Abstract
Bacterial flagellar motility and chemotaxis help cells to reach the most favorable environments and to successfully compete with other micro-organisms in response to external stimuli. Escherichia coli is a motile gram-negative bacterium, and the flagellar regulon in E. coli is controlled by a master regulator FlhDC as well as a second regulator, flagellum-specific sigma factor, σF. To define the physiological role of these two regulators, we carried out transcription profiling experiments to identify, on a genome-wide basis, genes under the control of these two regulators. In addition, the synchronized pattern of increasing CRP activity causing increasing FlhDC expression with decreasing carbon source quality, together with the apparent coupling of motility activity with the activation of motility and chemotaxis genes in poor quality carbon sources, highlights the importance of CRP activation in allowing E. coli to devote progressively more of its limited reserves to search out better conditions. In adaptation to a variety of carbon sources, the motile bacteria carry out tactical responses by increasing flagellar operation but restricting costly flagellar synthesis, indicating its capability of strategically using the precious energy in nutrient-poor environments for maximizing survival.
INTRODUCTION
The DNA-dependent RNA polymerase of bacteria is the sole enzyme capable of producing messenger, transfer and ribosomal RNA by catalyzing the 5′ to 3′ synthesis of phosphodiester bonds between adjacent ribonucleoside triphosphates. In E. coli, core DNA-dependent RNA polymerase consists of four different subunits and has the composition α2ββ′ω. It was discovered that an additional and separable factor, the sigma factor (σ70), was required for transcription of certain types of DNA (1). Numerous sigma factors have been described in E. coli and other prokaryotic organisms (2–5) since then. The seven known E. coli sigma factors are σ70, σ54, σ32, σS, σF, σE and σfecI. The holoenzyme complex (Eσ) (1,6) constituted by core RNA polymerase (E) together with a sigma factor (σ) is required to initiate transcription at specific DNA sequences termed promoters.
Through the specificity of its σ subunit, holoenzyme is directed to two conserved DNA hexamers centered ∼10 and 35 bp upstream of the transcription start to initiate transcription. Each sigma factor recognizes and directs RNA polymerase to a different set of promoters. While sigma factors provide the primary interaction with those consensus two-block promoter DNA sequences and play a central role in the regulation of global gene expression, additional transcriptional activators such as FIS (7,8) and CRP (9,10) can be utilized to strengthen the promoter–holoenzyme interaction.
Flagellar, chemotaxis and motility genes are organized into large complex units. The genes that constitute the flagellar regulon are expressed in a cascade that closely parallels the assembly hierarchy of the flagellar structure (11,12). The flagellar operons are divided into three gene classes with respect to this transcriptional hierarchy (classes I, II and III or early, middle and late classes, respectively). At the top of the hierarchy is the FlhDC master operon that encodes FlhDC complex as a positive transcriptional activator of σ70-dependent transcription from class II promoters which also include the fliA promoter. σF is made, binds to core RNA polymerase, and then transcribes from class III promoters. The synthesis of the flagellar system can be tightly regulated by a master regulator, the transcriptional activator FlhDC complex, as well as by the secondary regulator, the alternative sigma factor, σF.
In the previous characterizations of the flagellar regulon, genetic mutagenesis analysis was used as a main approach to study those non-flagellate mutants which included spontaneous, Mu phage-induced, and some transposon-induced flagellar mutants to define genes involved in the formation of a functional flagellar apparatus (13–17). Although several new flagellar genes have been recently identified in Salmonella enterica (18), no additional experiment has been published so far to systematically study the FlhDC and σF regulons in E. coli. Due to the high transcriptional and translational level of these two regulators in log-phase, our basic strategy was to delete these two regulators as well as to minimally perturb steady-state cell growth by moderate induction of these genes in defined MOPS minimal medium. We then monitor global RNA transcript abundance change using Affymetrix GeneChip E. coli Antisense Genome Arrays. While transcription by σF is known to be modulated by the anti-sigma FlgM (19), we will not consider effects of FlgM here. We believe that the genes which are dependent on σF can be identified by the σF deletion or induction experiments presented.
In addition, to characterize how the activity of motility is regulated by different carbon sources, we measured the transcriptional level of FlhDC and FliA as well as the level of several well-known genes under the direct control of these two regulators in a range of carbon source conditions by a quantitative RT-PCR analysis. The correlation between the activity of CRP and the motility of cells indicates that activation of CRP plays an important role in triggering E. coli ‘foraging’-like behavior (20–22) to actively search for better conditions as the quality of the available substrate(s) decreases. On the basis of these first systematic studies of the FlhDC complex and σF regulons in E. coli, including the effects of carbon source conditions on motility of cells, we gain insight into the complex networks regulated by these two regulators and how they contribute physiological adaptation to the changes in the external environment.
MATERIALS AND METHODS
Reagents, strains and plasmids
All reagents were purchased from Sigma Chemical Company (St Louis, MO) unless otherwise indicated. 10X MOPS minimal media was prepared as described in Neidhardt et al. (23). The media was filter sterilized through a 0.2 μm filter and stored at 4°C. The defined media for log-phase cell growth contained 1 X MOPS minimal media, 0.1% glucose, 0.66 mM K2HPO4.
Because the E. coli Genechip probe set is based on the sequenced E. coli K-12 strain MG1655 (λ− F− ilvG− rfb−50 rph-1, prototroph) (24), we chose this bacterial strain for use in our study. In order to disrupt the expression of FlhDC/σF in E. coli, we used a simple and highly efficient method (25,26) to prepared in-frame deletion strains for FliA (σF) and for the master regulator FlhDC. In this procedure (as shown in supplemental material Figure S1), we generated PCR products by using primers with 60- to 70-nt extensions that are homologous to regions adjacent to flhDC or fliA gene(s) and a template plasmid carrying kanamycin-antibiotic resistance genes. Using linear DNA to do homologous recombination in E. coli requires the phage lambda Red recombinase (λRed system), which is synthesized under the control of an inducible promoter on a low copy number plasmid. Deletion mutants of the fliA gene or flhDC operon were isolated as kanamycin antibiotic-resistant colonies after the introduction the respective PCR products into bacteria carrying a λRed expression plasmid. The replication of the temperature-sensitive plasmid pKD46 was inhibited and the loss of this plasmid in mutant strains occurred when clones were grown at 43°C. For controllable induction of individual regulators in vivo, we used the PLtet promoter which is controlled by the repressor TetR to construct these overexpression vectors as described previously (27). A downstream gene can be induced in the presence of inducer, aTc. All strains used in this study were derivatives of E. coli K12 MG1655.
