Abstract
Genomic variations deep in the intronic regions of pre-mRNA molecules are increasingly reported to affect splicing events. However, there is no general explanation why apparently similar variations may have either no effect on splicing or cause significant splicing alterations. In this work we have examined the structural architecture of pseudoexons previously described in ATM and CFTR patients. The ATM case derives from the deletion of a repressor element and is characterized by an aberrant 5′ss selection despite the presence of better alternatives. The CFTR pseudoexon instead derives from the creation of a new 5′ss that is used while a nearby pre-existing donor-like sequence is never selected. Our results indicate that RNA structure is a major splicing regulatory factor in both cases. Furthermore, manipulation of the original RNA structures can lead to pseudoexon inclusion following the exposure of unused 5′ss already present in their wild-type intronic sequences and prevented to be recognized because of their location in RNA stem structures. Our data show that intrinsic structural features of introns must be taken into account to understand the mechanism of pseudoexon activation in genetic diseases. Our observations may help to improve diagnostics prediction programmes and eventual therapeutic targeting.
INTRODUCTION
RNA secondary structure is increasingly recognized as a powerful modifier of splicing events (1). At the local level, RNA conformations have been shown to regulate the splicing process by affecting the basic identifying features of an exon. Numerous examples of such a regulatory role have been recently reported to occur for the donor site of Tau exon 10 (2), SMN2 exon 7 (3), the branch/acceptor sites in Drosophila Adh gene (4), the internal splicing enhancer region in the fibronectin gene (5) or to silencer regions in the presenilin 2 gene (6). In addition, RNA secondary structures have been shown to involve interactions between very distant regions of the pre-mRNA such as in Saccharomyces cerevisiae (7,8), humans (9–11) and Drosophila (12–14). Recently, complementary intron sequence motifs have also been proposed to mediate the peculiar phenomenon of exon repetition (15). Finally, RNA secondary structures are increasingly shown to play a part in other facets of mRNA biology such as in mantaining its stability (16), regulating translation (17) or transport (18). Taken together, these examples are consistent with the indications provided by in silico analyses which predict the existence of a vast array of conserved structural features both in selected human protein coding RNA transcripts (19) and in the human genome in general (20). From the latter has emerged the recent small RNAs revolution of functional non-coding RNAs (20,21).
Interestingly, RNA secondary structures have also been proposed to play a role in helping the splicing machinery to distinguish between real exons and pseudoexon sequences (22). Pseudoexon sequences are loosely defined as intronic sequences between 50 and 200 nt in length that are flanked by apparently good-to-consensus acceptor and donor-site signals. These sequences, however, are apparently never recognized by the splicing machinery (23) although this definition may contain many exceptions. In fact, it has recently been proposed that many members of this class may indeed be used to regulate the relative abundance of different pre-mRNA isoforms by selective Nonsense-Mediated Decay of alternatively spliced exons (24). In addition, a distinct class of pseudoexon sequences derived from transposable elements known as Alu sequences has been recently established as a major source of ‘real’ coding exonic sequences (25–27).
Nevertheless, estimates regarding pseudoexon sequence abundance in a typical pre-mRNA molecule has shown they may outnumber ‘real’ exons by an entire order of magnitude (23). It is therefore clear that, no matter how many exceptions to the exclusion rule there may be, avoiding the insertion of these sequences during the normal splicing process would be essential for correct pre-mRNA processing. To achieve this, recent research has proposed that their exclusion may be achieved by a combination of factors (23), including an enrichment within their sequence of inhibitory elements (28,29) or through the in silico indication that pseudoexons flanking regions have a distinct tendency to form double-stranded structures that include the pseudo exon itself (22).
Regarding human diseases, pseudoexon inclusion events have increasingly been described to occur as a result of a single-point mutation deep within intronic regions. In general, these mutations have the effect of creating either a very good acceptor or donor splice site followed by the selection of ‘opportunistic’ complementary sites. A few exceptions that are worth noting have been described elsewhere where the intronic mutation affects a regulatory element within the pseudoexon itself (30–32). For a general review of this topic see Buratti et al. (33) and Supplemetary Table 1 for an updated list of pathological pseudoexon inclusion events.
This kind of aberrant insertional events have never had any coding potential, as they all originate from the chance occurrence of a triggering mutation. Therefore, no selection pressure may have been applied during evolution to regulate their inclusion as far as codon usage/regulatory sequences are concerned (34,35). Interestingly, one characteristic feature of these pseudoexons is their reduced length with respect to the normal length spread of exons which is reported to exist in the human and mouse genomes (Supplementary Figure S1) (36). This reduced length, intriguingly similar to the previous estimates of the ‘window’ of naked RNA available for folding after transcription (37), suggests that intrinsic structural features of these RNAs could represent a major determinant in their ‘exonization’ process. In this work, we have used representative examples of these pseudoexon inclusion events to test the importance of RNA secondary structures in their splicing regulation.
MATERIALS AND METHODS
Plasmids carrying the ATM pseudoexon wild-type and mutant sequences
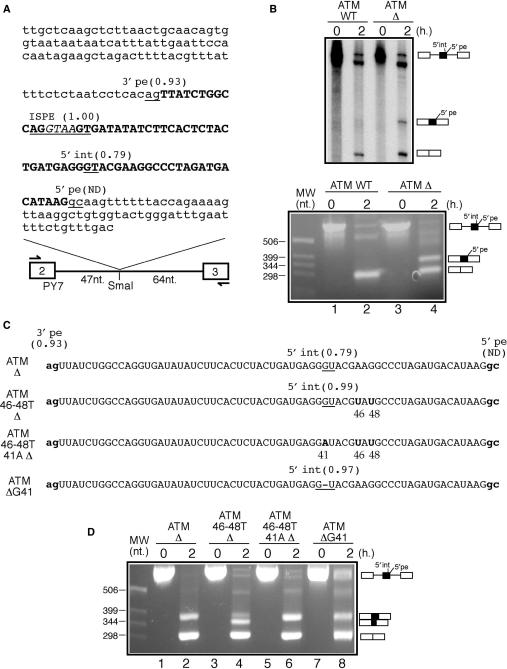
The PY7 plasmid has been described in detail elsewhere (38). For our purposes, we have inserted two SmaI and NdeI unique cloning sites at positions 44 and 50, respectively, in its 111-nt long intron. Both the wild-type (wt) and gtaa-deleted (Δ) ATM sequences were amplified from the pATM plasmids used in the original report (32) and inserted in the SmaI site of PY7 (Figure 1A). The oligos used for this task were the following: 5′-ttgctcaagctcttaactgcaacagtggt-3′ (s) and 5′-gtcaaacagaaaattcaaatcccag-3′ (as). In order to obtain the ATM 46-48T Δ mutant a two-step PCR method was used with the following primers 5′-tgagggtacgtatgccctagatg-3′ (s) and 5′-catctagggcatacgtaccctca-3′ (as). From this mutant we obtained ATM 46-48T 41A Δ using primers 5′-cactctactgatgaggatacg-3′ (s) and 5′-cgtatcctcatcagtagagtg-3′ (as). Finally, the primers used to insert the 21–23 substitution in both the ATM Δ and ATM 46-48T Δ mutants were the following: 5′-gtgatataccctcactctac-3′ (s) and 5′-gtagagtgagggtatatcac-3′ (as). On the other hand, to obtain the ATM 5′-new Δ mutant we created a unique StuI site by joining together nucleotides 38–40 (agg) with nucleotides 51–53 (cct). The ATM 5′-new Δ mutation was then introduced by ligating in this site the 5′-gtaggtaagtacgaaggc-3′ (s) and 5′-gccttcgtacttacctac-3′ (as) oligos. Donor splice-site scores were calculated according to the NNSplice 0.9 program available at http://www.fruitfly.org/seq_tools/splice.html (39).
