Abstract
The polyadenylation of messenger RNAs is mediated by a multi-subunit complex that is conserved in eukaryotes. Among the most interesting of these proteins is the 30-kDa-subunit of the Cleavage and Polyadenylation Specificity Factor, or CPSF30. In this study, the Arabidopsis CPSF30 ortholog, AtCPSF30, is characterized. This protein possesses an unexpected endonucleolytic activity that is apparent as an ability to nick and degrade linear as well as circular single-stranded RNA. Endonucleolytic action by AtCPSF30 leaves RNA 3′ ends with hydroxyl groups, as they can be labeled by RNA ligase with [32P]-cytidine-3′,5′-bisphosphate. Mutations in the first of the three CCCH zinc finger motifs of the protein abolish RNA binding by AtCPSF30 but have no discernible effects on nuclease activity. In contrast, mutations in the third zinc finger motif eliminate the nuclease activity of the protein, and have a modest effect on RNA binding. The N-terminal domain of another Arabidopsis polyadenylation factor subunit, AtFip1(V), dramatically inhibits the nuclease activity of AtCPSF30 but has a slight negative effect on the RNA-binding activity of the protein. These results indicate that AtCPSF30 is a probable processing endonuclease, and that its action is coordinated through its interaction with Fip1.
INTRODUCTION
Messenger RNA 3′ end formation is an essential step in the biogenesis of mRNAs in eukaryotic cells. This step consists of a series of events that culminate in the addition of a finite tract of poly(A) to the 3′-end of a processed pre-mRNA. These events are mediated by a complex of multi-subunit factors, the composition of which is generally conserved in eukaryotes (1–3). This complex recognizes specific sequence elements in a pre-mRNA, processes the RNA and adds the poly(A) tract. In animals, the canonical poly(A) signal AAUAAA is recognized by the 160-kDa subunit of cleavage and polyadenylation specificity factor (CPSF; the 160-kDa subunit is termed CPSF160), the U + G-rich downstream sequence element is bound by CstF64, the 64-kDa subunit of cleavage stimulatory factor (CstF) and other auxiliary elements situated 5′ to (or upstream of) the polyadenylation site are recognized by CFIm25, the 25-kDa subunit of cleavage factor I (CFIm). Two other subunits of the complex, hFip1 and CPSF30, also bind RNA (4,5), but the roles of these activities in the process remain poorly defined. These RNA–protein interactions, as well as a number of protein–protein interactions between other subunits of the subcomplexes, serve to coalesce the complex and position the RNA for cleavage and polyadenylation. Two different subunits of CPSF, namely CPSF73 and CPSF30, have been suggested as the endonuclease responsible for the cleavage of the mRNA at the polyadenylation site subsequent to polyadenylation (6–8). Subsequent to RNA processing, poly(A) polymerase (or PAP) is brought to the RNA, presumably through its interaction with animal orthologs of Fip1 (5), so as to add the polyadenylated tract. This final step in the reaction is modulated by a nuclear poly(A)-binding protein (PabN) that serves to promote processive poly(A) addition and to regulate the final length of poly(A) that is added to the mRNA (9,10).
In the budding yeast Saccharomyces cerevisiae, mRNA 3′ end formation is directed by a tripartite polyadenylation signal, the elements of which have been termed as the efficiency element, positioning element and polyadenylation site, respectively. The RNA-binding subunits Hrp1, Rna15 and Yth1 associate with these elements; Hrp1 binding to the efficiency element, Rna15 to the positioning element and Yth1 to the polyadenylation site (11–13). Hrp1 and Rna15 are subunits of the so-called cleavage factor I (CFIy, the subscript ‘y’ serving to distinguish this factor from the mammalian CFIm) (12,13) while Yth1 is a subunit of the cleavage and polyadenylation factor (CPF) (14). Rna15 and Yth1 are orthologs of CstF64 and CPSF30, respectively, while Hrp1 has no strict counterpart in the animal polyadenylation complex. Presumably as is the case in animals, these RNA–protein interactions that involve different multi-subunit factors serve to promote the assembly of the complete complex on a pre-mRNA, as well as to position the pre-mRNA for processing and subsequent polyadenylation. The nature of the processing endonuclease in yeast has not been explored, but both of the animal proteins (CPSF73 and CPSF30) that have been associated with suggestive nuclease activities have yeast counterparts—Ysh1 is the ortholog of CPSF73, and Yth1 is the counterpart of CPSF30. As in animals, PAP is tethered to the processed RNA through its interaction with Fip1 (15). While yeast has no formal counterpart of PabN, two other PabNs, Nab2 and Pab1, contribute to poly(A) length control in this organism (16–18).
