild-type (hTot) and mutated form (h▵2e). The left, central and right panels show the immunoprecipitation profiles obtained from each RNA using mAb 96 (specific against SF2/ASF), mAb 1H4 (specific against the phosphorylated RS domain) and an anti-SC35 antibody.
The SR protein family (14–16) has been predominantly studied in relationship with its involvement in alternative and constitutive splicing control (17–22) and indeed may have played a decisive role in the evolution of this process (23,24). However, it has also been recently shown to participate in a very wide range of functions that include the maintenance of genomic stability (25–27), mRNA export (28–31), mRNA surveillance (32) and protein translation (33,34).
In splicing regulation, SR proteins are generally considered to bind exonic splicing enhancer (ESE) sequences (35–37) and in this way they generally promote exon inclusion in the pre-mRNA molecule that is processed by the spliceosome. This enhancement is achieved in a variety of ways: by antagonizing the effect of negative regulators such as hnRNP proteins (38–41), by directly recruiting basic splicing factors such as U1 and U2snRNPs to the exon acceptor and donor sites (42,43), and by promoting spliceosome assembly through their RS domains (20,21). Because of all these functions, SR proteins represent one of the most important factors that promote exon inclusion (44) and is not surprising that an excess of SR proteins can compensate for complete U1snRNP inactivation and rescue correct splicing (45,46).
In general, most SR proteins share rather common enhancer properties despite they have different sequence binding abilities (47), protein domain compositional differences (15) or nucleo cytoploasm shuttling properties (48). However, this is by no means a rule. In fact, some SR protein family members have also been recently identified in connection with splicing repression. For example, a novel SR protein designated SRp38 has been recently demonstrated to posses splicing inhibitory activity in mitotic cells or following heat shock treatment (49–51). In addition, another SR-protein like factor (SR-15) has recently been described to possess general splicing inhibitory activity in the HSV1 virus (52).
Most importantly, there are many examples of factors that display either enhancer or repression activity in one system can display the opposite behavior in different pre-mRNAs. The SR proteins family is no exception to this observation. Indeed, past research has led to the discovery of a small number of splicing systems in which normally enhancing SR proteins display a inhibitory activity on the splicing process (53–60).
As previously mentioned, to this short list of examples we have to add the reports that describe SR proteins as general inhibitors of CFTR exon 9 splicing (11,61). To this date, however, no clear identification/mapping or functional binding sites for these inhibitory SR proteins has been provided. In this work, we have aimed to cover this gap and investigate the functional reasons that underlie this particular SR inhibitory activity.
EXPERIMENTAL PROCEDURES
Plasmid construction
Plasmid TG11T5 has been previously described by Niksic et al. (7). Plasmid pES was obtained by deleting part of the original IVS9 sequence in TG11T5 and inserting in its place a PstI/KpnI linker just before the NdeI cloning site (Figures 2A and 3A). All the other plasmids used in this study were obtained by cloning the sequence of interest in the pES plasmid either in the PstI/KpnI (PK series) or PstI site (P series). This was achieved by annealing two complementary oligonucleotides containing the sequence of interest and ligating according to standard protocols (sequence of the oligonucleotides is available upon request). In order to mutate the cryptic 3′ss sequence in the TG11T5 context (mutant IVS9del3′ss) we used the two following oligos: 5′ctctttttttttctaatttgtagtg3′ sense and 5′cactacaaattagaaaaaaaaagag3′ antisense.
Figure 2.
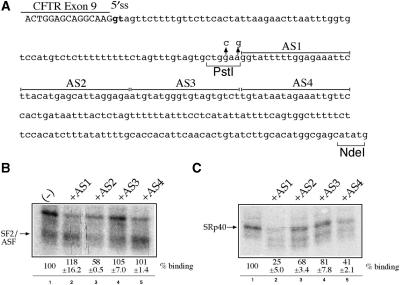
(A) Shows the sequence of the entire IVS9 (h3′int) sequence from the last nucleotides of exon 9 (in capital letters) to the NdeI site that was cloned in the TG11T5 original minigene. Also shown in this diagram are the positions of the two mutated nucleotides that create a unique PstI site. In the ISS region, the sequences targeted by each of the four antisense oligos (AS1–AS4) are underlined. (B) and (C) shows the immunoprecipitation profiles of the h3′int sequence using mAb 96 (B) and mAb 1H4 (C) monoclonal antibodies in the presence of each of the four antisense oligos. The migration of SF2/ASF and of SRp40 are marked with an arrow.
Figure 3.