Figure 1.
(A) A schematic diagram of the ATM WT construct. White boxes represent tropomyosin exons 2 and 3 whilst the single line represents the intron. The arrows indicate the primers used for the RT-PCR analyses of the processed RNA templates. The ATM intronic region is in small letters whilst pseudoexon sequences are in bold capital letters. The ISPE sequence is underlined and the GTAA nucleotides deleted in the ATM Δ construct are in italic. Within the ATM sequence all potential donor and acceptor splice-site sequences together with their scores calculated according to the NNSplice program are underlined (ND indicates that the splice site is not detected as a viable donor site by the program). It should be noted that although the ISPE sequence scores very well as a potential donor site it is never used by the splicing machinery. (B) In vitro splicing analysis of the ATM WT and ATM Δ RNAs both using radioactive templates (upper panel) and by RT-PCR (lower panel). In order to investigate 5′int/5′pe donor-site usage a number of mutants were engineered in the ATM Δ construct (C shows a schematic diagram). In mutant ATM 46-48T Δ the 46A and 48G nucleotides were replaced by a T in the ATM Δ context. This improved 5′int donor site was then inactivated by introducing an A for a G substitution in position 41 (mutant ATM 46-48T 41A Δ). Cryptic 5′int donor-site improvement was also obtained by deleting the G in position 41 (mutant ATM Δ G41). (D) In vitro splicing analysis of these mutants.
Plasmids carrying the CFTR 1811 + 1.6kbA > G wild-type and mutant sequences
The human genomic fragment flanking the CFTR pseudoexon region was cloned into the SmaI site in the modified PY7 vector (CFTR WT) following amplification with oligos: 5′-attggtttttaaaaaaatttttaaattggc-3′ (s) and 5′-ccatattaaatagaaatgagataatttc-3′ (as). In order to obtain the following mutants the CFTR WT construct was subjected to two-step PCR method of mutagenesis using the following primers: 5′-atataagttaggtaactaacaa-3′ (s) and 5′-ttgttagttacctaacttatat-3′ (as) for CFTR WT2, 5′-atataactaaggttagtaacaa-3′ (s) and 5′-ttgttactaaccttagttatat-3′ (as) for CFTR WT3, 5′-gatataacttaggtaagtatcaat-3′ (s) and 5′-attgatacttacctaagttatatc-3′ (as) for CFTR WT4, 5′-tacttgagatgtaagtaaggt-3′ (s) and 5′-accttacttacatctcaagta-3′ (as) for CFTR MUT, 5′-tttattacagcaacaattac-3′ (s) and 5′-gtaattgttgctgtaataaa-3′ (as) for CFTR Del1, 5′-agaatcctatgagatgtaag-3′ (s) and 5′-cttacatctcataggattct-3′ (as) for CFTR Del2. Finally, in order to construct the CFTR Rep mutant a two-step PCR on CFTR MUT mutant plasmid was performed first by using the following oligos: 5′-agaatcctatcatgaagagatgtaag-3′ (s) and 5′-cttacatctcttcatgataggattct-3′ (as) and then the resulting mutant (CFTR Dis) was subjected to the same methodology with the following primers: 5′-tttattacagttcatgcaacaattac-3′ (s) and 5′-gtaattgttgcatgaactgtaataaa-3′ (as) to make the CFTR Rep mutant.
In vitro and in Hep3B splicing analysis
Splicing reactions were performed in vitro using capped, SP-6 transcribed RNAs. Standard reactions were carried out in a 25 μl volume at 30°C for 2 h. Each reaction contained 15 μl of Nuclear Extract from HeLa cells (CilBiotech, Mons, Belgium, approx. concentration 10 μg/μl), 5 μl of 13% (w/v) polyvinyl alcohol, 1 μl of 80 mM MgCl2, 1 μl of 12.5 mM ATP, 1 μl of 0.5 M creatine phosphate and 1.25 μl of 0.4 M Hepes-KOH pH = 7.3 and 2 μl of in vitro transcribed pre-mRNA at 200 μg/ml. Therefore, the final concentrations of the various components in a standard processing reaction were as follows: 3.2 mM MgCl2, 500 μM ATP, 20 mM creatine phosphate, 2.7% (w/v) PVA, 20 mM Hepes (pH = 7.3), 6 μg/μl of Hela nuclear extract and 16 μg/ml of in vitro transcribed pre-mRNA. The processed RNAs were then extracted from the reaction mix using RNAwiz (Ambion, Inc.) and analysed by RT-PCR using a set of primers at the beginning of tropomyosin exons 2 and 3, respectively: 5′-gaatacaagcttgtcgaggaggac-3′ (s) and 5′-agaccggaattcggatcctctagag-3′ (as). In order to insert the various PY7-based sequences in the eukaryotic expression vector pcDNA3 the inserts were amplified using the following oligos T2F 5′-agggtaccagcttgtcgaggaggacatctcag-3′ and T3R 5′-cctctagagtcgatcgacctgcagg-3′ and inserted in the KpnI and XbaI sites of pcDNA3. Liposome-mediated transfections of 3 × 105 human hepatocarcinoma Hep3B cells were performed using DOTAP Liposomal Transfection Reagent (Alexis Biochemicals) according to manufacturer instructions. After 18 h the transfectiom medium was replaced with fresh medium and 24 h later the cells were washed with PBS and RNA was purified using RNAwiz (Ambion). In order to rescue the splicing of CFTR WT and CFTR WT2 we have also expressed a variant U1snRNP molecule (C > G U1) that have been described in a previous work from our lab (40).
DNA bands from EtBr-stained (1 mg/ml) agarose gel (1.5%) were photographed under UV illumination and acquired using a Kodak EDAS 290 apparatus. Quantification of band intensities in the scanned images has been performed using the ImageJ software freely available at http://rsb.info.nih.gov/ij.
RNA secondary structure determination
RNA secondary structure determination with the use of limited V1 RNAse (Ambion), T1 RNAse (Ambion) and S1 nuclease (Fermentas) digestion has been described in detail elsewhere (5). Briefly, 1 μg aliquots of ATM Δ RNA were digested in 100 μl final volume with 0.002 U of RNAse V1, 0.05 U of RNAse T1 and 19 U of S1 nuclease for 10 min at 30°C. An enzyme-free aliquot was processed together and used as a control. The cleaved RNAs were retrotranscribed according to standard protocols using the following antisense primers labelled with 32P-end-labelled oligonucleotide primers: 5′-gtcaaacagaaaattcaaatccc-3′ for the ATM pseudoexons and 5′-ccatattaaatagaaatgagataatttc-3′ for CFTR pseudoexons. In silico secondary structure predictions were performed using the mFold program (41,42).
RESULTS
Peculiar donor-site usage in the ATM pseudoexon
Recently, we have described the inclusion of a 65-nt long pseudoexon between ATM exons 20 and 21 in a patient affected by ataxia-telengiectasia (32). The activation event consisted of a 4 nt deletion (GTAA) that occurred within a high-affinity U1snRNP-binding site that surprisingly was not a masked 5′ss but an internal splicing repressor. This region was termed ISPE, for Intron-Splicing Processing Element (32,43). Figure 1A shows the nucleotide sequence of the pseudoexon (in bold), together with the ISPE sequence (underlined, the GTAA sequence is in italic) and its intron flanking regions.