Plant genomes possess genes with the potential to encode virtually the complete set of polyadenylation factor subunits that are found in animals and yeast (19). Moreover, as is the case in animals and yeast, the plant polyadenylation signal consists of several distinct cis elements—in plants, the so-called Near-Upstream Element, Far-Upstream Element and Cleavage Element, or NUE, FUE and CE, respectively (20–22). The plant polyadenylation complex has not been as extensively studied as its animal and yeast counterparts. However, recent studies have established that several of the plant counterparts of CPSF subunits (specifically, CPSF160, CPSF100 and symplekin) reside in a nuclear complex that also includes the ortholog of the yeast polyadenylation factor subunit Pfs2 (the plant protein has been termed FY) (23), and most of the plant subunits may be linked conceptually with PAP (24,25).
Eukaryotic CPSF30 proteins are among the most interesting of the subunits of the polyadenylation complex. As stated in the preceding paragraph, the yeast ortholog (Yth1) associates with the cleavage/polyadenylation site (11). The Drosophila ortholog possesses nucleolytic activity (26,27), leading some to propose that CPSF30 is a processing endonuclease in the polyadenylation reaction (8). In animal cells, CPSF30 has been shown to be the focus of regulatory events in virus infections (28,29). The animal and yeast proteins share an array of five tandemly arranged CCCH-type zinc finger motifs (typically, Cys-X8-Cys-X5-Cys-X3-His) that have been implicated in RNA binding by these proteins (11), in nucleolytic activity (26) and in interactions with other polyadenylation factor subunits (14). Moreover, animal proteins have additional zinc knuckle motifs that are absent from Yth1 (27), and the zinc knuckle motif of the bovine protein enhances the RNA-binding activity of the core zinc finger domain (26).
Among the set of plant genes that encode polyadenylation factor subunits are those that encode orthologs of CPSF30/Yth1. The Arabidopsis gene (as well as its counterparts in other plants; unpublished data) encodes two mRNAs (30); the smaller of the two (termed herein as AtCPSF30) specifies a ca. 250-amino acid polypeptide that is analogous to CPSF30/Yth1, whereas the larger is translated to yield a polypeptide in which all but the C-terminal 13 amino acids of AtCPSF30 are fused to a protein domain found in mammalian splicing associated factors [the so-called YT521B motif; (31)]. Both polypeptides can be detected in nuclear extracts, and the smaller resides in a nuclear complex with the Arabidopsis ortholog of CPSF100 (30). AtCPSF30 also interacts with an Arabidopsis ortholog of Fip1 (24), placing it in a network of interactions that includes PAP (among other polyadenylation factor subunits). Like its other eukaryotic counterparts, AtCPSF30 is an RNA-binding protein; interestingly, the RNA-binding activity of AtCPSF30 is inhibited by calmodulin in a calcium-dependent fashion (30), suggestive of a regulatory link between signaling and RNA processing in plants.
In our ongoing studies of AtCPSF30, it was noticed that a persistent nuclease activity co-purified with the recombinant protein, an activity that confounded attempts to define RNA-binding sites on substrate molecules. In this report, the properties of this nuclease activity are described. Specifically, we show that AtCPSF30 is itself an endonuclease, that the RNA products of endonucleolytic cleavage possess 3′-hydroxyl groups, and that an Arabidopsis Fip1 ortholog inhibits the AtCPSF30-associated nuclease activity. Together with other studies, these results suggest that AtCPSF30 is a processing endonuclease, and that the pre-mRNA cleavage step in plants is mediated by more than one protein.