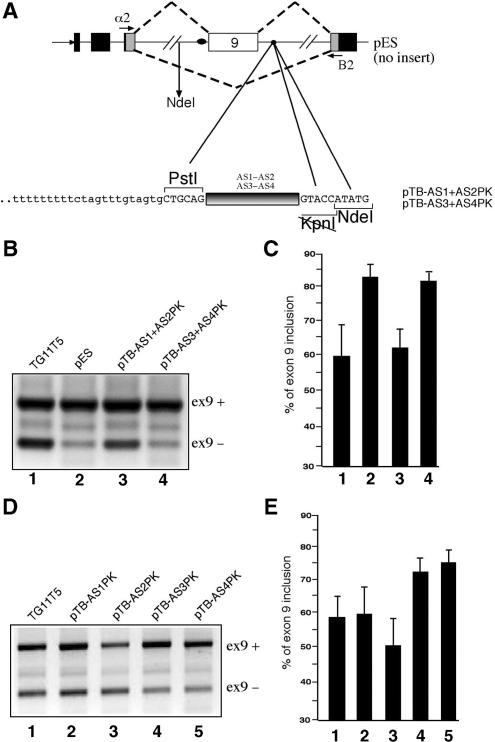
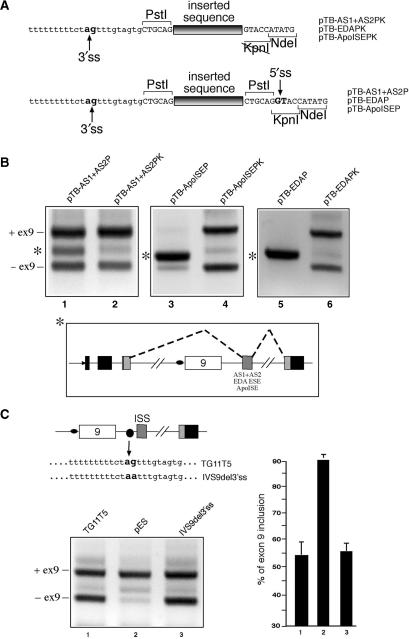
(A) Upper panel shows a schematic representation of the pES hybrid minigene carrying the CFTR exon 9 along with intronic flanking sequence. Black, shaded and white boxes represent alpha globin, fibronectin and CFTR exons, respectively. The arrows indicate the position of the primers used to amplify the processed mRNA species. The dotted lines represent the expected splicing pattern that determines exon 9 inclusion/exclusion. Lower panel shows a close-up view of the nucleotide composition of the IVS9 sequence in the PstI/KpnI/NdeI cloning region. The black box represents the exact position in which the AS1 + AS2 (pTB AS1 + AS2PK) and AS3 + AS4 (pTB AS3 + AS4PK) sequences were inserted. (B) Shows a comparison of the RT–PCR splicing profiles of these two constructs together with the pES and TG11T5 reference minigenes following transfection in Hep3B cells. The position of the transcripts including exon 9 (ex9+) and lacking exon 9 (ex9−) are marked on the right. The results of three independent experiments were quantified using radioactive PCR and are reported in (C) with SD values. (D) Shows a set of analogous transfections in Hep3B cells using a set of plasmids in which each individual AS1, AS2, AS3 and AS4 sequences was inserted in the PstI/KpnI sites of the pES plasmid (pTB-AS1PK to pTB-AS4PK). A quantification of the effect of these sequences on CFTR exon 9 inclusion is reported in (E).
In vitro transcription, UV cross-linking and immunoprecipitation analysis
To generate the RNA probe of h3′int a corresponding pBluescript II KS plasmid containing this sequence was linearized with NdeI and transcribed with T7 RNA Polymerase (Pharmacia Biotech) in the presence of α32P-UTP, according to standard procedures. The UV cross-linking assay was performed by incubating 1 × 106 c.p.m.-labeled RNA probes with 100 μg of total HeLa nuclear extracts (CilBiotech, Mons, Belgium) and 100 μg heparin in a 20-μl final reaction volume containing 20 mM HEPES pH = 7.9, 72 mM KCl, 1.5 mM MgCl2, 0.78 mM magnesium acetate, 0.52 mM dithiothreitol, 3.8% glycerol, 0.75 mM ATP and 1 mM GTP for 15 min at 30°C. Samples were transferred to HLA plate (Nunc, InterMed) on ice and irradiated with 0.8 J UV light for 5 min by using a BIO-LINK apparatus (Euroclone). Unbound RNA was digested with 30 μg of RNase A (Sigma) for 30 min at 37°C and incubated for 2 h at 4°C on a rotator wheel with 150 ml of IP buffer (20 mM Tris pH 8.0, 300 mM NaCl, 1 mM EDTA, 0.25% NP-40) and 1 ml of monoclonal antibodies anti-SF2/ASF (mAb 96) (Zymed Laboratories Inc), anti-RS phosphorylated domain (mAb 1H4) (Zymed Laboratories Inc) and an anti-SC35 monoclonal antibody (Sigma). Each mixture was then incubated with 30 ml of Protein A/G-Plus Agarose (Santa Cruz Biotechnologies) at 4°C overnight. Beads were collected by centrifugation, washed four times with 1.5 ml of IP buffer and then loaded onto a SDS-10% PAGE gel. Gels were run at a constant 30 mA for ∼3.5 h, dried under vacuum, and exposed for 4 days with a BioMax Screen (Kodak).
Transient transfection minigene splicing analysis in Hep3B cells
Liposome-mediated transfections of 3 × 105 human hepatocarcinoma Hep3B cells were performed using DOTAP Liposomal Transfection Reagent (Alexis Biochemicals) according to manufacturer instructions. After 18 h the transfectiom medium was replaced with fresh medium and 24 h later the cells were washed with PBS and RNA was purified using RNAwiz (Ambion). RT–PCR reactions to specifically amplify the minigene transcripts was performed as previously reported (11). In order to quantify the amplified fragments, the PCR reaction was performed in the presence of α32dCTP and the samples run on a 5% denaturing polyacrylamide gel. Radioactive intensity was measured using a Cyclone (Packard). Transfection of a siRNA reagent against TDP-43 were performed as previously published (62).
RESULTS
Characterization of SR proteins binding to the ISS element in IVS9
In order to better define which SR proteins are binding to the ISS region we performed immunoprecipitation analysis using, as substrate, the entire h3′int intronic region (Figure 1A). Figure 1C shows an immunoprecipitation analysis with HeLa nuclear extract of this RNA (h3′int) together with two control RNAs from the fibronectin EDA exon, one bearing a well-characterized ESE sequence (hTot) and one where this sequence has been deleted (h▵2e) (63). Each RNA was labeled using α32P-UTP and incubated with ∼150 μg of Hela nuclear extract before being subjected to UV-crosslinking and digestion with RNAse A. Samples were then run on a 10% SDS–PAGE gel and exposed using BioMax autoradiographic films. Immunoprecipitation was performed using equal amounts of each UV-crosslinked sample and following the addition of specific monoclonal antibodies against SF2/ASF (Figure 1C, left panel, mAb96), against the phosphorylated RS domain (Figure 1C, center panel, mAb 1H4) and against the SC35 protein (Figure1C, right panel). The mobility of the SR proteins is indicated by arrows. The results show that SF2/ASF and SRp40 are the major SR protein family members binding to the h3′int region.