Microarray experiments
Escherichia coli strains were grown overnight in MOPS minimal media at 37°C in an air shaker with vigorous aeration (225 rpm). Two milliliters of the overnight culture was used to inoculate 100 ml of fresh MOPS minimal medium. For preparing the total RNA for microarray experiments, 15 ml samples of culture (corresponding to 7.5 × 109 cells) were taken for wild-type and deletion mutant strains when the culture density OD600 value reached 0.2 and the same amount of culture was taken before and 5 min after induction in FlhDC or σF overexpression strains [based on our previous results from σ32 time-course experiments (27), we choose 5 min after induction in this assay because it is a reasonable time point that provides sufficient time to induce sigma-dependent genes and also reduce potential post-transcription or other indirect effects]. RNA was stabilized immediately by mixing with a double volume of RNAprotect Bacterial Reagent (Qiagen) and incubated at room temperature for 10 min. Cells were centrifuged at 5800 g for 20 min and cell pellets were stored at −80°C prior to RNA extraction. Preparation of labeled probes and microarray procedures were performed exactly as described previously (27) and in Supplemental Material.
Real-time quantitative RT-PCR
Quantitative reverse transcription (RT)-PCR primers were designed using Primer Express software (Applied Biosystems) and were synthesized by the UW Biotechnology Center. Two steps of real-time quantitative RT-PCR are performed. Five micrograms of the DNase-treated total RNA was reverse transcribed for first strand cDNA by using Superscript II system (Invitrogen). Reactions were then performed using 1 ng cDNA and 100 nM of each primer in a 50 µl volume with 1XSYBR Green I mixture. Controls lacking AmpliTaq Gold DNA Polymerase or template were used. Reactions were run on an ABI 7700 instrument (Applied Biosystems) using the following cycling parameters: 95°C for 10 min, 40 cycles of denaturation at 94°C for 15 s and extension at 60°C for 1 min. Relative gene expression data analysis was carried out with the standard curve method (28). Assays were performed in triplicate.
Multiple-round in vitro transcription following by filter-binding assay for radioactive incorporation
The incorporation of radioactivity in the newly synthesized RNA can be measured by the DE81 filter-binding assay. The DNA fragments used for in vitro transcription assays were amplified by PCR with/without the upstream sequence (∼350 bp) of those candidate σF-dependent genes with high confidence identified in our microarray data. The DNA fragment (∼50 nM) was incubated with ∼15 nM purified σF-associated holoenzyme in a buffer containing 50 mM Tris–HCl (pH 7.9), 150 mM K Glu, 150 µM 3 NTP's (CTP, ATP, GTP) and 20 μM UTP plus 1 μl [α-32P] UTP (∼10 µCi), 100 μg/ml BSA, 1 mM EDTA, 10 mM MgCl2, 1 mM DTT in a total volume of 20 µl. The mixtures were incubated at 37°C and transcription was stopped by addition of 0.5 M EDTA (final 100 mM) 30 min after reactions were initiated. The samples were loaded directly onto DE81 filter discs of 22.5-mm diameter (Whatman). The 32P-labeled transcripts were bound to DE81 filters by absorbing the total reaction mixture on the filters. Unincorporated nucleoside triphosphates were removed by washing the filters three times with 500 mM Na2HPO4 buffer, pH 7.6 and twice with 95% ethanol. The filter discs were dried, and nucleotide incorporation was quantified by Cerenkov counting. Incorporation was corrected for background radioactivity by measuring the amount of apparent incorporation in the absence of holoenzyme.
Electrophoretic mobility-shift assays (EMSA)
The DNA fragments (∼350 bp) used for gel mobility shift assays were amplified by PCR from the upstream sequence of flhDC operon. The DNA fragment was 32P-labeled at the 5′ end using T4 polynucleotide kinase. Samples of <40 ng of the labeled DNA fragments were included in 20-μl reaction mixtures containing DNA-binding buffer (10 mM Tris–HCl, pH 7.5, 50 mM KCl, 0.5 mM EDTA, 5% glycerol, 1 mM dithiothreitol), 500 µg/ml bovine serum albumin (BSA) and 25 µg/ml herring sperm DNA. The CRP was added at the following concentrations: 0, 5, 10 and 25 nM, respectively. cAMP was included in all reaction mixtures at a final concentration of 2 mM. Reaction mixtures were incubated for 15 min at 37°C and then were stopped by the addition of 1 μl of loading buffer (0.1% xylene cyanol and 50% glycerol in H2O). The samples were loaded on a 4–10% native Tris–glycine Novex Gel (Invitrogen) and dried on a Slab Dryer (BioRad) as described previously (27). Biomax MS film (Kodak) was used for autoradiography. The gels were scanned using a PhosphorImager (Molecular Dynamics), and the intensities of the bands were determined using ImageQuant version 5.2 software.
Additional experimental procedures are provided in Supplemental Material.
RESULTS
The inactivation and overexpression of flhDC and fliA in E. coli MG1655 strains
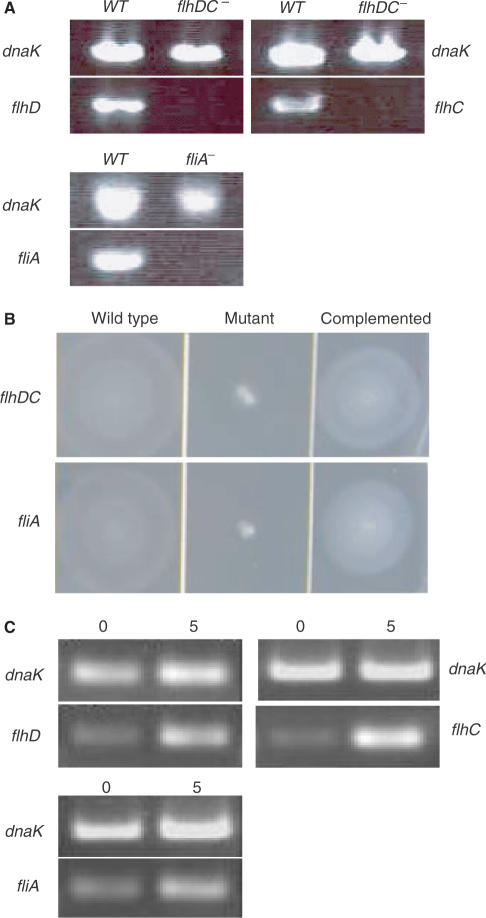
The sequenced E. coli K-12 strain MG1655 (λ− F− ilvG− rfb−50 rph-1, prototroph) (24), on which E. coli Affymetrix Genechip probe design is based, was chosen for our studies. E. coli FlhDC or σF in-frame deletion strains as well as FlhDC or σF overexpression strains were constructed as described in Zhao et al. (27) and in Experimental Procedures. RT-PCR was used to examine the expression of flhD, flhC and fliA in the respective deletion mutants before microarray analysis. As expected, the FlhDC mutant did not express flhD and flhC, and the σF mutant failed to express fliA (Figure 1A), confirming inactivation of these genes.