METHODS
Protein purification
The purification of MBP-AtCPSF30, m4 and m9 proteins (Figure 4) has been described elsewhere (30). The zinc finger mutants of AtCPSF30 (Figure 4) were generated by using quick-change site-directed mutagenesis kit (Stratagene) using the pMALC2-AtCPSF30 plasmid (30) as template as per manufacturer's instructions. The results of mutagenesis were confirmed by DNA sequencing; accordingly, the C-terminus of the first zinc finger was changed from DACGFLHQF to DASTFLYQ, the second zinc finger from QDCVYKHTN to QDSTYKYTN and the third from PDCRYRHAK to PDSTYRYAK. The oligonucleotide primers that were used for the mutagenesis are given in Table 1. The cloning of coding sequence corresponding to the N-terminal 137 of the chromosome V-encoded Arabidopsis Fip1 protein into bacterial protein expression vectors pGEX-2T (Pharmacia) and pMAL-2C for making GST and MBP fusion proteins, respectively, was as described (24). The coding sequence corresponding to the N-terminal 483 amino acids was cloned into pDEST17 vector (Invitrogen) from the corresponding entry clone (24) to produce histidine-tagged fusion protein. Histidine-tagged and GST fusion proteins were purified as described by Forbes et al. (24) and MBP fusion proteins purified as described in Delaney et al. (30). It should be pointed out that the purification of MBP and GST fusion proteins included a wash of proteins bound to the affinity media with buffers containing 2 M NaCl, a step intended to remove almost all non-specifically bound bacterial proteins from the MBP fusion protein preparations. Control proteins (GST, MBP and histidine-tagged β-glucuronidase) were prepared as described (24,30). Protein concentrations were estimated by comparing the purified preparations with BSA standards by SDS–PAGE and staining of the gels with Coomassie Brilliant Blue.
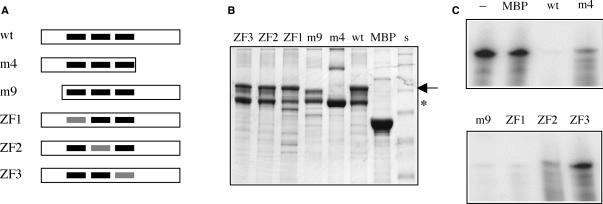
Figure 4.
Mutational analysis of the nuclease activity. (A) Illustration of AtCPSF30 and the various mutants. For the wild-type protein (‘wt’), the three zinc fingers are represented as black bars. The parts of the protein present in the m4 and m9 deletion mutants (30) are shown. Mutated zinc fingers for each of the three zinc finger mutants are depicted as gray bars. (B) Stained gel showing the various purified protein preparations.Three microliter of each purified protein preparation was separated on a 10% gel, which was subsequently stained with Coomassie Brilliant Blue. The identity of each preparation is indicated above the scan of the gel; the full-sized MBP-AtCPSF30 is denoted with an arrow besides the gel, and the previously described breakdown product (30) noted with an *. The high molecular weight bands seen in the MBP and m4 lanes are not consistently seen in all preparations. s—SeeBlue2 (Invitrogen) pre-stained protein size standards. (C) Nuclease activity of the AtCPSF30 variants. Uniformly labeled RNA (2 pmol = 200 nM) was incubated with purified MBP-AtCPSF30 mutant proteins or with MBP for 30 min at 30°C and the remaining RNA recovered and analyzed on a sequencing gel. The identity of the AtCPSF30 variant is indicated above the gel image (‘-’: no added protein); the quantities of the enzyme preparations were: MBP—12 pmol; ‘wt’ (wild-type AtCPSF30)—6 pmol; m4—12 pmol; m9—10 pmol; ZF1—10 pmol; ZF2—10 pmol; ZF3—10 pmol.
Table 1.