Mapping of the SR protein-binding regions in h3′int using small antisense oligos
In order to map the binding sites of each SR protein in h3′int we used in immunoprecipitation analysis a set of antisense oligos which targeted the original ISS region (Figure 2A, AS1–AS4). Figure 2A shows a schematic diagram of the entire IVS9 sequence inserted in the original TG11T5 plasmid (7) together with the sequences targeted by the AS1–AS4 antisense oligos that cover the originally mapped ISS region (11). Also shown in this figure are the two single-point mutations introduced in h3′int to create a unique PstI cloning site that can be used to remove this entire region from the template minigene plasmid. Based on the results obtained in Figure 1C, each oligo was then used in immunoprecipitation analysis with mAb 96 and mAb 1H4 together with the h3′int labeled RNA. As shown in Figure 2B lane 3, oligo AS2 was the most efficient in inhibiting SF2/ASF binding to h3′int whilst oligo AS1 was very efficient in blocking SRp40 binding to this region (Figure 2C, lane 2). A lesser amount of inhibition for SRp40 binding could also be detected in the presence of the AS4 antisense oligo (Figure 2C, lane 5). However, as the AS1 and AS4 sequence share considerable similarity (almost 50%, especially in the ‘agaaatt’ central region) the AS4 oligo may have cross-hybridized with the AS1 sequence blocking partially its interaction with SRp40 in vitro. This hypothesis is consistent with the observed lack of functional effects of the AS4 sequence alone on CFTR exon 9 inclusion.
Functional importance of the different AS1 to AS4 sequences on CFTR exon 9 inhibition
Figure 3A shows a schematic representation of the CFTR exon 9 hybrid minigene construct lacking the ISS sequence (pES). In this construct, the IVS9 sequence was shortened by exploiting the creation of a novel PstI site which was directly joined to NdeI through a small linker that also provided a unique KpnI site. The unique PstI/KpnI sites could then be used to insert different combination of the AS1–AS4 sequences in the ISS position (Figure 3A, lower panel, has a detailed scheme of the inserted fragment position in the pES plasmid). Based on the results of the immunoprecipitation analyses it was then decided to insert in pES the two combinations of AS1 + AS2 (pTB AS1 + AS2PK) and AS3 + AS4 (pTB AS3 + AS4PK). Figure 3B shows the RT–PCR assays of these plasmids (lane 3 and lane 4, respectively) following transfection in Hep3B cells together with two control minigene constructs: the original TG11T5 (lane 1) and the pES minigene (lane 2). The upper and lower bands correspond to exon 9 inclusion (ex9+) and exclusion (ex9−), respectively. Quantification of these bands from three independent experiments following radioactive RT–PCR (Figure 3C) demonstrated that the SR-binding AS1 + AS2 region could entirely recover the ISS inhibitory activity displayed by the original TG11T5 minigene construct (compare lanes 1 and 3). On the other hand, the AS3 + AS4 region that had no apparent SR-binding ability could not display any inhibitory activity with respect to the original pES plasmid (compare lanes 2 and 4).
A similar pattern could also be observed when each individual sequence was inserted in the pES plasmid (Figure 3D). A quantitation of the inhibitory activity of each sequence (Figure 3E) confirmed that only the AS1 and AS2 sequences (Figure 3D, lanes 2 and 3) could inhibit CFTR exon 9 inclusion whilst AS3 and AS4 did not cause any drop in CFTR exon 9 splicing efficiency (Figure 3D, lanes 4 and 5). Moreover, the fact that the AS1 and AS4 showed very distinct inhibitory activities supports the conclusions drawn from the immunoprecipitation experiments in Figure 2C.
Inhibitory effects of other SR-binding sequences on CFTR exon 9 inhibition
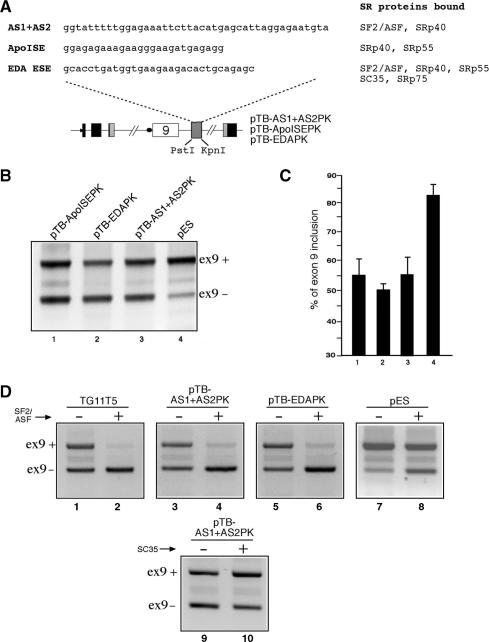
In consideration of the fact that ISS activity strongly correlates with AS1 + AS2 it was of interest to determine whether different SR protein-binding sequences were also capable of inhibiting CFTR exon 9 inclusion. To this end, it was decided to test the activity of two unrelated SR-binding sequences that have been extensively characterized by our laboratory: the ESE region of the fibronectin EDA exon (63) and the ISE region (ApoISE) of the Apo AII intron 3 (64) (Figure 4A). The advantage of using these sequences is represented by the fact that they all possess different combinations of SR protein-binding abilities (Figure 4A) and also that in their respective contexts they functionally behave as ESE and ISE elements, respectively. These additional polypurinic sequences were then cloned in the PstI and KpnI sites of the pES plasmid and transfected in Hep3B cells. Figure 4B and C show that both the EDA ESE sequence (pTB-EDAPK, lane 1), and the ApoISE sequence (pTB ApoISEPK, lane 2) were capable of inhibiting CFTR exon 9 inclusion in a way comparable with that observed with pTB AS1 + AS2PK (lane 3) and higher than the pES plasmid alone (lane 4).