The major peculiarity of this event is represented by the observation that although the ISPE itself is an excellent 5′ss it is never used by the splicing machinery. The reason for this probably resides in its vicinity to the well-defined 3′ss sequence of the ATM pseudoexon (3′pe) that possesses a score of 0.93 according to the NNSplice program (39). In fact, recent experiments using modified U1snRNA molecules binding to consecutive positions along this pseudoexon have suggested that the U1snRNP molecule has to be placed 40 nt away from the 3′ss before pseudoexon inclusion can be observed (43). Even more surprisingly, the only donor site (5′pe) used in vivo is represented by a AAGgcaagt non-consensus sequence (32,43). The GC dinucleotide is a fairly rare donor-site sequence which accounts for only 0.56% of all donor-site junctions in humans (44). Moreover, in this specific context this choice is made even more peculiar due to the presence inside the pseudoexon itself of another reasonable potential donor-site sequence (Figure 1A, the AGGGTACGA sequence, designated throughout this work as 5′int) in addition to the ISPE itself. In fact, the 5′int site possesses a well above cut-off score of 0.79 by the NNSplice program whilst 5′pe is not even considered as a potential donor site by this in silico prediction program. This difference in predicted strengths in favour of 5′int is also reflected by several other donor-site scoring methods such as MaxEntScan (ME, MDD, MM and WMM scores for 5′int are 7.38/10.38/7.61/6.10, respectively compared to the scores of 3.24/7.72/4.43/4.95 obtained for 5′pe) (45) or Spliceview (46) where 5′int has a predicted score of 81 whereas 5′pe is not detected at all.
Setting up an in vitro splicing system to analyse ATM pseudoexon inclusion
In order to clarify this peculiar mechanism of donor-site usage it was decided to clone part of this sequence in the PY7 splicing system for in vitro analysis (Figure 1A). Two versions of this construct were initially analysed, the first one (ATM WT) containing the wild-type intronic sequence whilst the second one (ATM Δ) carrying the 4-nt (GTAA) deletion that had been previously characterized to cause pseudoexon activation (32). The transcribed RNAs from these plasmids were then used in a standard in vitro splicing assay and analysed both by polyacrylamide gel electrophoresis and by RT-PCR in a time-course analysis (Figure 1B, upper and lower panels, respectively). As shown in both Figure 1B panels the ATM Δ RNA could successfully induce pseudoexon inclusion whilst, as expected, no pseudoexon inclusion could be detected during the processing of the ATM WT RNA. It should also be noted that there is a strong similarity between the results obtained using the radioactively labelled RNAs and the RT-PCR products. Subcloning and sequencing of the RT-PCR reaction in Figure 1B lower panel, lane 4 confirmed that only the 5′pe sequence was used in the pseudoexon inclusion event and failed to detect any use of the 5′int donor site (data not shown). The in vitro data, therefore, wholly confirmed the previous results obtained using minigenes and patient's transcript analyses (32,43).
Investigating donor-site choice in the ATM pseudoexon
It was then decided to investigate the eventual activation of the 5′int donor site by introducing two single nucleotide substitutions in position 46 and 48 (ATM 46-48T Δ mutant). As shown in Figure 1C, this had the effect of improving the predicted NNSplice donor-site score from 0.79 to 0.99. The ATM 46-48T Δ mutant was then spliced in vitro and the results showed that this improvement caused the almost complete switching from normal pseudoexon donor-site usage to this internal sequence (Figure 1D, lane 4). Interestingly, inactivating this improved cryptic 5′gt sequence in the presence of the 46–48 substitutions by mutating it to a 5′at sequence (Figure 1C, ATM 46-48T 41A Δ mutant) restored splicing to the ATM Δ RNA pattern (Figure 1D, lane 6). However, Figure 1D lane 8 shows that a mutation (ATM Δ G41) which strongly improves the predicted donor-site score in the 5′int position from 0.79 to 0.97 (Figure 1C) did not result in preferential usage of this splice site (compare Figure 1D lane 4 and lane 8). It was therefore evident that additional factors besides predicted scores were playing a role in defining donor-site usage within the ATM pseudoexon. In consideration that RNA secondary structure represents a potentially very powerful modifier of splice-site usage it was then decided to investigate the potential folding of this sequence.
The ATM pseudoexon can efficiently fold upon itself to mask splice-site usage
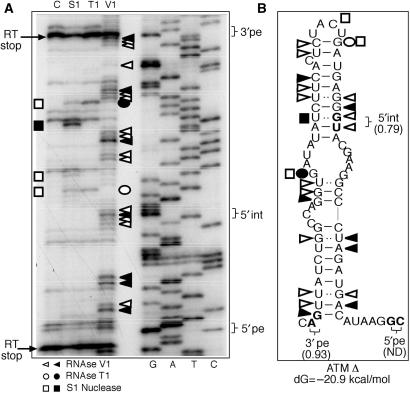
Preliminary in silico analyses obtained using the mFold program (41,42) suggested that the ATM Δ pseudoexon sequence could fold upon itself to form a very compact, double-stranded structure (Figure 2B). Its existence was thus tested experimentally by partial RNA digestion using single and double-strand-specific RNAses such as V1 (which cleaves double-stranded RNA and stacked RNA regions), T1 (which cleaves single-stranded guanosines) and S1 nuclease (which cleaves single-stranded RNA without sequence specificity) (Figure 2A). The overall position of the different cleavages shows that the ATM Δ pseudoexon sequence does indeed display a structure that is consistent with the model proposed in Figure 2B. In addition, it is interesting to note that two particularly strong RT stops (indicated by arrows in Figure 2A) seem to enclose the pseudoexon sequence, possibly reflecting the difficulty of the RT to travel across a compact structure.
Figure 2.
In vitro transcribed ATM Δ RNA enzymatically digested with S1 nuclease, T1 and V1 RNases (A: lanes S1, T1 and V1). No enzyme was added to the RNA in a control reaction mixture (A: lane C). The RNA substrate on which this analysis was perfomed consisted of the entire RNA transcript from the ATM Δ RNA plasmid (∼700 nt). By providing an extensive background of flanking RNA molecule we have aimed to minimize any folding bias that may have derived from analysing the pseudoexon sequence alone. The cleaved fragments were detected by performing a RT reaction using a labelled 32P-end labelled antisense oligo and separating them in a denaturing 6% polyacrylamide gel. A sequencing reaction performed with the same RT primer was run in parallel to the cleavages in order to determine the cleavage sites (A: lanes G, A, T, C). Squares, circles and triangles indicate S1, T1 and V1 cleavage sites. Black and white symbols indicate high and low cleavage intensity, respectively. RT stops are indicated by arrows. The positions of the 3′pe, 5′int and 5′pe splice sites are indicated on the right. The observed cleavages were then compared with the ATM folding predictions by mFold (B).
Finally, V1 cleavages can be observed to cluster in the exact position of the 5′int cryptic donor site, suggesting that the double-stranded structure in this region may inhibit use of this donor site. It is also interesting to note that a prominent S1 cleavage (black square) is also present in this region but on the opposite strand to the 5′int splice site, and this may represent a tendency for the U of the wobble U–G base pair to remain outside the stem region. Alternatively, the V1 sensitivity of the G in this wobble U–G base pair may be owing to the fact that it is stacked in a run of five purines, making a small internal bulge in this predicted stem structure. It should be noted, however, that the overall way in which cleavages are distributed also provides a general indication regarding how any approaching molecule will see that particular RNA region. In this case, the much greater number of V1 cleavages that occur on both sides of the stem in correspondence to 5′int would suggest that this whole region predominantly looks like a compact, double-stranded structure to any approaching molecule.
Taken together, these results allow us to propose a working model of splice-site usage in this pseudoexon. In this model, once the repression provided the ISPE sequence is removed the 5′pe sequence is chosen as donor site because the RNA secondary structure hides the internal 5′int sequence. This is consistent with previous data which showed that short artificial hairpins have been long known to be able to inhibit usage of a donor site embedded in their stem (47). Moreover, the stem configuration may also provide a competitive advantage by approximating the 3′pe to the 5′pe sequence.