Figure 1.
Confirmation of FlhDC and σF deletion and overexpression in respective strains. (A) RT-PCR analysis of flhD, flhC and fliA expression in deletion strains. RNA isolated from each strain was converted to cDNA and PCR-amplified with primers specific for flhD, flhC and fliA. DnaK served as positive control. Expression of DnaK can be detected by RT-PCR in the wild-type strain as well as in flhDC and fliA mutant strains. Expression of flhD, flhC or fliA can only be detected in wild-type strain, but the expression of these genes is absent in the respective mutants. (B) Motility in E. coli wild-type strain and flhDC and fliA mutant strains. The flhDC and fliA deletion strains lose their motility on soft tryptone swarm agar. The motility can be recovered from these mutant strains by in vivo expression of FlhDC or σF from a plasmid-borne flhDC or fliA gene, respectively. (C) RT-PCR analysis of flhD, flhC and fliA expression in overexpression strains. Compared with the transcript level of control (dnaK), a significant increase of transcripts of flhD, flhC and fliA expression can be seen after a 5-min induction.
Soft tryptone swarm agar plates (29) were used to evaluate motility or swimming ability of the strains. In the tryptone swarm agar (Figure 1B), the flhDC and fliA deletion mutants were totally non-motile and did not form any swarm rings compared with the wild type. When the flhDC or fliA mutant was complemented with a cloned flhDC or fliA gene on a low-copy plasmid, the motility was restored as shown in Figure 1B. The empty vector pACYC184 served as a control and had no influence on the motility of the flhDC or fliA mutant (data not shown).
RT-PCR (semi-quantitative) was also performed to detect the mRNA level of flhDC or fliA genes before and after induction of a plasmid-borne flhDC or fliA genes in E. coli. Instead of 35 PCR cycles as performed earlier to test gene disruption, the semi-quantitative PCR was performed for 24 cycles. This reduces the chance of saturation of final PCR product. The synthesized DNA was loaded onto an agarose gel and stained with ethidium bromide. A significant increase of RNA level from target genes was observed after a 5-min induction as shown in Figure 1C, confirming induction of those genes.
FlhDC regulon
To characterize the effects of the decreasing or increasing FlhDC protein level in vivo on gene expression, global RNA transcript abundance was monitored in the deletion mutant strain and the overexpression strain 5 min after FlhDC induction with cells grown in log-phase (OD600 = 0.2) in MOPS minimal medium at 37°C. Transcription profiles were obtained as described in ‘Materials and Methods’ section.
Expression profiling of transcripts corresponding to the complete set of ORFs in the E. coli genome revealed that the response to deletion of FlhDC in vivo was quite broad. There are 117 genes (2.7% of the genome) downregulated 2-fold or more in the flhDC deletion mutant strain. The wide distribution of FlhDC-dependent genes in E. coli genome (as shown in Figure 2) indicates that FlhDC might play a larger role in the global gene transcription regulation than just to serve as a master regulator for the flagellar regulon. There are 53 genes in E. coli known to be directly involved in flagellar structure and motor function (11,12). Compared with the transcriptional level of genes in the wild-type strain, DNA microarray results showed the transcriptional level of all these genes are significantly downregulated in the FlhDC deletion strain (see Supplemental Material, Table S1). Most of these known genes in the flagellar regulon were initially identified through genetic mutagenesis analysis and can be divided into two functional groups: (1) chemotaxis and mobility; (2) surface structures.
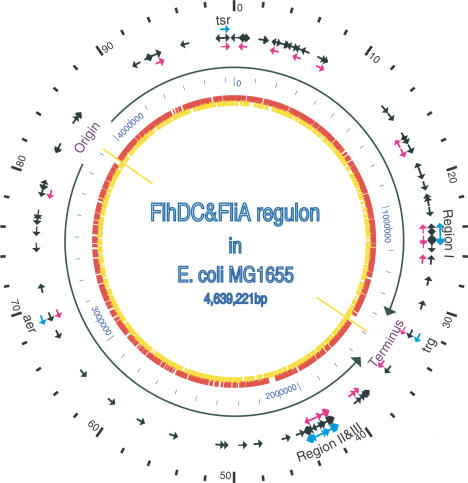
Figure 2.
The location and transcription direction of genes in the FlhDC and FliA regulons on the map of the E. coli genome. The outer scale point circle indicates the coordinates in minutes (100 equals intervals of DNA). Blue arrows show the locations and directions of transcription of 53 known flagellar regulon genes. Most of these genes are located in regions I, II and III (i.e. at 24, 42 and 43 min in centisomes, respectively), except three chemotaxis genes trg, aer and tsr, which are at 32, 69, 98 min, respectively. The black arrows show all the genes (117) in FlhDC regulon that were identified by our microarray studies, which include all the previously known flagellar-related genes. The pink arrows show 37 σF-dependent genes identified in our microarray studies. These include all 21 previous known σF-controlled genes in flagellar system. The origin and terminus of replication are shown as yellow lines, with black arrows indicating replichores. The next point circle in light blue gives the scale in base pairs. The inner circle shows ORFs distribution on two complement DNA strands as presented in (24).
Comparing the DNA microarray results before and 5 min after FlhDC induction in the overexpression strain, there are no significant changes (no RNA level changed more than 2-fold compared to the uninduced control) of RNA level of genes in the flagellar regulon (data not shown). We are not too surprised with this result. Compared with previous experiments (27) where initial σ32 protein level is low and then is induced almost 8-fold after a 5-min induction, the fold induction of FlhDC under the same inducible PLtet promoter control must be limited due to the high initial abundance of this protein. Therefore, we expect the reason for no significant increased transcription of FlhDC-dependent genes is due to the high initial protein level of FlhDC before induction. The low fold increase of FlhDC in this short time period (5 min) is not enough to further increase transcription of FlhDC-dependent genes using σ70-associated holoenzyme.