Figure 4.
(A) Upper panel shows a comparison between the AS1 + AS2 sequence and the heterologous polypurinic sequences from the EDA ESE and the Apo AII ISE elements. The column on the right shows the SR protein-binding specificity of all these sequences as previously determined by immunoprecipitation analysis. The EDA ESE and Apo ISE sequences were then inserted in the pES plasmid (as shown in the lower panel), thus creating constructs pTB EDAPK and pTB ApoISEPK, respectively. The levels of CFTR exon 9 inclusion displayed by these three plasmids in Hep3B cells and compared with that of the pES and pTB AS1 + AS2PK constructs is reported in (B). A quantification of three independent experiments is reported in (C). (D) Shows the response of the TG11T5, pTB AS1 + AS2PK, pTB EDAPK and pES plasmids to SF2/ASF overexpression, and of the response of the pTB AS1 + AS2PK construct to SC35 overexpression, following transfection in Hep3B cells.
For the sequences that can bind SF2/ASF, the functional specificity of SR protein binding on the inhibitory activity was also investigated by overexpressing this particular SR protein in transfected cells (Figure 4D). As shown in this figure, the increased levels of CFTR exon 9 inhibition for constructs pTB AS1 + AS2PK (Figure 4D, lanes 3 and 4) and pTB-EDAPK (Figure 4D, lanes 5 and 6) is comparable to that observed for the TG11T5 plasmid (Figure 4D, lanes 1 and 2) and greater than the one observed for the pES plasmid alone (Figure 4D lanes 7 and 8). In addition, SC35 overexpression does not result in increased levels of CFTR exon 9 skipping in the pTB AS1 + AS2PK plasmid (Figure 4D, lanes 9 and 10). This result is consistent with the immunoprecipitation results shown in Figure 1. We have also tried overexpression of SRp55 and SRp75 that also induced CFTR exon 9 skipping to different degrees (data not shown), in keeping with the SR protein response profile obtained in the original study by Pagani et al. (11). The binding experiments did not show reproducible direct interaction of these proteins with h3′int, although at times bands compatible with SRp55 and SRp75 were visible (Figure 1C, middle panel). The lack of correlation between this absence of binding in IP assays and functional experiments may be due to the well-known non-specific effects of SR protein overexpression or to indirect interactions with the ISS sequence. Further studies will be needed to clarify these differences. In any case, taken together, these data further reinforce our conclusion that the AS1 + AS2 sequence contains all the functional properties of the originally mapped ISS element.
The results in Figure 4 conclusively show that both the EDA ESE and the Apo AII ISE sequences were capable of acting as ISS elements in the CFTR exon 9 context. Therefore, it was of interest to determine whether the exon 9 AS1 + AS2 ISS sequence and the Apo ISE sequence were capable of acting as ESEs in a heterologous splicing context. To this end, we cloned the AS1 + AS2 and ApoISE sequences in the dsx-XH reporter system (65,66) (Figure S1). This is a well-known in vitro splicing system in which processing of the IVS3 intron from the Drosophila doublesex (dsx) gene is dependent on the sequences inserted at the 3′ end of the construct. As shown in Figure S1, both the AS1 + AS2 and ApoISE sequence display higher ESE activity than the control AS3 + AS4 sequence.
Changing ISS/ISE to ESE activities in the pES context by providing a heterologous donor site
In order to further test these potential enhancer activities of AS1 + AS2 and ApoISE in the CFTR exon 9 context it was then decided to insert in the pES plasmid a donor sequence that could be used as a viable 5′ss to promote ‘exon’ inclusion. This was achieved by simply cloning the AS1 + AS2, ApoISE, and EDA ESE sequences in the PstI site of the pES plasmid. Figure 5A shows that this cloning procedure, as opposed to cloning in the PstI/KpnI sites, provided any PstI-inserted sequence with a downstream donor site sequence possessing a score of 0.76 according to the NNSPLICE predictor program (67). The results of this cloning procedure on the resulting pTB AS1 + AS2P, pTB ApoISEP and pTB EDAP plasmids are reported in Figure 5B, lanes 1, 3 and 5, respectively. In all three cases, the cloning in the PstI site alone resulted in the appearance of an extra band which, when sequenced, was shown to consist in a ‘mini-exon’ sequence that exploited the newly inserted 5′ss and a cryptic 3′ss in the h3′int sequence (Figure 5B, lower panel). Interestingly, the intensity of this extra band in the pTB AS1 + AS2P and pTB ApoISEP is markedly different, with the pTB AS1 + AS2P splicing profile still retaining some of the ex9+/ex9− splicing forms. This is consistent with the different ESE abilities displayed in the dsx-XH plasmid (Figure S1), where the ApoISE sequence is a much more efficient ESE (66%) than the AS1 + AS2 sequence (20%).
Figure 5.