Analysis of the role played by the ATM RNA secondary structure on 5′int/5′pe usage
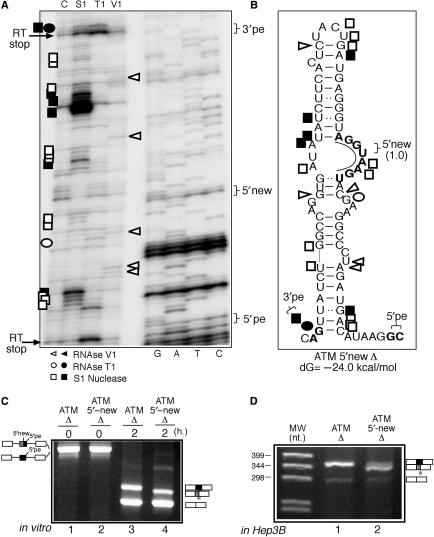
The importance of internal 5′ss availability was then tested experimentally by engineering a mutant ATM sequence (ATM 5′-new Δ) that carried a 5′ss sequence designed to remain in an open structure/internal bulge conformation (Figure 3B). It should be noted that these sequence changes also abolish the original 5′int donor like sequence according to the NNsplice prediction program although not according to the MaxEntScan program (data not shown). The fact that the ATM 5′-new Δ mutant has a more open structure also in the mutated 5′int donor site is suggested by the increase of S1 single-strand cleavages in this region of the ATM 5′-new mutant Δ when compared with the ATM Δ mutant (Figures 2B and 3B). The structural probing of this mutant shown in Figure 3A displays a marked reduction in the number of V1 cleavages (double-strand specific) and an increase in the S1 single-stranded cleavages around the central bulge region carrying the new donor site. Therefore, with respect to the structural analysis shown in Figure 2A for the ATM Δ mutant, the overall picture of the ATM 5′-new Δ mutant shows a predominantly open configuration (and thus presumably becomes more accessible for splicing factors).
Figure 3.
(A) The structural analysis of the ATM 5′new Δ mutant. Nomenclature of the enzymes and cleavage site intensities are as described in the legend to Figure 2. (B) A schematic diagram of this mutant. The inserted bases with respect to the ATM Δ sequence are underlined and in bold. The in vitro splicing analysis of this 5′new Δ RNA compared with the ATM Δ RNA is reported in (C). The activation of the novel internal 5′ss during ATM pseudoexon splicing is indicated as a box with vertical lines. The ATM 5′new Δ mutant sequence has also been transferred to the pcDNA3 eukaryotic expression vector and transiently transfected in Hep3B cells (D).
In keeping with this, when the activity of this mutant was tested in vitro (Figure 3C) the results demonstrated that this time the new internal 5′ss are preferentially used with respect to the 5′pe sequence (Figure 3C, compare lanes 3 and 4). Interestingly, direct sequencing of the amplified product has showed that although most fragments (∼75%) derive from the use of the 5′-new donor site whilst the remaining are derived from 5′int donor-site usage (data not shown). This result further highlights the importance of an ‘open’ structure on donor-site sequence availability. In addition, because the efficiency of pseudoexon recognition when transfected in eukaryotic cells using minigene systems was significantly higher than in vitro (32) the behaviour of this mutant was also tested following its introduction in pcDNA3 and transfection in Hep3B cells. Also in this case the ATM 5′-new Δ mutant was shown to splice predominantly to the internal donor sites when transfected in Hep3B cells (Figure 3D, compare lanes 1 and 2).
Effects of secondary structure stability on 5′int site usage in the ATM pseuodexon
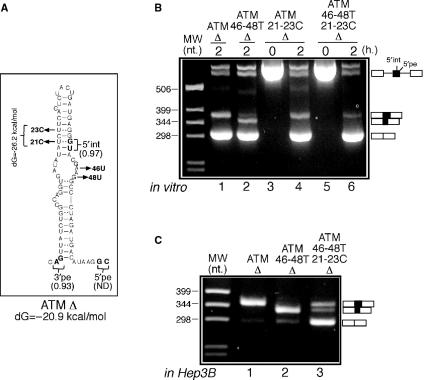
The functional importance of RNA secondary structure was also tested experimentally by performing a more subtle mutational analysis based on the proposed RNA secondary structure of the ATM pseudoexon in Figure 2B. According to this model, it should be possible to reduce 5′int activation in the ATM 46-48T Δ mutant by ‘tightening’ the upper stem (Figure 4A). In order to do this, the thymidines in position 21 and 23 were replaced by cytosine both in the original ATM Δ context as control (mutant ATM 21-23C Δ) and in the ATM 46-48T Δ context (mutant ATM 21-23C 46-48T Δ). The rationale of this change was to replace the natural UG ‘wobble’ base-pairings in these positions with two more canonical CG base pairs. This change, according to secondary structure predictions, would have the effect of raising the free energy of the ATM pseudoexon structure from –20.9 to –26.2 kcal/mol (Figure 4A). Figure 4B shows that the introduction of the 21–23C substitutions had no effect in donor-site usage of the ATM Δ context (Figure 4B, lane 4). However, the same substitutions in the ATM 46-48T Δ context were capable of obtaining the full inhibition of 5′int usage (Figure 4B, lane 6). Interestingly, and in keeping with the results obtained by the ATM 46-48T 41A Δ mutant (compare Figure 4B, lane 6 with Figure 1D, lane 6), the inactivation of the 5′int donor site also partially restored splicing to the 5′pe natural donor sequence.
Figure 4.
(A) The two T > C substitutions in positions 21 and 23 that have the effect of tightening the upper stem by replacing the two ‘wobble’ UG base pairs with two CG bonds. These two substitutions were introduced both in the ATM Δ context (mutant ATM 21-23C Δ) and in the ATM 46-48T Δ (mutant ATM 46-48T 21-23C Δ) context. The in vitro and in Hep3B splicing analyses of these mutants is shown in (B) and (C).
The inhibitory behaviour of the 21-23C substitutions in the 46-48T context with regards to 5′int usage are also clearly visible when the same mutants are cloned in pcDNA3 and transfected in Hep3B cells (Figure 4C, compare lanes 2 and 3), although a marked reduction in splicing efficiency was also observed in this case.
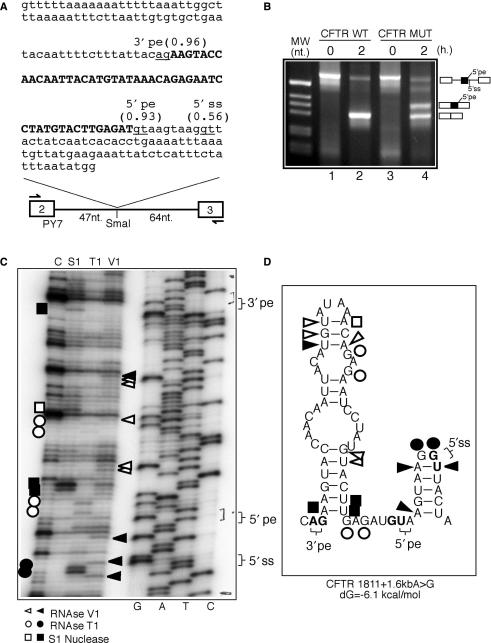
Donor-site usage in the CFTR 1811 + 1.6kbA > G pseudoexon
Splice-site mutation 1811 + 1.6kbA > G occurs in intron 11 of the CFTR gene and creates a novel 5′ splice site. This mutation results in the inclusion of a cryptic 49 bp exon in the final mature mRNA (48). As shown in Figure 5A, an interesting feature of this pseudoexon is represented by the fact that near to the newly created donor site (5′pe) there is a second, not optimal, but viable donor-site sequence (/guuacu, 5′ss) detected by the NNSplice prediction program. This was an interesting finding, as other likely donor-site sequences had already been identified in the original study (48). Indeed, an interesting feature of this CFTR sequence is represented by its containing several downstrean donor-like sequences after the activating mutation (/guaa/guaag/guuacu). However, at the time no indication could be obtained that any of these additional donor-site sequences besides the one created by the A > G mutation were ever used in normal CFTR pre-mRNA splicing (48). Using our PY7 in vitro system we have confirmed this data. In fact, as shown in Figure 5B the CFTR WT sequence that lacks the A > G substitution does not give rise to any pseudoexon insertion event originating from the 5′ss natural sequence (lane 2). In addition, when the A > G substitution was inserted in the sequence (mutant CFTR MUT) only one pseudoexon inclusion event could be observed (Figure 5B, lane 4) that originated from this newly created 5′pe donor site as determined by sequencing of the upper band (data not shown).