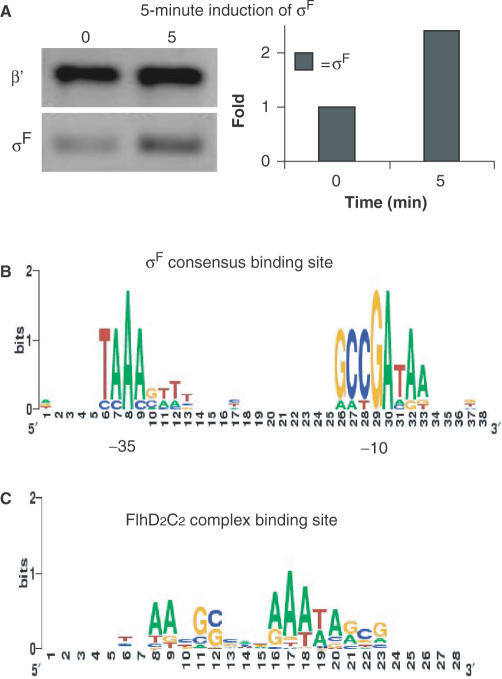
In Table S2 (Supplemental Material), we show a group of 12 new candidate genes whose transcriptional level decreased more than 3-fold as well as a non-flagellar gene, yecR, known to be regulated by FlhDC complex in E. coli (30). Computer prediction of FlhDC-related binding element (27) shows that the consensus is represented as tNAAcGCc(N)2AAATAgcg (Figure 3C), where lowercase letters indicate less highly conserved sites. This consensus agrees well with the previously reported FlhDC binding consensus that was aligned from several published FlhDC-dependent genes (31). Note, the general height of these consensus element displayed on SEQUENCE LOGO is not high. This indicates that FlhDC consensus binding sequence might not be as strict as sigma factor-binding sites.
Figure 3.
Determination the σF protein level and the consensus-binding sites of σF and FlhDC complex. (A) Left: Western blot of β' and σF before and 5 min after induction. β′-subunit of core RNA polymerase served as an internal control. Right: quantification of Western blot. The σF protein level increases approximately 2.3-fold after 5 min of induction. Signal intensities are determined using ImageQuant version 5.2 software. (B) The two-block promoter element recognized by σF is aligned using BioProspector (36) from the upstream sequence of σF-dependent genes identified in our microarray data and displayed using SEQUENCE LOGO (59). (C) The FlhDC-related DNA-binding element is aligned using BioProspector from the upstream sequence of FlhDC-dependent genes identified in our microarray data and displayed using SEQUENCE LOGO. The height of each column reflects the non-random bias of particular residues at that position, the size of each residue letter reflects its frequency at that position.
σF regulon
There are 21 known σF-dependent genes in E. coli involved in flagellar synthesis and function (11,12). In Table 1, we can see that the transcriptional level of these genes is significantly downregulated in the fliA− strain and is slightly increased in σF overexpression strain. These results are consistent with our previous hypothesis that a change of the intracellular level of a given sigma factor will cause a change of the transcriptional level of genes dependent on this sigma factor. Jishage (32) reported that the intracellular level of σF is maintained at 50% the level of σ70 during log and stationary phase growth and σF is thought to be the second most abundant sigma factor among seven sigma factors in E. coli. Loss of σF in cells will greatly decrease the transcription of σF-dependent genes; especially those genes that can only be transcribed by σF-associated holoenzyme (such as fliC in Table 1). Compared with our previous study where the induction of σ32 caused high induction of σ32-dependent genes, induction of σF in this study did not show a large increase in the transcriptional level of σF-dependent genes. Using a specific monoclonal antibody for σF, we determined that the protein level of σF increased about 2.3-fold after a 5-min induction (Figure 3A), lower than the 8-fold increase in the previous σ32 induction experiments.
Table 1.
All 21 known σF-dependent genes in flagellar system are downregulated (or induced) after FliA deletion (or overexpression)
| b no.a | Gene | Product | Function | Deletion | Overexpression |
|---|---|---|---|---|---|
| b1923 | fliCb | Flagellar biosynthesis; flagellin, filament structural protein | Surface structures | −78.8c | 3.9d |
| b1924 | fliD | Flagellar biosynthesis; filament capping protein, enables filament assembly | Surface structures | −9.9 | 3.2 |
| b1071 | flgM | Anti-FliA (anti-sigma) factor; also known as RflB protein | Surface structures | −6.9 | 2.0 |
| b1082 | flgK | Flagellar biosynthesis; hook-filament junction protein 1 | Surface structures | −5.5 | 2.2 |
| b1083 | flgL | Flagellar biosynthesis; hook-filament junction protein | Surface structures | −4.9 | 1.8 |
| b1925 | fliS | Flagellar biosynthesis; repressor of class 3a and 3b operons (RflA activity) | Surface structures | −4.6 | 2.2 |
| b1070 | flgN | Flagellar biosynthesis; believed to be export chaperone for FlgK and FlgL | Surface structures | −4.6 | 1.9 |
| b1926 | fliT | Flagellar biosynthesis; putative export chaperone for FliD | Surface structures | −2.2 | 3.4 |
| b3072 | aer | Aerotaxis sensor receptor, senses cellular redox state or proton motive force | Degradation of small molecules: | −2.0 | 2.8 |
| b1889 | motB | Enables flagellar motor rotation, linking torque machinery to cell wall | Chemotaxis and mobility | −46.9 | 3.5 |
| b1881 | cheZ | Chemotactic response, CheY protein phosphatase | Chemotaxis and mobility | −40.8 | 2.9 |
| b1887 | cheW | Purine-binding chemotaxis protein; regulation | Chemotaxis and mobility | −26.0 | 1.8 |
| b1888 | cheA | Chemotactic sensory histidine kinase (soluble) in two- component regulatory system with CheB and CheY, senses chemotactic signal | Chemotaxis and mobility | −16.1 | 3.2 |
| b1883 | cheB | Chemotactic response regulator; methylesterase, in two-component regulatory system with CheA, regulates chemotactic response | Chemotaxis and mobility | −14.5 | 3.7 |
| b1890 | motA | Proton conductor component of motor, torque generator | Chemotaxis and mobility | −14.0 | 4.5 |
| b1886 | tar | Methyl-accepting chemotaxis protein II, aspartate sensor receptor | Chemotaxis and mobility | −13.2 | 2.4 |
| b1882 | cheY | Chemotactic response regulator in two-component regulatory system with CheA, transmits signals to FliM flagelllar motor component | Chemotaxis and mobility | −12.0 | 3.1 |
| b1885 | tap | Methyl-accepting chemotaxis protein IV, peptide sensor receptor | Chemotaxis and mobility | −6.7 | 2.3 |
| b4355 | tsr | Methyl-accepting chemotaxis protein I, serine sensor receptor | Chemotaxis and mobility | −5.4 | 3.1 |
| b1884 | cheR | Chemotactic response, glutamate methyltransferase | Chemotaxis and mobility | −5.0 | 2.3 |
| b1421 | trg | Methyl-accepting chemotaxis protein III, ribose and galactose sensor receptor | Chemotaxis and mobility | −2.0 | 2.2 |
ab no. indicates Blattner number.
bIt is possible that one gene has several different gene names.
cNumbers indicate fold decrease to wild-type strain.
dNumbers indicate fold increase to pre-σF induction.
Comparative analysis of the microarray data from the set of genes whose transcription is downregulated in the fliA deletion strain (decrease of σF) and the set of genes with increased transcription at 5 min after σF induction (increase of σF) allow us to assign many additional genes to the σF regulon. In Table 2, there are 13 new candidate genes in σF regulon. The transcriptional level changes of three reported σF-dependent genes (33,34) which were not assigned into traditional flagellar regulon before (flxA, b1194 and yhjH) are also listed in Table 2. These three genes are non-flagellar genes or have unknown function in the flagellar system.