(A) Shows a comparison of the different splice site composition when the AS1 + AS2, EDA and ApoISE sequences are cloned in the PstI/KpnI sites (upper panel, PK plasmid series) as opposed to the PstI site alone (lower panel, P plasmid series). The differing splice site composition in the region surrounding the point of insertion is indicated by arrows, with the conserved 3′ss ‘ag’ and 5′ss ‘gt’ nucleotides highlighted in bold. (B) Shows a comparison of the splicing profiles between P and PK plasmids carrying the AS1 + AS2, EDA and ApoISE sequence following transfection in Hep3B cells. The asterisk shows the extra band that is observed in the plasmids carrying the heterologous donor site downstream of the inserted sequence (pTB AS1 + AS2P, pTB EDAP and pTB ApoISEP). The lower panel shows a schematic diagram of the new splicing event indicated by the asterisk whilst the position of the transcripts including exon 9 (ex9+) and lacking exon 9 (ex9−) are marked on the right. (C) Left panel shows a transfection analysis of a TG11T5 mutant (IVS9del3′ss) carrying a ‘ag’ to ‘aa’ mutation in the cryptic 3′ss sequence. The CFTR exon 9 inclusion levels are comparable to those detected for the TG11T5 wild-type plasmid (C, right panel).
These results also suggested that the action of the ISS element could be dependent on the presence of the naturally occurring cryptic 3′ss in CFTR IVS9, perhaps through the recruitment of a non-productive spliceosomal complex in this position. However, mutating this 3′ss sequence from ‘ag’ to ‘aa’ in the natural TG11T5 context (mutant IVS9del3′ss) did not significantly affect the efficiency of CFTR exon 9 inclusion (Figure 5C). This result ruled out the possibility that this acceptor-like sequence alone could be playing a role in wild-type ISS functioning.
Improving acceptor site recognition of CFTR exon 9 results in context-specific effects on the splicing pattern
To better assess the context-dependent effects of these different SR protein-binding sequences in the vicinity of CFTR exon 9 we then improved the definition of this exon by removing TDP-43 through siRNA treatment (Figure S2). As previously demonstrated, TDP-43 is a major inhibitory splicing factor that specifically recognizies the TGm polymorphic locus in IVS8 (6,12), and its specific removal from the transfected cell can offset many splicing inhibitory effects, including those mediated by the ISS (62). It was therefore interesting to test the effect of removing this factor on the splicing patterns of the pTB AS1 + AS2P, pTB ApoISEP and pTB EDAP minigenes. Figure S2 shows that removal of TDP-43 in these different contexts can have very different outcomes depending on the type of enhancer sequence present in the ISS position.
Inhibitory effects of a C/A-rich YB-1 binding sequence in the ISS position
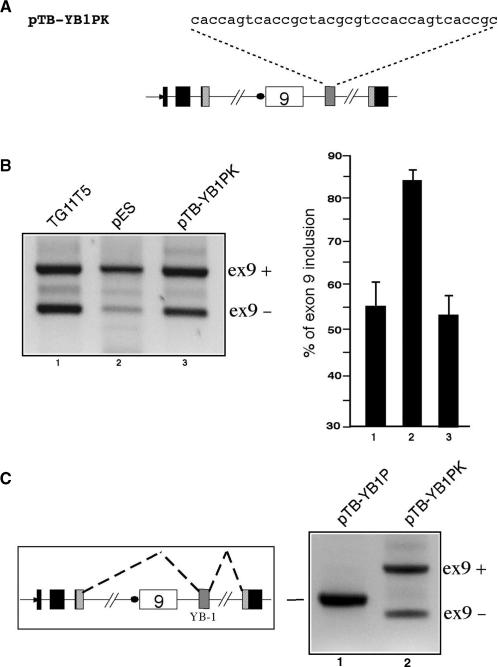
Finally, it was interesting to assess whether the inhibitory effects mediated by these different G/A-rich sequences in the ISS position could also be mimicked by a non-polypurinic sequence with well-known splicing enhancer effects. Therefore, an A/C-rich enhancer sequence from the alternatively spliced exon v4 of the CD44 gene was inserted in the ISS position (Figure 6A) (68). This sequence was identified by Stickeler et al. (68) as the binding site of YB-1, a member of the family of multifunctional cold shock domain proteins (CSD proteins). As shown in Figure 6B, cloning this YB-1 binding sequence in the PstI/KpnI sites of the pES plasmid (pTB YB-1PK) can successfully mimick the effect of the original ISS activity in TG11T5 (compare lanes 1 and 3). Analogously, when this sequence is cloned in the PstI site only of the pES plasmid to obtain the pTB YB-1P construct (thus providing a downstream donor site capable of supporting exon recognition) the splicing pattern is totally shifted towards the inclusion of a miniexon containing only the YB-1 sequence (Figure 6C, compare lanes 1 and 2).
Figure 6.
(A) Shows the sequence of the YB-1-binding motif inserted in the PstI/KpnI restriction sites of the pES plasmid (pTB YB1PK). (B) Left panel shows a comparison of the levels of CFTR exon 9 inclusion in the TG11T5, pES and pTB YB1PK plasmids following transfection in Hep3B cells. The position of the transcripts including exon 9 (ex9+) and lacking exon 9 (ex9−) are marked on the right. The results of three independent experiments as quantified by radioactive RT–PCR are reported in (B), right panel. The effect of cloning the YB-1 sequence only in the PstI site of the pES plasmid (pTB YB-1P) as opposed to the PstI/KpnI sites (pTB YB-1PK) is shown in (C). The schematic diagram on the left shows the composition of the unique splice product observed in lane 1 whilst the position of the transcripts including exon 9 (ex9+) and lacking exon 9 (ex9−) are marked on the right.
DISCUSSION
CFTR exon 9 splicing is a complex event in which several cis-acting elements located in both intronic and exonic sequences play an important role (6–8,10). In particular, an ISS sequence in IVS9 has been previously shown by Pagani et al. (11) to represent a negative element towards its inclusion in the final mRNA molecule.
Our work has shown that this region specifically binds two members of the SR protein family, SF2/ASF and SRp40, which are normally associated with binding to enhancer elements that promote exon inclusion. Because of this, the CFTR ISS is one of the relatively few splicing systems in which SR proteins behave as suppressors of splicing. It is therefore interesting to compare its functioning with other systems in which SR proteins have also been identified as negative splicing regulators (53–60) (Figure 7).