Figure 5.
(A) The sequence of the CFTR intronic region inserted in the SmaI site of PY7 that contains the A > G mutation responsible for creating a high score donor site (5′pe) (highlighted in bold). The positions and scores of the acceptor (3′pe) and additional donor splice site (5′ss) that occur within this sequence are also shown. (B) In vitro splicing reaction of two transcripts, one that did not contain the A to G substitution (CFTR WT) and the other carrying the A to G pseudoexon-creating mutation (CFTR MUT). (C) The structural analysis of CFTR MUT using limited RNAse digestion with the S1, V1 and T1 enzymes (nomenclature of the cleavages and of the enzymes used is as described in Figure 2 legend) and the in silico predicted structure on which the individual cleavages have been reported (D).
As in the ATM pseudoexon example, in silico analysis of this sequence (Figure 5D) suggested that avoidance of 5′ss usage in normal splicing could be ascribed to a particular folding of this RNA. An alternative explanation for the lack of 5′ss usage may also reside in the peculiar arrangement of the CFTR sequence (see earlier) where tandem donor-like sequences (/guaag/guuacu) are closely grouped together, an event that may lead to splicing inhibition probably at the level of the A and B complexes due to sterical hindrance of closely binding U1snRNP molecules, as proposed by Nelson and Green in previous studies (49). In this CFTR system, however, this possibility is unlikely as U1snRNP cannot be detected as binding to this position in the normal CFTR WT context, at least as determined by super-shift analysis using a anti-U1A monoclonal antibody (data not shown).
Therefore, in order to confirm the structural hypothesis we first performed RNAse digestion analysis on the PY7 MUT transcript. As shown in Figure 5C, the cleavages were consistent with the secondary structure depicted in Figure 5D and in particular the inactive 5′ss donor site appears to be embedded in a tightly folded structure, as highlighted by the two strong V1 cleavages that immediately flank the two T1 cuts in correspondence to the GG nucleotides. Although this would seemingly make an energetically very unfavourable, 2 nt loop configuration, it should be noted that in order to cleave the V1 RNAse does not necessarily require a bonded interaction but only a sufficiently close distance between two nucleotides. On the other hand, the newly created 5′pe (Figure 5D) is localized in the apparently more open region joining two stem-loop structures.
Investigating repression of 5′ss usage in the CFTR WT context
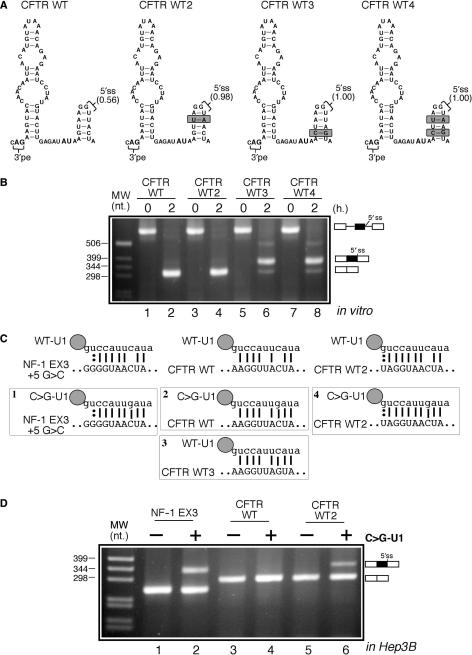
In order to better understand the molecular mechanisms that repress 5′ss usage in the CFTR WT transcript it was then decided to test the importance of predicted donor-site strengths on CFTR WT pseudoexon inclusion efficiency. As shown in Figure 6A, three mutants (WT2, WT3 and WT4) were created in which we substantially improved the predicted donor-site scores (0.98, 1.00 and 1.00, respectively) according to NNSplice without changing the structure and base-composition of the lower stem. The improvement in predicted strengths for these mutants was very evident using also other in silico prediction programs such as MaxEnt scan that focus only on the 9 nt consensus stretch itself. In this case, the predicted 5′ss donor-site strengths according to the ME, MDD, MM and WMM scoring models were 1.25/6.68/2.34/3.92 for WT, 6.43/10.08/5.09/7.20 for WT2, 8.54/13.58/7.21/7.76 for WT3 and 9.66/13.68/9.96/11.04 for WT4. In vitro splicing analysis of these mutants (Figure 6B, compare lanes 2, 4, 6 and 8) clearly showed that only WT3 and WT4 were capable of promoting pseudoexon inclusion from their mutated 5′ss sequences. This difference between the splicing behaviour of the WT2 and WT3 mutants (Figure 6B, compare lanes 4 and 6) prompted us to focus on the importance of the +5 position.
Figure 6.
(A) A schematic diagram of the three CFTR WT mutants (WT2, WT3 and WT4). The stem-conservative mutations introduced in these constructs are highlighted in gray and the relative predicted strengths of the resulting 5′ss sequences are also indicated. The splicing pattern displayed by these mutants in vitro is shown in (B). In (C) there is a schematic diagram of the predicted base-pairing between the 5′ end of wild-type U1snRNP (WT U1) and of the mutant U1snRNP (C > G U1) with the three different constructs tested: the NF-1 exon 3, the CFTR WT and CFTR WT2 5′ss sequences. (D) The effects of overexpressing the mutant C > G U1 in the presence of each of the three constructs in Hep3B cells. It should be noted that U1 rescue is more efficient in other cell types such as HeLa cells.
Therefore, we decided to improve the CFTR WT-U1snRNP matching region in this position by co-transfecting a modified U1snRNP molecule (C > G U1) that has been previously used to recover splicing from a +5G > C mutated donor site in exon 3 of the NF-1 gene (see Figure 6C and D, lanes 1 and 2) (40). The results of this experiment showed that CFTR WT could not display pseudoexon inclusion even when cotransfected with the C > G U1 (Figure 6D, lanes 3 and 4) whilst only a very small amount of pseudoexon inclusion could be observed when the CFTR WT2 mutant was cotransfected with this C > G U1 (Figure 6D, lane 6). Interestingly, addition of the C > G U1 to the CFTR WT 5′ss donor site gives a complementarity pattern that was identical to the one obtained by the CFTR WT3 mutant with wild-type U1 molecule (Figure 6C, compare boxes 2 and 3). However, in the first case no pseudoexon inclusion was observed (Figure 6D, lane 4) whilst the CFTR WT3 mutant could naturally promote inclusion (Figure 6B, lane 6). This finding suggests that the critical importance of the +5 position might be unrelated to U1snRNP binding and the most likely possibility is represented by the interaction with U6snRNP, that would remain impaired in the CFTR WT+ C > G U1 system whilst it would be unimpeded in the CFTR WT3 mutant. This hypothesis cannot, however, explain the recovery of CFTR WT2 pseudoexon recognition (although at very low levels) in the presence of C > G U1. However, in this case it may well be found that the very extended complementarity between the CFTR WT2 sequence and the C > G U1 (Figure 6C, lower right schematic diagram) may still promote donor-site usage. It should also be noted that the level of splicing recovery is considerably less than the one obtained for the NF-1 exon 3 donor site in the presence of C > G U1 (Figure 6D, compare lanes 2 and 6) although there is an even greater complementarity between CFTR WT2 with the C > G U1 sequence (Figure 6C, compare boxes 1 and 4). Taken together, all these differences suggest that the secondary structure may represent an additional constraint to the use of this splicing system.