Plasmids and oligonucleotides used in this study
| Designation | Sequence (5′ ->3′) or description | Use | Reference (for plasmids) |
|---|---|---|---|
| T7-E9 | TAATACGACTCACTATAGGGAGTATTAT GGCATTGGGAA | PCR production of rbcS-E9 in vitro transcription template | |
| E9 61–80 | AAATGTTTGCATATCTCTTA | PCR production of rbcS-E9 in vitro transcription template | |
| T7-7/26 | TAATACGACTCACTATAGGGA GAATTAATATTATTTGTTTT | PCR production of ‘DE’ in vitro transcription template | |
| rbcS-17/6 | TGAGAATGAACAAAAGGACC | PCR production of ‘PC’ in vitro transcription template | |
| BS-T3 | CAATTAACCCTCACTAAAGGGAA | PCR production of the ‘GFP’ transcription template (Figure 3A) | |
| GFP-3′ | TTATTTGTATAGTTCATCCATGCCATGTG | PCR production of the ‘GFP’ transcription template (Figure 3A) | |
| ZF1-A | GTATGAAAGGTGACGCCAGTACTTTTCT CTATCAGTTCGATAAAGCTC | Site-directed mutagenesis to produce the ZF1 mutant | |
| ZF1-B | GAGCTTTATCGAACTGATAGAGAAAAGT ACTGGCGTCACCTTTCATAC | Site-directed mutagenesis to produce the ZF1 mutant | |
| ZF2-A | GGTGAATGTCGAGAGCAGGATAGTACTT ATAAATATACCAATGAAGATATCAAAG AATGC | Site-directed mutagenesis to produce the ZF2 mutant | |
| ZF2-B | GCATTCTTTGATATCTTCATTGGTATATTT ATAAGTACTATCCTGCTCTCGACATTCACC | Site-directed mutagenesis to produce the ZF2 mutant | |
| ZF3-A | GTCCCAATGGTCCTGATAGTACTTACA GGTATGCAAAGCTTCCTGGAC | Site-directed mutagenesis to produce the ZF3 mutant | |
| ZF3-B | GTCCAGGAAGCTTTGCATACCTGTAAGTACT ATCAGGACCATTGGGAC | Site-directed mutagenesis to produce the ZF3 mutant | |
| rbcS-E9 | plasmid | PCR template for ‘wt’, ‘PC’ and ‘DE’ in vitro transcription substrates | (32) |
| rbcS-Δ120/61 | plasmid | PCR template for ‘ΔFUE’ in vitro transcription substrate | (32) |
| rbcS-Δ−60/+41 | plasmid | PCR template for ‘ΔNUE’ in vitro transcription substrate | (32) |
| pBlueScript-GFP* | plasmid | PCR template for ‘GFP’ transcription substrate (Figure 3A) | This report |
Preparation of RNA substrates
Labeled RNA was derived from the pea rbcS-E9 polyadenylation signal, and contained nucleotides extending from 145 nt upstream to 80 nt downstream from the site noted as ‘+1’ by Mogen et al. (32). Various mutant forms of the polyadenylation signal lacking one or more cleavage sites or sequence elements were also used for making the transcripts. Templates for in vitro transcription were prepared by PCR, using the plasmid templates and primers listed in Table 1. Uniformly labeled and unlabeled RNAs were prepared by in vitro transcription using Ampliscribe kit from Epicentre as per manufacturer's instructions, and purified using MegaClear kits (Ambion).
In addition to rbcS-E9-derived RNAs, a control RNA from a GFP gene was also used in some experiments. Transcription template for this RNA was produced by PCR using the primers indicated in Table 1 (‘BS-T3’ and ‘GFP-3′’) and a truncated GFP plasmid as template. This plasmid was produced by subcloning the GFP gene in KY80-GFP (a gift from Dr Randy Dinkins, USDA/ARS, Lexington, KY) into pBluescript vector as an XbaI–SacI fragment. The resulting recombinant plasmid was digested with HincII and ligated to release the first 490 bases of GFP.
Circular RNA was made from uniformly labeled linear RNA. Initially, linear RNA was dephosphorylated with calf intestinal alkaline phosphatase (Promega) by incubating 24 µg of RNA with 10 units of enzyme at 37°C for 30 min. The reaction was stopped by heating at 94°C for 5 min, and RNA was recovered by phenol–chloroform extraction followed by ethanol precipitation. The precipitated RNAs were re-suspended in water and re-phosphorylated by phosphonucleotide kinase (Ambion) at 37°C for 30 min. Re-phosphorylated RNA was purified by another round of phenol–chloroform extraction and ethanol precipitation. Ligation of 5′ and 3′ ends were carried out by incubating the re-phosporylated RNA with T4 RNA ligase (Ambion) at 37°C for 40 min as per manufacturer's instructions. The reaction was stopped by heating at 94°C for 5 min and RNAs were extracted with phenol–chloroform. After ethanol precipitation, the circular RNAs so obtained were re-dissolved in water.