Table 2.
Additional genes for the σF regulon
| b no.a | Gene | Product | Function | Deletion | Overexpression |
|---|---|---|---|---|---|
| b1566 | flxAb | Qin prophage | Unknown | −28.4c | 8.2d |
| b1194 | b1194 | Putative regulator | Unknown | −17.5 | 3.3 |
| b4109 | yjdA | Conserved hypothetical protein | Not classified | −12.3 | 5.3 |
| b3525 | yhjH | Conserved protein | Unknown | −9.6 | 1.9 |
| b1760 | ynjH | Conserved hypothetical protein | Unknown | −6.9 | 7.9 |
| b1742 | ydjR | Conserved hypothetical protein | Unknown | −4.7 | 4.5 |
| b0250 | ykfB | CP4-6 prophage | Unknown | −3.7 | 4.0 |
| b0764 | modB | Molybdate transport protein | Transport of small molecules | −3.2 | 2.0 |
| b0763 | modA | Molybdate transport protein | Transport of small molecules | −3.0 | 1.3 |
| b0154 | hemL | Glutamate-1-semialdehyde aminotransferase (aminomutase), PLP-dependent | Biosynthesis of cofactors, carriers: Heme, porphyrin | −3.0 | 2.3 |
| b0315 | yahA | Putative transcriptional repressor | Unknown | −2.5 | 1.1 |
| b0049 | apaH | Diadenosine tetraphosphatase | Salvage of nucleosides and nucleotides | −2.5 | 1.1 |
| b4110 | yjcZ | Unknown | Unknown | −2.3 | 4.2 |
| b1126 | potA | Spermidine/putrescine transport protein | Transport of small molecules: | −2.1 | 1.4 |
| b4315 | fimI | Fimbrial protein, internal segment | Surface structures | −2.0 | 2.6 |
| b0757 | galK | Galactokinase | Degradation of small molecules | −2.0 | 2.6 |
ab no. indicates Blattner number.
bIt is possible that one gene has several different gene names.
cNumbers indicate fold decrease relative to wild-type strain.
dNumbers indicate fold increase relative to pre-σF induction.
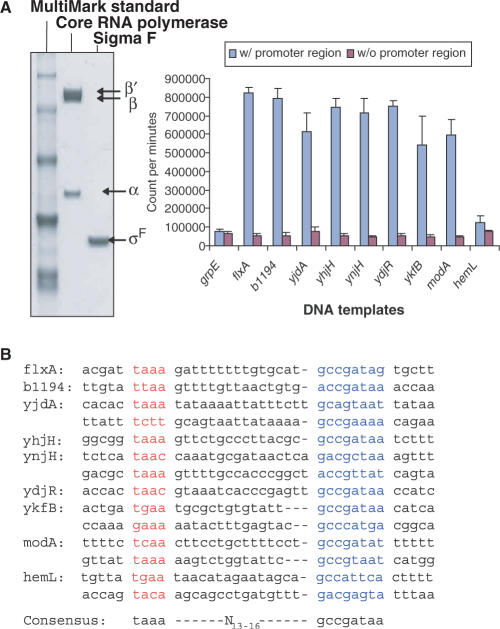
We chose the top 10 genes in Table 2 for in vitro transcription assays because no experiment has been performed so far to test if these genes can be directly transcribed by σF-associated holoenzyme. The upstream sequence of grpE gene, encoding a heat shock protein, was chosen as a negative control for the in vitro transcription assay because transcription of this gene is dependent on σ32 and is not a σF-dependent gene (35). In vitro transcription assay results (Figure 4A) show that most of these 10 genes can be directly transcribed by σF holoenzyme. Note both modA and modB genes are in one operon. The modB gene can be co-transcribed by σF-dependent promoter located in the upstream region of the modA gene. Results from promoter region consensus analysis using the algorithms BioProspector (36) and HMMER (37) revealed σF holoenzyme-binding sites in the upstream regulatory sequences of these genes (Figure 4B). Note that several of these genes have two putative promoters. Based solely on the negative results from our in vitro transcription assay, we are not sure if the hemL gene belongs to the σF regulon. While these possible two-block DNA consensus sequences might provide the primary interaction with holoenzyme, additional transcriptional activators such as FIS and CRP might be utilized to strengthen the promoter–holoenzyme interaction in vivo which are not available in our in vitro transcription assay. Due to the virulence role of pathogenic bacteria flagella system in adhesion, biofilm formation and colonization of host organisms and in secretion of virulence determinants to host (38–40), the homologous counterparts of the newly identified genes presented here might be potentially associated with these functions, especially with that of the pathogenicity island SPI-1 TTSS (type three secretion system) which appears to be most commonly found in pathogenic bacteria.
Figure 4.
In vitro transcription assays to test the transcription by σF holoenzyme of the DNA fragments carrying putative promoter element(s). (A) SDS–PAGE gel shows purified core RNA polymerase and σF as well as MultiMark Standard. In vitro transcription assays are performed using purified σF-associated holoenzyme with DNA fragments from the upstream sequence of candidate σF-dependent genes. The upstream sequence of grpE served as a negative control. Error bars represent standard deviation in three different experiments. (B) Potential σF consensus-binding sites of each gene are predicted and aligned by computer program. Previously known σF two-block promoter element consensus is shown below.
Motility control by carbon source: CRP binds to and scalably activates FlhDC operon in alternative carbon sources
Recently, Liu and coworkers (20) proposed a new model for carbon source foraging strategy by E. coli. By growing E. coli in several different carbon sources, they discovered, as carbon substrate quality declines (defined by growth rate), cells systematically increase the number of genes expressed in a hierarchical manner. Concomitantly, cells also increase their motility. They proposed a RNA polymerase (RNAP) reapportioning model to explain the expansion of genes expression. But the mechanism of increasing the motility in a low-quality carbon source remains unknown.
The gradual increase of motility activity with decreased carbon quality was unanticipated because motility by means of flagella is very expensive for cellular economy in terms of the number of genes and the energy required for flagellar biosynthesis and functioning (11). Using energy-intensive flagella in poor nutrient environments would trigger a high risk of more rapidly exhausting the sole energy supply. In Liu's paper (20), a strategy known as risk-prone foraging (21,22) has been proposed for this behavior; that bacteria take a risk and use the flagellar system to actively search out better conditions. It was observed many years ago that carbon catabolite repression affects flagellar biosynthesis (41,42). This led us to think that the relief from the carbon catabolite repression might be a key factor involved in the effects on motility activity inversely correlated with different carbon source quality.