Further investigating the relationship between secondary structure and this 5′ss donor-site usage would nonetheless represent a very difficult task as modifications introduced to change the folding of the stem structure involved would also affect donor-site strength and possibly activate the nearby donor consensus elements. Therefore, it was decided to focus our attention on the major structural element of this particular sequence.
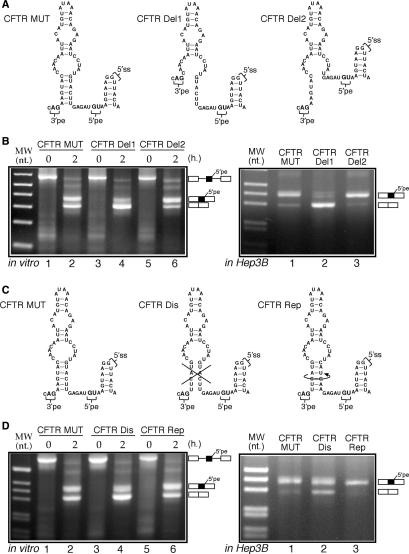
Importance of RNA secondary structure on CFTR 1811 + 1.6kbA > G inclusion
Indeed, disruption of the major structural features of this RNA region should affect the efficiency of CFTR pseudoexon inclusion. Thus, we began testing this by selectively deleting the supporting lower stem in either of the two strands (mutants CFTR Del1 and Del2, see Figure 7A for a schematic diagram). In parallel, in order to test the importance of sequence conservation within the pseudoexon the structure was either ‘disrupted’ and then ‘repaired’ by engineering mutants CFTR Dis and CFTR Rep (see Figure 7C for a schematic diagram). These mutants were then tested either in vitro or following transfection in Hep3B cells for ability to recognize the pseudoexon (Figure 7B and D, respectively).
Figure 7.
(A) A schematic diagram of the two deletion mutants (CFTR Del1 and CFTR Del2) inserted in the PY7 system starting from the CFTR MUT plasmid. The splicing pattern displayed by these mutants either in vitro (left panel) or when transfected in Hep3B cells (right panel) is shown in (B). (C) The schematic diagram of an additional set of CFTR mutants (CFTR Dis and CFTR Rep) that were subjected to in vitro splicing (D: left panel) and transfected in Hep3B cells (D, right panel).
For the first set of constructs, the results demonstrate that pseudoexon inclusion in vitro is reduced from 50% inclusion in the CFTR MUT construct to 22% in the Del1 mutant (Figure 7B, left panel, lane 4). Even more strikingly, pseudoexon inclusion is almost completely abolished in the CFTR Del1 mutant when transfected in Hep3B cells (Figure 7B, right panel, lane 2). It should also be noted that the Del1 deletion changes only slightly the predicted strength of the 3′splice site of the CFTR pseudoexon (from a score of 0.96 to 0.91 according to the NNSplice program), ruling out the possibility that this deletion may have simply affected the quality of this site. On the other hand, no effect on splicing efficiency with respect to the CFTR MUT could be observed for the Del2 mutant in transfected Hep3B cells (Figure 7B, right panel, lane 3) whilst a small increase (to 68% inclusion) was observed in vitro (Figure 7B, left panel, lane 6).
For the second set of constructs it was observed that the pseudoexon in the CFTR Dis mutant was recognized with reduced effiency both in vitro and in Hep3B cells (Figure 7D, left panel lane 4 and right panel lane 2). In fact, pseudoexon inclusion in the CFTR Dis mutant fell to 30% in the in vitro splicing assay (from a 50% starting value of CFTR MUT in vitro) and to 64% inclusion in Hep3B cells (from a starting value of 93% inclusion for CFTR MUT in Hep3B cells). On the other hand, in keeping with the structural predictions, the CFTR Rep mutant fully recovered splicing efficiency with respect to the CFTR MUT construct (Figure 7D, left panel lane 6 and right panel lane 3). These results were thus in keeping with the proposed regulatory role of RNA structure on CFTR pseudoexon inclusion. Furthermore, they demonstrated that conservation of the primary nucleotide sequences was not required.
Nonetheless, on the basis of secondary structure considerations alone (Figure 7A) we also would have expected the Del1 and Del2 mutants to have similar effects on splicing efficiency. It was therefore of interest to check whether the splicing changes detected could be correlated with unforeseeable differential structural rearrangements introduced by these two mutations.
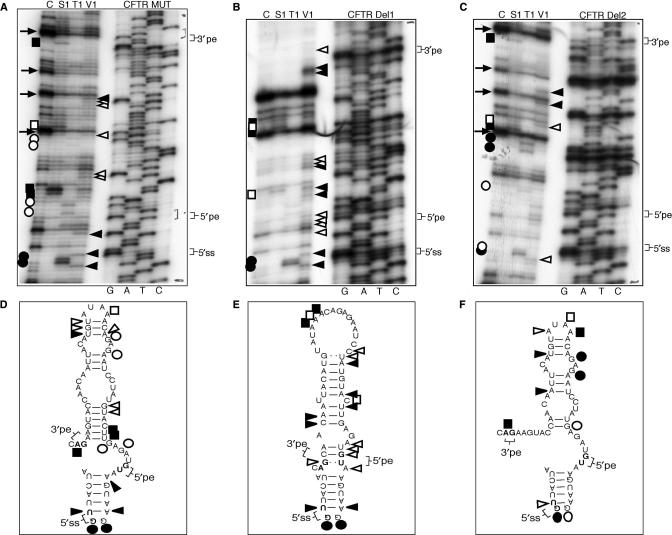
Structural analysis of the CFTR Del1-Del2 mutants
The results of the structural analysis performed on the Del1 mutant (Figure 8B) showed that this RNA displayed a markedly different RNAse profile with respect to the CFTR MUT profile (Figure 8A) and CFTR Del2 (Figure 8C). In particular, with the exception of the 5′ss region, there is very little conservation in cleavage pattern profiles between the CFTR Del1 mutant and the CFTR MUT, CFTR Del2 mutants. As shown in the schematic diagrams below, these results are in keeping with the predicted Del1 structure (Figure 8E) that includes the 5′pe, 5′ss, and 3′pe in a stem configuration, presumably making them less available to the splicing machinery. On the other hand, the cleavage pattern obtained for the Del2 mutant is very similar to that obtained for the MUT sequence (compare Figure 8A and C), up to the conservation of several RT stops indicated by arrows. Finally, in support of the structural changes detected in the Del1/Del2 mutants it has to be noted that the cleavages in correspondence to the 5′ss position are mostly conserved in all three structures (Figure 8A, B and C, 5′ss region), demonstrating that the differences in cleavage efficiencies for the different mutants do not represent an artifactual occurrence owing to different RNAse activities/efficiencies in the three analyses.