Figure 7.
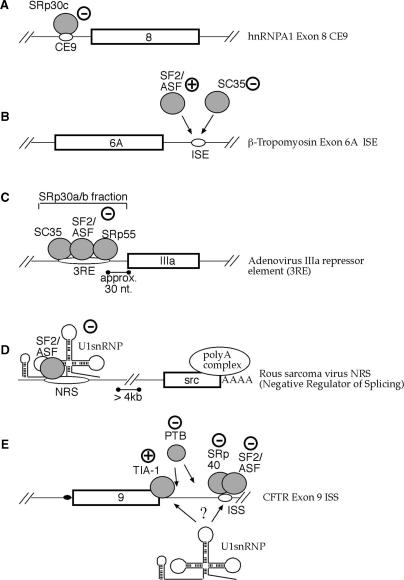
This figure shows a schematic diagram of different splicing systems in which SR proteins have been observed to behave as inhibitors of exon inclusion. (A) Shows the CE9 element characterized in the hnRNP A1 gene, (B) to the ISE element observed downstream of b-tropomyosin exon 6A, (C) to the 3RE element present in Adenovirus and (D) to the NRS element present in Rous Sarcoma Virus. Finally, a working model of the way SR proteins may inhibit CFTR exon 9 recognition is reported in (E).
In the hnRNPA1 gene ISS, a particular SR protein (SRp30c) can recognize the silencer element (CE9) and directly down-regulate exon recognition (53) (Figure 7A). Most importantly, CE9 cannot compromise the assembly of U2-dependent complexes on the 3′ss of hnRNP A1 exon 8 (69) and cannot function as an enhancer element in a heterologous system (53). Although the molecular mechanism of CE9 functioning remains unclear, this represents an important functional difference with respect to the CFTR exon 9 ISS, because the AS1 + AS2 sequence can function as an enhancer sequence in a heterologous splicing system.
A second type of inhibition has also been found in the case of what is really a ISE element localized downstream of b-tropomyosin gene exon 6A (57). In this case, competition occurs between a SR protein enhancer factor (SF2/ASF) and another SR protein that has no enhancer effect in this system (SC35) (Figure 7B). Also this model does not seem to apply to the CFTR exon 9 ISS working model for a number of reasons. First of all, mapping of the SF2/ASF and SRp40-binding sites on the CFTR ISS sequence has shown that they are physically distinct, making competition between the two highly unlikely (unlike the case of the ISE element of b-tropomyosin exon 6A where the binding sites of SF2/ASF and SC35 actually overlap). In addition, our observation that heterologous SR-binding sequences with rather different splicing specificities (EDA ESE and Apo ISE elements) and even a YB-1 binding sequence can restore CFTR ISS function in the absence of its natural sequence would tend to rule out any specific effects by particular SR proteins.
A higher degree of similarity can be found between CFTR exon 9 ISS and the action of the IIIa repressor element (3RE) originally described in Adenovirus (58) (Figure 7C). In fact, just like CFTR exon 9 ISS, also the 3RE sequence was observed to function as an enhancer element when inserted in a heterologous splicing system, showing that the mechanisms of action of 3RE is wholly dependent on context. From a functional point of view, the inhibitory mechanism mediated by 3RE was initially thought to reside in the physical inhibition of U2snRNP binding to the IIIa 3′ss, because of its nearness to the IIIa acceptor site (∼30 nt) (58). More recently, the inhibitory activity of SF2/ASF on IIIa splicing has been specifically identified as residing in its second RNA binding domain, RBD2, although the exact mechanism still remains to be defined (70). It is difficult to determine whether physical hindrance may also represent the mode of action for CFTR ISS as this sequence is localized rather far away from the CFTR exon 9 5′ss (∼80–100 nt). Given this limitation, the potential physical hindrance between U1snRNP binding to the natural donor site and SF2/ASF and SRp40 binding to AS1 + AS2 seems unlikely.
Another SR-mediated inhibitory situation is represented by the Negative Regulator of Splicing (NRS) of the Rous Sarcoma Virus (Figure 7D). In this case, together with SR protein SF2/ASF (71), the NRS can also bind a U1snRNP molecule (60), a U11 snRNP molecule (72) and hnRNP H (73). Interestingly, also this sequence can function as an enhancer element when inserted in the dsx-HX in vitro splicing system (59). However, the mechanism of action of the NRS sequence cannot be explained by physical hindrance because it is localized very far away from the 3′ss that is inhibited. Recent research has shown that the exact mechanism through which splicing inhibition occurs may probably reside in the formation of non-productive complexes between the NRS inhibitory splicing complex, the 3′ss of the src exon, and the polyadenylation process (60,74). In addition, it has been suggested that RNA polymerase II can ‘tether’ emerging splice sites in the pre-mRNA (75). In this case the NRS might also act as a disturbing presence for the recognition of the natural 5′ss.
The CFTR exon 9 ISS is an excellent example of the importance of sequence-context in determining the action of cis-acting sequences (76). In the natural situation, the heterologous 5′ss sequence is absent hence no IVS9 intron sequences may be ‘exonized’. Nonetheless, the presence of the exon enhancer-like complexes formed by SR proteins or YB-1 in the ISS may create a situation in which the U1snRNP molecule approaching CFTR exon 9 would remain ‘undecided’ between binding to the wild-type suboptimal site and waiting for an indication from the SR/YB-1 proteins present in the ISS of a better target immediately downstream (Figure 7E). This is consistent with the fact that the outcome of TDP 43 removal depends on the strength of the ISS–SR interactions. Weak SR interactions and removal of TDP 43 lead to complete recovery of CFTR exon 9 inclusion while strong SR interaction results in the inclusion of a super-exon 9 sequence by selecting exclusively the new 5′ss downstream of the ISS (see Fig. S2). This data suggests that the ISS may act as a sort of ‘decoy’ system hampering recognition of the exon 9 5′ss. The result of this stalemate would be a net decrease in CFTR exon 9 donor site recognition and, consequently, in a lesser inclusion of CFTR exon 9 in the mature mRNA. In this respect, therefore, a critical issue might be represented by the processing speed of the RNA polymerase II molecule in presenting the ISS sequence after having transcribed the CFTR exon 9 region, and these issues are currently being investigated in our laboratory.