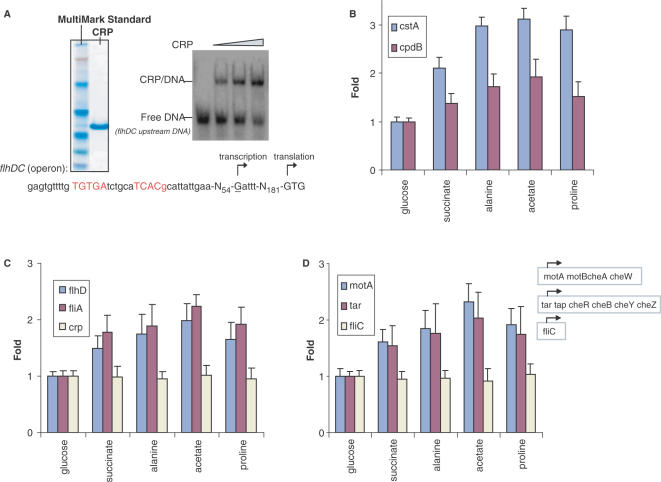
Relief from the carbon catabolite repression is a complex regulatory circuit that triggers reprogramming of global gene expression patterns to adapt the changes in external environment. This mechanism will activate the cyclic AMP receptor protein (CRP) (43), a global transcriptional factor that positively regulates most carbon catabolic pathways. While it is known that carbon catabolite repression affects the flagellar synthesis and the CRP activation might be involved in alleviating this repression (41,42,44,45), much less is known regarding the role of CRP in motility regulation under a range of carbon source conditions. To determine the effect of different carbon sources on E. coli CRP activity as well as the functional relevance between the active CRP level and the expression of FlhDC operon in a range of conditions with the sequenced E. coli strain MG1655, we performed the following assays. CRP protein was purified using the pET expression system (Supplemental Material, Figure S2) for in vitro assays. In electrophoretic mobility-shift assays as shown in Figure 5A, the upstream DNA fragment of flhDC operon can be shifted by purified CRP protein. CRP is a dimer of identical subunits. The consensus tandem DNA-binding site for CRP dimer has been identified by in silico analysis as shown in red color which is approximately palindromic and provides two very similar recognition sites, one for each subunit of the dimer. For sequenced E. coli strain MG1655, there are 250 bp between the CRP-binding sites and FlhDC translational start site, which has a rare translation start codon with GTG that is different from another E. coli strain studied by Soutourina et al. (44). We then measured the transcriptional levels of two well-known CRP positively controlled genes (cstA and cpdB) (to represent the level of active CRP in the cell) as well as the expression level of flhDC operon by the quantitative RT-PCR approaches (Figure 5B and C). Compared with cells grown in glucose, the transcriptional levels of CRP-dependent genes gradually increased in succinate, alanine and acetate grown cells, with a slight decline in proline compared with the level in acetate. The same change in the transcriptional pattern of the FlhDC operon in cells grown in alternative carbon sources relative to that in glucose-grown cells can be observed as shown in Figure 5C. In addition, we made a cyaA in-frame deletion strain. cyaA encodes adenylate cyclase that is required to catalyze the formation of cyclic AMP (46). The pattern of increasing the transcriptional levels of CRP-dependent genes in low-quality carbon sources, as was observed in the wild-type strain, disappeared in the cyaA deficient strain (as shown in Supplemental Material, Figure S3). This indicates that CRP-cAMP plays an important role in promoter activation in our assays. No significant change of the transcriptional level of CRP was observed (Figure 5C), suggesting that the induction of CRP-dependent genes might be mainly due to the activation of CRP rather than the increase in CRP expression (47–49).
Figure 5.
Carbon source effects on cell motility through activation of CRP. (A) SDS–PAGE gel shows purified CRP protein as well as MultiMark Standard and native gel shift assays show the binding of CRP to upstream DNA fragment of FlhDC operon. The palindromic consensus DNA-binding site for CRP dimer is shown in red. (B) The transcript abundance of two CRP-dependent genes, cstA and cpdB, in cells grown on different carbon sources. (C) The transcript abundance of flhD, fliA and crp in cells grown on different carbon sources. (D) The transcript abundance of σF-dependent genes, motA, tar and fliC, in cells grown on different carbon sources. Note, these carbon sources are of differing quality as defined by the resulting log-phase growth rates which are 0.97 generation h−1 in glucose, 0.50 generation h−1 in succinate, 0.34 generation h−1 in alanine, 0.21 generation h−1 in acetate and 0.13 generation h−1 in proline.
Genes for motility and chemotaxis are systematically induced
In an intact flagellar system, the mechanism by which cells control flagellar operation involves genes that are under the direct control of alternative sigma factor, σF. In E. coli motility control, the cell has a family of transmembrane proteins with receptor functions (50,51) termed methyl-accepting chemotaxis proteins, or MCPs (52). MCPs mediate responses to external environmental stimuli. These receptors bind stimulatory ligands and undergo conformational changes that regulate the activities of a network of signal transduction proteins within the cytoplasm. There are six cytoplasmic signal transduction proteins, the products of the Che genes: cheA, cheB, cheR, cheW, cheY and cheZ in E. coli. The signal transduction pathways then deliver information to flagellar apparatus to mediate cell filament (product of fliC) as a propeller to do rotation and switching. Genomic organization of these genes in respective operons is presented in Figure 5D.
The apparent coupling of motility activity and flhDC operon activation in these experiments prompted us to further measure the σF synthesis rate as well as σF-dependent genes synthesis rates in each culture, since these parameter are known to be correlated with motility control (11). We choose the first gene in each operon for the RT-PCR experiments to test the activity of these genes as a function of various carbon sources. Compared with the transcriptional levels of these motility and chemotaxis genes in the fastest growing culture (glucose), a significant increase of the transcriptional level of these genes in slow growing cultures with alternative carbon sources (succinate, alanine, acetate, proline) can be seen in our assays as shown in Figure 5D. The upregulation of motility genes correlated with increasing motility of cells in poor quality carbon sources, together with the synchronized pattern of increasing CRP activity causing increasing FlhDC transcription, further suggests CRP is a factor (or at least one of multiple factors) to play a positive role in triggering E. coli ‘foraging’-like behavior to actively search for better conditions as the quality of the available substrate(s) decreases. Interestingly, the transcriptional level of the costly cell filament encoding gene fliC did not have any significant changes across different carbon sources. Recently, the Kelly Hughes group (53,54) reported studies on the transcriptional and translational control of the fliC gene in promoter and 5′ untranslated regions in Salmonella. Our observations here suggest, in a poor nutrient environment, that bacteria may strategically use the precious energy on the basis of fine tuning of flagellar-gene expression in response to environmental challenges, which it possibly does by sequestering a transcriptional repressor or other factor(s) to inhibit expensive fliC gene expression. The biological implications of this finding are discussed later.