Figure 8.
(A, B and C) The enzymatic structure determination profiles of the CFTR MUT, CFTR Del1 and CFTR Del2 RNAs using RNAse digestion with the S1, V1 and T1 enzymes. Nomenclature of the cleavages and of the enzymes used is as described in Figure 2 legend. (D, E and F) The predicted RNA secondary structure of each RNA together with the corresponding cleavages observed in each probing assay. The positions of the acceptor (3′pe), pseudoexon donor (5′pe) and additional (5′ss) donor sites are indicated.
Therefore, the result of this analysis were also consistent with a proposed regulatory role of the major stem-loop element in CFTR pseudoexon inclusion. In addition, these results also highlight the ‘unpredictability’ of mutations aimed to modify RNA structural elements. In fact, based just on the 2D structural predictions the Del1 and Del2 mutations should have resulted in similar effects. The fact that limited RNAse digestion analysis showed this not to be the case underscores the need for direct experimental testing of secondary structures whenever evaluating the potential effects of introduced mutations.
DISCUSSION
Pre-mRNA secondary structure is increasingly recognized as a general modifier of splicing (1). However, evaluating its influence on the processing of individual exons is often a very difficult task and any research on RNA structures within coding regions is complicated by the need to rule out additional sources of bias. These include conservation of coding potential (35,50–52), the vast array of positive and negative cis-acting sequences that are now known to be present in most coding sequences (33,34,53), and that the RNA structure itself can be heavily influenced by RNA–protein interactions (54). Indeed, to this date very few examples exist regarding the existence of splicing-regulatory secondary structures residing entirely within exonic coding regions (5,6). It is difficult to assess if this scant representation of secondary structure is a feature of exons or the fact that they have never been extensively searched for, particularly in view of the laborious task of confirming unreliable computer predictions in naked RNA and the almost impossibility up to now of probing structure in vivo. Hopefully, the recent increase in sequencing data from many different organisms coupled with ever more refined folding algorithms and new technologies for gene expression visualization will allow researchers to identify additional likely examples of regulatory structural elements within mRNA molecules (19). It is also very possible that the often highly intricate network of regulatory processes embedded in exons/introns (33,55–58) and the need for the processed mRNA molecule to be translated, have all contributed to keep the 5′ss and 3′ss of real exons in regions of limited RNA structure.
Many of these limitations, of course, do not apply to intronic or non-coding sequences. Indeed, conserved stem-loop regions within introns have been recently shown to play a role in the pre-mRNA splicing processes of human SMN exon 7 (59), human COL2A1 exon 2 (60), in the highly conserved insect homeothorax gene (61) and in the yeast YRA1 gene (62). In addition, the structure-forming ability of introns has been very useful to explain a whole range of splicing phenomena that would otherwise be difficult to explain if the pre-mRNA molecule was considered as existing in a predominantly linear form (11–13,63).
In this work we have studied the importance of RNA secondary structure in pseudoexon inclusion events that are involved in human disease. These pseudoexons originate from the inclusion of apparently random intronic sequences following the introduction of an activating mutation, in general the creation of a novel acceptor or donor splice site although new observations also include modifications within splicing regulatory elements (see Supplementary Table 1 for a comprehensive list of these events). Our investigation of an event that concerns the deletion of a splicing suppressing element in the ATM gene (32) or of a splice-site creation event in the CFTR gene (48) has shown that in both cases RNA secondary structure plays a major role on donor-site usage and splicing efficiency.
Moreover, our results concerning the reasons that underlie the lack of use of an apparently viable 5′ss splice site in the CFTR genomic sequence has suggested that the RNA secondary structure may play a role of inhibiting post U1snRNP interactions, such as that with U6snRNP, by providing an additional inhibitory influence on the suboptimal +5 position. In this respect, therefore, our observations confirm previous suggestions that RNA secondary structures may have a general inhibitory effect on pseudoexon sequences (22). In fact, the presence of potentially fully functional splice-site sequences within the intronic sequences of most genes clearly shows that at least some of these sequences would not really be expected to wait for an activating mutation in order to be partially included in the final mRNA molecule. However, if these splice-site sequences find themselves bracketing sequences containing binding sites for splicing inhibitor molecules or, like in our cases, embedded into unfavourable RNA structures, then an explanation can be offered regarding why additional requirements have to be satisfied in order to obtain their ‘upgrade’ to exon status (for example, their presence in an ‘open’ structural configuration). The difficulty in evaluating these events on the basis of the primary sequence alone can also be appreciated from the performance of 5′ss scoring programs used in this work. In fact, beside the MaxEnt Scan evaluation methods that consider the MAG|GURAGU (M is A or C; R is purine) consensus sequence even methods such as NNsplice that consider in their evaluation sequences beyond the strict consensus fail to accurately predict the exact donor-site usage observed in our experimental systems. Our results will hopefully provide further information to facilitate the fine tuning of these splice-site prediction algorithms.
It should be noted, however, that RNA secondary structure may not represent the only determining factor in the ATM or CFTR pseudoexons, as highlighted by some of our experiments (i.e. Figure 4C) where the structure-affecting mutations also cause a drop in overall splicing efficiency of the pseudoexon. These results raise the possibility that splicing controlling elements may also be affected by these changes. In the future, it will be interesting to study in selected examples the potential interplay between RNA structural features and potential enhancer and silencer splicing controlling regions present in the pseudoexon of the type already demonstrated in the fibronectin EDA exon (5). Indeed, the role played by splicing factors in pseudoexon inclusion is a promising issue that remains to be fully investigated in the splicing field. In fact, in at least a minority of cases pseudoexon inclusion has been strongly correlated with creation of splicing regulatory enhancer elements (30–32). A similar situation may be present for nucleotide variations that remove the binding site for splicing inhibitory factors (32).
Nonetheless, our work suggests that taking into account structural features may represent a useful strategy to understand the functional mechanisms that underlie the activation of pseudoexon sequences in other genes causing human genetic diseases. This may be important also for attempting novel molecular therapy approaches to modify the regulation of pseudoexon inclusion events and for refining computer algorithms for the prediction of the splicing effect of any genomic variation. In fact, from a molecular therapy point of view the mapping of pseudoexon RNA secondary structures may provide useful indications regarding the development of inhibitory molecules such as antisense nucleotide that have already been suggested to possess increased working efficiency when targeted against open structural regions (64).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We wish to thank C.W. Smith for his kind gift of the PY7 plasmid. This work was supported by Telethon Onlus Foundation (Italy) (grant no. GGP02453 and GGP06147), FIRB (RBNE01W9PM) and by a European community grant (EURASNET- LSHG-CT-2005-518238). Funding to pay the Open Access publication charges for this article was provided by ICGEB.
Conflict of interest statement. None declared.