Nuclease assays
The nuclease assay reactions contained varying quantities (ranging from 0.12 to 12 pmol) of purified protein, 2 pmol of labeled RNA, 0.6 mM MgCl2 and RNasin (30 U, Eppendorf) in a volume of 10 μl of Tris–HCl buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl). After incubating the reaction mixture at 30°C for appropriate periods of time, reactions were stopped with phenol–chloroform. Three to five microliters of the aqueous phase was mixed with equal volume of gel loading buffer II (Ambion) and heated at 65°C for 15 min. The samples were subsequently chilled on ice for 2 min before conducting electrophoresis on 8–10% polyacrylamide–urea gels. Following electrophoresis at 7–8 mA constant current, the gels were dried and analyzed by autoradiography. Autoradiographs were analyzed using ImageJ analysis software.
RNA-binding reactions
For RNA binding, the electrophoretic mobility shift assay described elsewhere (30) was used. It should be pointed out that the conditions used in these assays are such that appreciable RNA, including partial breakdown products, remains after the 15 min incubation time (Figure 1A, lanes 8 and 12), and thus that RNA binding can be measured for forms of AtCPSF30 that possess nuclease activity.
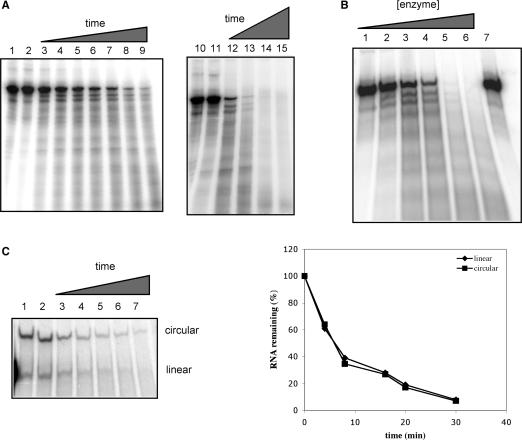
Figure 1.
Nuclease activity of AtCPSF30. (A) Time course of the activity. Uniformly labeled RNA (200 nM) was incubated with MBP fusion protein of At CPSF30 (600 nM), samples were taken out at regular time intervals and added to phenol–chloroform to stop the reaction. The RNAs were recovered from the aqueous phase and subjected to urea-PAGE as described in Methods section. Lanes 2 and 11 are reactions that were halted before AtCPSF30 was added. Time points are: lanes 3–9—1, 2, 5, 7.5, 10, 15 and 20 min, respectively; lanes 12–15—15, 30, 60 and 120 min, respectively. Lanes 1 and 10 show the results of reactions performed with purified MBP instead of AtCPSF30 for 20 (lane 1) or 120 min (lane 10). (B) Concentration dependence of the nuclease activity. Uniformly labeled RNA (200 nM) was incubated with serial dilutions of purified protein (final AtCPSF30 concentrations of 1.2, 12, 120, 300, 600 and 960 nM, in lanes 1, 2, 3, 4, 5 and 6, respectively) for 30 min. The sample in lane 7 shows the results of incubating the RNA in the absence of protein. (C) AtCPSF30 acts on circular RNAs. Circular RNA was prepared from uniformly labeled RNA as described in the Methods section. The resultant mixture of circular and linear RNAs (so indicated in the figure) was incubated with MBP-At CPSF30 for 0, 4, 8, 16, 20 and 30 min(lanes 2, 3, 4, 5, 6 and 7, respectively) and the products were recovered and separated on a sequencing gel. Lane 1 shows the RNA mixture incubated for 30 min in the presence of purified MBP. The results are plotted on the right; in this panel, the percentage of the RNA substrate (circular or linear) remaining was plotted as a function of time, with the quantity in lane 2 being set as 100%.
Analysis of 3′ termini
The procedure is similar to the one followed by Ivanov et al. (33). Unlabeled RNA was blocked at 3′ end using yeast PAP (USB) and cordycepin 5′-triphosphate as per manufacturer's instructions. The RNA so obtained was treated with MBP-AtCPSF30 fusion protein for 15 min and the resulting breakdown products recovered by phenol–chloroform and ethanol precipitation. Subsequently, the RNAs were dissolved in water and end-labeled with T4 RNA ligase (6 U) and [32P] cytidine 3′,5′-bisphosphate at 4°C for 20 h. The RNAs were subsequently precipitated with ethanol, re-suspended in gel-loading buffer II (Ambion) and analyzed by 10% denaturing polyacrylamide–urea gel electrophoresis and autoradiography. Autoradiographs were analyzed using ImageJ analysis software.