Finally, from a pathological point of view, the importance of having mapped exactly the binding sites for the SR trans-acting factors that are responsible for this inhibitory action can provide researchers with a useful target to inhibit their action, for example, by antisense oligonucleotide approaches (77,78).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The study was funded by Telethon Onlus Foundation (Italy) (grant no. GGP02453 and GGP06147), FIRB (RBNE01W9PM) and by a European community grant (EURASNET- LSHG-CT-2005-518238). We also wish to thank Albrecht Bindereif for kindly providing the dsx-XH plasmid. Funding to pay the Open Access publication charges for this article was provided by ICGEB.
Conflict of interest statement. None declared.
REFERENCES
- 1.Disset A, Michot C, Harris A, Buratti E, Claustres M, Tuffery-Giraud S. A T3 allele in the CFTR gene exacerbates exon 9 skipping in vas deferens and epididymal cell lines and is associated with Congenital Bilateral Absence of Vas Deferens (CBAVD) Hum. Mutat. 2005;25:72–81. doi: 10.1002/humu.20115. [DOI] [PubMed] [Google Scholar]
- 2.Groman JD, Hefferon TW, Casals T, Bassas L, Estivill X, Des Georges M, Guittard C, Koudova M, Fallin MD, et al. Variation in a repeat sequence determines whether a common variant of the cystic fibrosis transmembrane conductance regulator gene is pathogenic or benign. Am. J. Hum. Genet. 2004;74:176–179. doi: 10.1086/381001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuppens H, Lin W, Jaspers M, Costes B, Teng H, Vankeerberghen A, Jorissen M, Droogmans G, Reynaert I, et al. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J. Clin. Invest. 1998;101:487–496. doi: 10.1172/JCI639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rave-Harel N, Kerem E, Nissim-Rafinia M, Madjar I, Goshen R, Augarten A, Rahat A, Hurwitz A, Darvasi A, et al. The molecular basis of partial penetrance of splicing mutations in cystic fibrosis. Am. J. Hum. Genet. 1997;60:87–94. [PMC free article] [PubMed] [Google Scholar]
- 5.Chu CS, Trapnell BC, Curristin S, Cutting GR, Crystal RG. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nat. Genet. 1993;3:151–156. doi: 10.1038/ng0293-151. [DOI] [PubMed] [Google Scholar]
- 6.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niksic M, Romano M, Buratti E, Pagani F, Baralle FE. Functional analysis of cis-acting elements regulating the alternative splicing of human CFTR exon 9. Hum. Mol. Genet. 1999;8:2339–2349. doi: 10.1093/hmg/8.13.2339. [DOI] [PubMed] [Google Scholar]
- 8.Hefferon TW, Broackes-Carter FC, Harris A, Cutting GR. Atypical 5′ splice sites cause CFTR exon 9 to be vulnerable to skipping. Am. J. Hum. Genet. 2002;71:2. doi: 10.1086/341664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuccato E, Buratti E, Stuani C, Baralle FE, Pagani F. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 2004;279:16980–16988. doi: 10.1074/jbc.M313439200. [DOI] [PubMed] [Google Scholar]
- 10.Pagani F, Buratti E, Stuani C, Baralle FE. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 2003;278:26580–26588. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- 11.Pagani F, Buratti E, Stuani C, Romano M, Zuccato E, Niksic M, Giglio L, Faraguna D, Baralle FE. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a non-evolutionary conserved intronic element. J. Biol. Chem. 2000;275:21041–21047. doi: 10.1074/jbc.M910165199. [DOI] [PubMed] [Google Scholar]
- 12.Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am. J. Hum. Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 Binds Heterogeneous Nuclear Ribonucleoprotein A/B through Its C-terminal Tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 14.Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- 15.Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 16.Sanford JR, Ellis J, Caceres JF. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem. Soc. Trans. 2005;33:443–446. doi: 10.1042/BST0330443. [DOI] [PubMed] [Google Scholar]
- 17.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graveley BR, Hertel KJ, Maniatis T. SR proteins are ‘locators’ of the RNA splicing machinery. Curr. Biol. 1999;9:R6–R7. doi: 10.1016/s0960-9822(99)80032-3. [DOI] [PubMed] [Google Scholar]
- 19.Hertel KJ, Graveley BR. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem. Sci. 2005;30:115–118. doi: 10.1016/j.tibs.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 21.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 22.Tacke R, Manley JL. Determinants of SR protein specificity. Curr. Opin. Cell. Biol. 1999;11:358–362. doi: 10.1016/S0955-0674(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 23.Ram O, Ast G. SR proteins: a foot on the exon before the transition from intron to exon definition. Trends Genet. 2007;23:5–7. doi: 10.1016/j.tig.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Webb CJ, Romfo CM, van Heeckeren WJ, Wise JA. Exonic splicing enhancers in fission yeast: functional conservation demonstrates an early evolutionary origin. Genes Dev. 2005;19:242–254. doi: 10.1101/gad.1265905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Manley JL. New talents for an old acquaintance: the SR protein splicing factor ASF/SF2 functions in the maintenance of genome stability. Cell Cycle. 2005;4:1706–1708. doi: 10.4161/cc.4.12.2210. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 29.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y, Steitz JA. SRprises along a messenger's journey. Mol. Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl Acad. Sci. USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol. Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 33.Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 35.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 36.Ladd AN, Cooper TA. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-reviews0008. reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng ZM. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 40.Fu XD. Towards a splicing code. Cell. 2004;119:736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 41.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 42.Lavigueur A, La Branche H, Kornblihtt AR, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 43.Eperon IC, Ireland DC, Smith RA, Mayeda A, Krainer AR. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 45.Tarn WY, Steitz JA. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 1994;8:2704–2717. doi: 10.1101/gad.8.22.2704. [DOI] [PubMed] [Google Scholar]