REFERENCES
- 1.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donahue CP, Muratore C, Wu JY, Kosik KS, Wolfe MS. Stabilization of the Tau Exon 10 stem loop alters pre-mRNA splicing. J. Biol. Chem. 2006;281:23302–23306. doi: 10.1074/jbc.C600143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Stephan W. Compensatory evolution of a precursor messenger RNA secondary structure in the Drosophila melanogaster Adh gene. Proc. Natl Acad. Sci. USA. 2003;100:11499–11504. doi: 10.1073/pnas.1932834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buratti E, Muro AF, Giombi M, Gherbassi D, Iaconcig A, Baralle FE. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higashide S, Morikawa K, Okumura M, Kondo S, Ogata M, Murakami T, Yamashita A, Kanemoto S, Manabe T, et al. Identification of regulatory cis-acting elements for alternative splicing of presenilin 2 exon 5 under hypoxic stress conditions. J. Neurochem. 2004;91:1191–1198. doi: 10.1111/j.1471-4159.2004.02798.x. [DOI] [PubMed] [Google Scholar]
- 7.Spingola M, Grate L, Haussler D, Ares M. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howe KJ, Ares M. Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl Acad. Sci. USA. 1997;94:12467–12472. doi: 10.1073/pnas.94.23.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miriami E, Margalit H, Sperling R. Conserved sequence elements associated with exon skipping. Nucleic Acids Res. 2003;31:1974–1983. doi: 10.1093/nar/gkg279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lian Y, Garner HR. Evidence for the regulation of alternative splicing via complementary DNA sequence repeats. Bioinformatics. 2005;21:1358–1364. doi: 10.1093/bioinformatics/bti180. [DOI] [PubMed] [Google Scholar]
- 11.Baraniak AP, Lasda EL, Wagner EJ, Garcia-Blanco MA. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 2003;23:9327–9337. doi: 10.1128/MCB.23.24.9327-9337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreahling JM, Graveley BR. The iStem, a long-range RNA secondary structure element required for efficient exon inclusion in the Drosophila Dscam pre-mRNA. Mol. Cell. Biol. 2005;25:10251–10260. doi: 10.1128/MCB.25.23.10251-10260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith CW. Alternative splicing–when two's a crowd. Cell. 2005;123:1–3. doi: 10.1016/j.cell.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Dixon RJ, Eperon IC, Samani NJ. Complementary intron sequence motifs associated with human exon repetition: a role for intragenic, inter-transcript interactions in gene expression. Bioinformatics. 2007;23:150–155. doi: 10.1093/bioinformatics/btl575. [DOI] [PubMed] [Google Scholar]
- 16.Chamary JV, Hurst LD. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Bor YC, Misawa Y, Xue Y, Rekosh D, Hammarskjold ML. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- 19.Meyer IM, Miklos I. Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs. Nucleic Acids Res. 2005;33:6338–6348. doi: 10.1093/nar/gki923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- 21.Mendes Soares LM, Valcarcel J. The expanding transcriptome: the genome as the ‘Book of Sand'. EMBO J. 2006;25:923–931. doi: 10.1038/sj.emboj.7601023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang XH, Leslie CS, Chasin LA. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun H, Chasin LA. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grellscheid SN, Smith CW. An apparent Pseudo-Exon acts both as an alternative exon that leads to nonsense-mediated decay and as a zero-length exon. Mol. Cell. Biol. 2006;26:2237–2246. doi: 10.1128/MCB.26.6.2237-2246.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorek R, Lev-Maor G, Reznik M, Dagan T, Belinky F, Graur D, Ast G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 26.Krull M, Brosius J, Schmitz J. Alu-SINE exonization: en route to protein-coding function. Mol. Biol. Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 27.Lei H, Day IN, Vorechovsky I. Exonization of AluYa5 in the human ACE gene requires mutations in both 3′ and 5′ splice sites and is facilitated by a conserved splicing enhancer. Nucleic Acids Res. 2005;33:3897–3906. doi: 10.1093/nar/gki707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, Giorda R, Pozzoli U. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32:1783–1791. doi: 10.1093/nar/gkh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishii S, Nakao S, Minamikawa-Tachino R, Desnick RJ, Fan JQ. Alternative splicing in the alpha-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype. Am. J. Hum. Genet. 2002;70:994–1002. doi: 10.1086/339431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King K, Flinter FA, Nihalani V, Green PM. Unusual deep intronic mutations in the COL4A5 gene cause X linked Alport syndrome. Hum. Genet. 2002;111:548–554. doi: 10.1007/s00439-002-0830-3. [DOI] [PubMed] [Google Scholar]
- 32.Pagani F, Buratti E, Stuani C, Bendix R, Dork T, Baralle FE. A new type of mutation causes a splicing defect in ATM. Nat. Genet. 2002;30:426–429. doi: 10.1038/ng858. [DOI] [PubMed] [Google Scholar]
- 33.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 35.Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 36.Sakharkar MK, Perumal BS, Sakharkar KR, Kangueane P. An analysis on gene architecture in human and mouse genomes. In Silico Biol. 2005;5:347–365. [PubMed] [Google Scholar]
- 37.Eperon LP, Graham IR, Griffiths AD, Eperon IC. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 38.Deirdre A, Scadden J, Smith CW. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;14:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 40.Baralle M, Baralle D, De Conti L, Mattocks C, Whittaker J, Knezevich A, Ffrench-Constant C, Baralle FE. Identification of a mutation that perturbs NF1, a gene splicing using genomic DNA samples and a minigene assay. J. Med. Genet. 2003;40:220–222. doi: 10.1136/jmg.40.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 42.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewandowska MA, Stuani C, Parvizpur A, Baralle FE, Pagani F. Functional studies on the ATM intronic splicing processing element. Nucleic Acids Res. 2005;33:4007–4015. doi: 10.1093/nar/gki710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burset M, Seledtsov IA, Solovyev VV. SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 2001;29:255–259. doi: 10.1093/nar/29.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 46.Rogozin IB, Milanesi L. Analysis of donor splice sites in different eukaryotic organisms. J. Mol. Evol. 1997;45:50–59. doi: 10.1007/pl00006200. [DOI] [PubMed] [Google Scholar]
- 47.Goguel V, Wang Y, Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol. Cell. Biol. 1993;13:6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chillon M, Dork T, Casals T, Gimenez J, Fonknechten N, Will K, Ramos D, Nunes V, Estivill X. A novel donor splice site in intron 11 of the CFTR gene, created by mutation 1811 + 1.6kbA–>G, produces a new exon: high frequency in Spanish cystic fibrosis chromosomes and association with severe phenotype. Am. J. Hum. Genet. 1995;56:623–629. [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson KK, Green MR. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988;2:319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- 50.Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl Acad. Sci. USA. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xing Y, Lee C. Alternative splicing and RNA selection pressure–evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 52.Parmley JL, Chamary JV, Hurst LD. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol. Biol. Evol. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- 53.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 54.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell. Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 55.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 56.Singh R, Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 57.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 58.Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 59.Miyaso H, Okumura M, Kondo S, Higashide S, Miyajima H, Imaizumi K. An intronic splicing enhancer element in survival motor neuron (SMN) pre-mRNA. J. Biol. Chem. 2003;278:15825–15831. doi: 10.1074/jbc.M209271200. [DOI] [PubMed] [Google Scholar]
- 60.McAlinden A, Havlioglu N, Liang L, Davies SR, Sandell LJ. Alternative splicing of type II procollagen exon 2 is regulated by the combination of a weak 5′ splice site and an adjacent intronic stem-loop cis element. J. Biol. Chem. 2005;280:32700–32711. doi: 10.1074/jbc.M505940200. [DOI] [PubMed] [Google Scholar]
- 61.Glazov EA, Pheasant M, McGraw EA, Bejerano G, Mattick JS. Ultraconserved elements in insect genomes: a highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 2005;15:800–808. doi: 10.1101/gr.3545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Preker PJ, Guthrie C. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA. 2006;12:994–1006. doi: 10.1261/rna.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwarze U, Hata R, McKusick VA, Shinkai H, Hoyme HE, Pyeritz RE, Byers PH. Rare autosomal recessive cardiac valvular form of Ehlers-Danlos syndrome results from mutations in the COL1A2 gene that activate the nonsense-mediated RNA decay pathway. Am. J. Hum. Genet. 2004;74:917–930. doi: 10.1086/420794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aartsma-Rus A, De Winter CL, Janson AA, Kaman WE, Van Ommen GJ, Den Dunnen JT, Van Deutekom JC. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15:284–297. doi: 10.1089/oli.2005.15.284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.