DISCUSSION
In this study, we used two different genetic approaches (1) moderately expressing FlhDC or σF from anhydrotetracycline (aTc) inducible and Tet repressor-controlled PLtet promoter in a plasmid-borne flhDC or fliA gene; (2) disrupting the expression of FlhDC or σF in flhDC or fliA deletion mutant strains, to efficiently and reliably study the regulon members of the two flagellar biosynthesis regulators, FlhDC and σF. Our results demonstrate that there are many more genes than previous known under the control of these two regulators. In our previous studies (27), we have demonstrated that our approaches have a significant advantage over those approaches for stimulons and regulons studies in various stress conditions. In this paper, we further discovered that the strength of these two different regulon study approaches depends mainly on the initial concentrations of sigmas or other regulators in vivo. Low initial concentration of a regulator can be induced to a high level after a short induction, which in turn increases transcription of its dependent genes to a significant level. Loss by deletion of the high initial concentration regulator might totally shut off the transcription of its dependent genes in vivo. Note that, for some important regulators, loss of their functions might cause severe growth problems. Currently, using our lab collection of the monoclonal antibodies for each sigma factor in E. coli, we are using the ChIP-chips assay (55,56) as a complementary approach to pull down the DNA fragment that is bound and crosslinked by a given sigma factor. Combination of two or several different regulon study approaches will give us more confidence in positive results.
In addition to extending the repertoire of FlhDC and σF regulon with new candidates, this work demonstrates that the carbon source utility and motility activity are interdependent. This study was motivated by the previous observation that E. coli increases its motility in poor nutrition environments. Much less is known about how the activation of CRP is affected as well as how this activation will affect cell motility by different carbon source supplies. Our results shed light on cell motility control in a range of different carbon sources and suggest intriguing hypotheses about its establishment and function during utilizing different energy compounds. The observations reported here have many notable features: in the defined MOPS minimum medium with different carbon sources, CRP is activated in poor quality (defined by growth rate) carbon sources relative to rich quality ones; this activated CRP associates with the flhDC operon encoding the master regulator with roles in flagellar regulation and development; the activation of CRP is correlated with the increase of expression of σF as well as the increase of cell motility.
Flagellar genes are organized into a transcriptional hierarchy that underlies temporal and spatial control of biogenesis program. A large amount of energy is required to synthesize flagella and a large part of this energy is used for filament synthesis. The filament consists of an assembly of around 20 000 subunits of a single protein, flagellin (fliC), and the amount of flagellin alone composes about 8% of the total cell protein when the flagellar operons are expressed optimally (57). Although the motility of cells gradually increase in low-quality carbon sources supplies, we found the transcription of the fliC gene, which encodes costly flagellin, does not increase in those minimum growth conditions. This may be a means of conserving energy. The cost to the cell of flagellar synthesis and flagellar operation is about 2% and about 0.1% of total energy expenditure under normal growth conditions, respectively (11). As the growth potential of an environment decreases, the ability to reach a potential food source ahead of siblings is a group competition behavior (58). Also the ability to efficiently utilize cellular resources to conserve energy for individual self-protection, such as hibernation, would provide a significant survival advantage. We show that motile bacteria carry out tactical responses to a variety of carbon sources by increasing flagellar operation but restricting costly additional flagellar synthesis. This may provide a paradigm for cost and benefit behaviors in prokaryotes, which result in maximum benefit for survival. The delicate balance between conserving more energy and using the energy-intensive flagella to search out better conditions is modulated by environmental conditions. The inverse correlation between the increasing motility of cell with carbon quality might be in a certain minimum nutrition range. As the growth condition becomes more and more harsh as shown in lowest-quality carbon source (proline), conserving precious energy outweighs the cost of expending energy for potential benefit of locating and utilizing good sources. In this situation, the cell will gradually turn off both of the costly flagellar synthesis (flagellin) and flagellar operation (motility and chemotaxis) and save more energy for a long time survival to passively wait for less adverse (natural) conditions. The downshift of the expression of flagellar genes as well as motility activity in proline might represent these behaviors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Julius Adler, Fred Blattner, Richard Gourse, Rodney Welch, Brian Haugen and Justin Lemke for critical reading of the manuscript and many useful discussions. We also thank Frederick Blattner group for technical advice in microarray experiments and assistance in data analysis and Sandra Splinter-BonDurant, Wayne Davis, John Leucke at the Gene Expression Center for excellent technical assistance.
Conflict of interest statement. The research was supported by a grant from NIGMS (GM28575) to R.R.B. In compliance with the University of Wisconsin-Madison's Conflict of Interest Committee Policy, RRB acknowledges financial interest in NeoClone, LLC (Madison, WI) which markets mAbs used in this study.