- 46.Crispino JD, Blencowe BJ, Sharp PA. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science. 1994;265:1866–1869. doi: 10.1126/science.8091213. [DOI] [PubMed] [Google Scholar]
- 47.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Caceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol. Cell. Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 50.Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 51.Nilsen TW. Too hot to splice. Nat. Struct. Mol. Biol. 2004;11:208–209. doi: 10.1038/nsmb0304-208. [DOI] [PubMed] [Google Scholar]
- 52.Li Q, Zhao H, Jiang L, Che Y, Dong C, Wang L, Wang J, Liu L. An SR-protein induced by HSVI binding to cells functioning as a splicing inhibitor of viral pre-mRNA. J. Mol. Biol. 2002;316:887–894. doi: 10.1006/jmbi.2001.5318. [DOI] [PubMed] [Google Scholar]
- 53.Simard MJ, Chabot B. SRp30c is a repressor of 3′ splice site utilization. Mol. Cell. Biol. 2002;22:4001–4010. doi: 10.1128/MCB.22.12.4001-4010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.ten Dam GB, Zilch CF, Wallace D, Wieringa B, Beverley PC, Poels LG, Screaton GR. Regulation of alternative splicing of CD45 by antagonistic effects of SR protein splicing factors. J. Immunol. 2000;164:5287–5295. doi: 10.4049/jimmunol.164.10.5287. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Wang J, Gao L, Lafyatis R, Stamm S, Andreadis A. Tau exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c.SRp55 complex that either recruits or antagonizes htra2beta1. J. Biol. Chem. 2005;280:14230–14239. doi: 10.1074/jbc.M413846200. [DOI] [PubMed] [Google Scholar]
- 56.Aznarez I, Chan EM, Zielenski J, Blencowe BJ, Tsui LC. Characterization of disease-associated mutations affecting an exonic splicing enhancer and two cryptic splice sites in exon 13 of the cystic fibrosis transmembrane conductance regulator gene. Hum. Mol. Genet. 2003;12:2031–2040. doi: 10.1093/hmg/ddg215. [DOI] [PubMed] [Google Scholar]
- 57.Gallego ME, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanopka A, Muhlemann O, Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre- mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 59.McNally LM, McNally MT. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol. Cell. Biol. 1998;18:3103–3111. doi: 10.1128/mcb.18.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNally LM, McNally MT. U1 small nuclear ribonucleoprotein and splicing inhibition by the rous sarcoma virus negative regulator of splicing element. J. Virol. 1999;73:2385–2393. doi: 10.1128/jvi.73.3.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nissim-Rafinia M, Chiba-Falek O, Sharon G, Boss A, Kerem B. Cellular and viral splicing factors can modify the splicing pattern of CFTR transcripts carrying splicing mutations. Hum. Mol. Genet. 2000;9:1771–1778. doi: 10.1093/hmg/9.12.1771. [DOI] [PubMed] [Google Scholar]
- 62.Ayala YM, Pagani F, Baralle FE. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett. 2006;580:1339–1344. doi: 10.1016/j.febslet.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 63.Buratti E, Muro AF, Giombi M, Gherbassi D, Iaconcig A, Baralle FE. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woerfel G, Bindereif A. In vitro selection of exonic splicing enhancer sequences: identification of novel CD44 enhancers. Nucleic Acids Res. 2001;29:3204–3211. doi: 10.1093/nar/29.15.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol. Cell. Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 68.Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20:3821–3830. doi: 10.1093/emboj/20.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simard MJ, Chabot B. Control of hnRNP A1 alternative splicing: an intron element represses use of the common 3′ splice site. Mol. Cell. Biol. 2000;20:7353–7362. doi: 10.1128/mcb.20.19.7353-7362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dauksaite V, Akusjarvi G. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem. 2002;277:12579–12586. doi: 10.1074/jbc.M107867200. [DOI] [PubMed] [Google Scholar]
- 71.McNally LM, McNally MT. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J. Virol. 1996;70:1163–1172. doi: 10.1128/jvi.70.2.1163-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McNally LM, Yee L, McNally MT. Two regions promote U11 small nuclear ribonucleoprotein particle binding to a retroviral splicing inhibitor element (negative regulator of splicing) J. Biol. Chem. 2004;279:38201–38208. doi: 10.1074/jbc.M407073200. [DOI] [PubMed] [Google Scholar]
- 73.Fogel BL, McNally MT. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 2000;275:32371–32378. doi: 10.1074/jbc.M005000200. [DOI] [PubMed] [Google Scholar]
- 74.Fogel BL, McNally LM, McNally MT. Efficient polyadenylation of Rous sarcoma virus RNA requires the negative regulator of splicing element. Nucleic Acids Res. 2002;30:810–817. doi: 10.1093/nar/30.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 76.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garcia-Blanco MA. Making antisense of splicing. Curr. Opin. Mol. Ther. 2005;7:476–482. [PubMed] [Google Scholar]
- 78.Wilton SD, Fletcher S. RNA splicing manipulation: strategies to modify gene expression for a variety of therapeutic outcomes. Curr. Gene. Ther. 2005;5:467–483. doi: 10.2174/156652305774329249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.