REFERENCES
- 1.Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 2.Wosten MM. Eubacterial sigma-factors. FEMS Microbiol. Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]
- 3.Haldenwang WG. The sigma factors of Bacillus subtilis. Microbiol. Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer U, Santangelo JD, Treuner A, Buchholz M, Durre P. Sigma factor and sporulation genes in Clostridium. FEMS Microbiol. Rev. 1995;17:331–340. doi: 10.1111/j.1574-6976.1995.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 5.Gomez JE, Chen JM, Bishai WR. Sigma factors of Mycobacterium tuberculosis. Tuber. Lung Dis. 1997;78:175–183. doi: 10.1016/s0962-8479(97)90024-1. [DOI] [PubMed] [Google Scholar]
- 6.Burgess RR, Travers AA. Escherichia coli RNA polymerase: purification, subunit structure, and factor requirements. Fed. Proc. 1970;29:1164–1169. [PubMed] [Google Scholar]
- 7.Aiyar SE, McLeod SM, Ross W, Hirvonen CA, Thomas MS, Johnson RC, Gourse RL. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol. 2002;316:501–516. doi: 10.1006/jmbi.2001.5390. [DOI] [PubMed] [Google Scholar]
- 8.McLeod SM, Aiyar SE, Gourse RL, Johnson RC. The C-terminal domains of the RNA polymerase alpha subunits: contact site with Fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter. J. Mol. Biol. 2002;316:517–529. doi: 10.1006/jmbi.2001.5391. [DOI] [PubMed] [Google Scholar]
- 9.Attey A, Belyaeva T, Savery N, Hoggett J, Fujita N, Ishihama A, Busby S. Interactions between the cyclic AMP receptor protein and the alpha subunit of RNA polymerase at the Escherichia coli galactose operon P1 promoter. Nucleic Acids Res. 1994;22:4375–4380. doi: 10.1093/nar/22.21.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blazy B, Takahashi M, Baudras A. Binding of CRP to DNA-dependent RNA polymerase from E. coli: modulation by cAMP of the interactions with free and DNA-bound holo and core enzyme. Mol. Biol. Rep. 1980;6:39–43. doi: 10.1007/BF00775753. [DOI] [PubMed] [Google Scholar]
- 11.Macnab RM. Flagella and Motility. Washington, DC: ASM Press; 1996. [Google Scholar]
- 12.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverman M, Simon M. Genetic analysis of flagellar mutants in Escherichia coli. J. Bacteriol. 1973;113:105–113. doi: 10.1128/jb.113.1.105-113.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silverman M, Simon M. Genetic analysis of bacteriophage Mu-induced flagellar mutants in Escherichia coli. J. Bacteriol. 1973;116:114–122. doi: 10.1128/jb.116.1.114-122.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman M, Simon M. Positioning flagellar genes in Escherichia coli by deletion analysis. J. Bacteriol. 1974;117:73–79. doi: 10.1128/jb.117.1.73-79.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman M, Matsumura P, Simon M. The identification of the mot gene product with Escherichia coli-lambda hybrids. Proc. Natl Acad. Sci. USA. 1976;73:3126–3130. doi: 10.1073/pnas.73.9.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komeda Y, Kutsukake K, Iino T. Definition of additional flagellar genes in Escherichia coli K12. Genetics. 1980;94:277–290. doi: 10.1093/genetics/94.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006;188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ, Blattner FR. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J. Biol. Chem. 2005;280:15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- 21.Hastjarjo T, Silberberg A, Hursh SR. Risky choice as a function of amount and variance in food supply. J. Exp. Anal. Behav. 1990;53:155–161. doi: 10.1901/jeab.1990.53-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houston AI. Risk-sensitive foraging theory and operant psychology. J. Exp. Anal. Behav. 1991;56:585–589. doi: 10.1901/jeab.1991.56-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blattner FR, Plunkett G., 3rd., Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 2004;186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao K, Liu M, Burgess RR. The global transcriptional response of Escherichia coli to induced sigma32 protein involves sigma32 regulon activation followed by inactivation and degradation of sigma32 in vivo. J. Biol. Chem. 2005;280:17758–17768. doi: 10.1074/jbc.M500393200. [DOI] [PubMed] [Google Scholar]
- 28.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 29.Tisa LS, Adler J. Cytoplasmic free-Ca2+ level rises with repellents and falls with attractants in Escherichia coli chemotaxis. Proc. Natl Acad. Sci. USA. 1995;92:10777–10781. doi: 10.1073/pnas.92.23.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stafford GP, Ogi T, Hughes C. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology. 2005;151:1779–1788. doi: 10.1099/mic.0.27879-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claret L, Hughes C. Interaction of the atypical prokaryotic transcription activator FlhD2C2 with early promoters of the flagellar gene hierarchy. J. Mol. Biol. 2002;321:185–199. doi: 10.1016/s0022-2836(02)00600-9. [DOI] [PubMed] [Google Scholar]
- 32.Jishage M, Iwata A, Ueda S, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ide N, Kutsukake K. Identification of a novel Escherichia coli gene whose expression is dependent on the flagellum-specific sigma factor, FliA, but dispensable for motility development. Gene. 1997;199:19–23. doi: 10.1016/s0378-1119(97)00233-3. [DOI] [PubMed] [Google Scholar]
- 34.Ko M, Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 35.Lipinska B, King J, Ang D, Georgopoulos C. Sequence analysis and transcriptional regulation of the Escherichia coli grpE gene, encoding a heat shock protein. Nucleic Acids Res. 1988;16:7545–7562. doi: 10.1093/nar/16.15.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Brutlag DL, Liu JS. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001:127–138. [PubMed] [Google Scholar]
- 37.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 38.Karlinsey JE, Lonner J, Brown KL, Hughes KT. Translation/secretion coupling by type III secretion systems. Cell. 2000;102:487–497. doi: 10.1016/s0092-8674(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 39.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 40.Cheng LW, Schneewind O. Type III machines of Gram-negative bacteria: delivering the goods. Trends Microbiol. 2000;8:214–220. doi: 10.1016/s0966-842x(99)01665-0. [DOI] [PubMed] [Google Scholar]
- 41.Adler J, Templeton B. The effect of environmental conditions on the motility of Escherichia coli. J. Gen. Microbiol. 1967;46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- 42.Yokota T, Gots JS. Requirement of adenosine 3′, 5′-cyclic phosphate for flagella formation in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 1970;103:513–516. doi: 10.1128/jb.103.2.513-516.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crasnier M. Cyclic AMP and catabolite repression. Res. Microbiol. 1996;147:479–482. doi: 10.1016/0923-2508(96)84002-2. [DOI] [PubMed] [Google Scholar]
- 44.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli. J. Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 46.Holland MM, Leib TK, Gerlt JA. Isolation and characterization of a small catalytic domain released from the adenylate cyclase from Escherichia coli by digestion with trypsin. J. Biol. Chem. 1988;263:14661–14668. [PubMed] [Google Scholar]
- 47.de Crombrugghe B, Busby S, Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984;224:831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- 48.Kim J, Adhya S, Garges S. Allosteric changes in the cAMP receptor protein of Escherichia coli: hinge reorientation. Proc. Natl Acad. Sci. USA. 1992;89:9700–9704. doi: 10.1073/pnas.89.20.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makman RS, Sutherland EW. Adenosine 3′,5′-Phosphate in Escherichia Coli. J. Biol. Chem. 1965;240:1309–1314. [PubMed] [Google Scholar]
- 50.Adler J. Chemoreceptors in bacteria. Science. 1969;166:1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- 51.Springer MS, Goy MF, Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc. Natl Acad. Sci. USA. 1977;74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Springer MS, Goy MF, Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979;280:279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- 53.Aldridge P, Gnerer J, Karlinsey JE, Hughes KT. Transcriptional and translational control of the Salmonella fliC gene. J. Bacteriol. 2006;188:4487–4496. doi: 10.1128/JB.00094-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosu V, Chevance FF, Karlinsey JE, Hirano T, Hughes KT. Translation inhibition of the Salmonella fliC gene by the fliC 5′ untranslated region, fliC coding sequences, and FlgM. J. Bacteriol. 2006;188:4497–4507. doi: 10.1128/JB.01552-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson BO. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 2005;187:6166–6174. doi: 10.1128/JB.187.17.6166-6174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raffaelle M, Kanin EI, Vogt J, Burgess RR, Ansari AZ. Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo. Mol. Cell. 2005;20:357–366. doi: 10.1016/j.molcel.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 57.Neidhardt FC, Umbarger HE. Chemical Composition of E. coli. Washington, DC: ASM Press; 1987. [Google Scholar]
- 58.Stragier P. To kill but not be killed: a delicate balance. Cell. 2006;124:461–463. doi: 10.1016/j.cell.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 59.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.