Yeast two-hybrid analysis
Two hybrid assays of the interactions between an Arabidopsis Fip1 ortholog and derivatives of AtCPSF30 were carried out as described (24). The different portions of AtCPSF30 were subcloned into pGEM as described (30), excised as BglII fragments and cloned into pGAD-C(1) and pGBD-C(1) (34) to yield for activation domain (AD) and binding domain (BD) clones, respectively. AD and BD plasmids were transformed into PJ69-4 (34) and dual transformants (identified as colonies growing on media lacking leucine and tryptophan, the selective markers for these two plasmids) subsequently tested on media lacking leucine, tryptophan and adenine (the latter being one of the scorable markers for interactions). Positive interactions were those in which all tested colonies (between 4 and 10) grew on the adenine-free media. Negative controls for these tests included transformations with combinations of plasmids that included unmodified pGAD-C(1) or pGBD-C(1). The positive control was that used by Forbes et al. (24), namely the set of plasmids that carried the Arabidopsis CstF77 and CstF64 orthologs.
RESULTS
AtCPSF30 is an endonuclease that leaves 3′-hydroxyl groups on the ends of cleaved RNAs
Prior work in this laboratory established that AtCPSF30 is an RNA-binding protein (30). Attempts to explore the RNA-binding properties of AtCPSF30 by mapping possible sites of binding on labeled RNA (11) were not successful owing to persistent nuclease activity that was insensitive to commercially available ribonuclease inhibitors (data not shown). Extensive and exhaustive purification, as well as characterizations of equally pure preparations of MBP (the tag used to purify recombinant AtCPSF30), led to the realization that AtCPSF30 itself was the source of this activity. In light of reports ascribing similar activities to the Drosophila counterpart of CPSF30 (26,27), the nuclease activity of AtCPSF30 was characterized in some detail. For these studies, an RNA containing a complete suite of polyadenylaton-associated cis elements was used (Figure 3A). This RNA is derived from the pea rbcS-E9 gene and includes the well-defined FUE)and NUEs (32,35) from this gene, and was chosen so as to best recapitulate the complex nature of plant polyadenylation signals. Initial studies using uniformly labeled RNA revealed an activity that yielded numerous breakdown intermediates at early times in a typical time-course study, and an eventual loss of all detectable poly- nucleotides or oligonucleotides after extended times (Figure 1A). Nuclease activity was dependent on enzyme concentration, and was not seen when comparable quantities of purified MBP were used in place of the MBP-AtCPSF30 protein (Figure 1B).
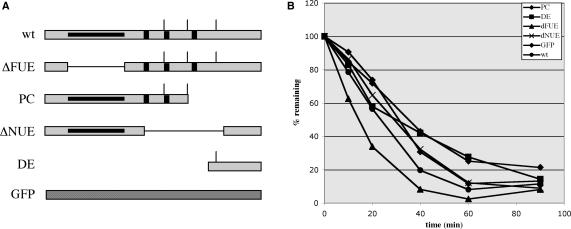
Figure 3.
Activities with different RNAs. (A) Structure of the rbcS-E9-related RNAs. The designations of substrate E9 RNAs lacking one or more of the polyadenylation signal sequence elements (32), and their respective structures, are shown. ΔFUE (dFUE in the inset of the graph shown in panel B)—RNA lacking the FUE in this RNA; PC—‘pre-cleavedRNA, one that ends at the principal poly(A) site of this gene; ΔNUE (dNUE in the inset of the graph in panel B)—RNA lacking the three NUEs present in this region; DE–RNA containing sequences spanning from the third poly(A) site to a point 80-nt downstream from the second site; GFP—a control RNA containing no known polyadenylation-related cis elements. The polyadenylation signal (gray box) includes the FUE (black line within the poly(A) signal region), NUEs (black vertical bars) and cleavage sites (vertical line above the signal). (B) The RNAs depicted in panel A were incubated with MBP-AtCPSF30 for the indicated times; in all cases, the initial RNA concentration was 200 nM, and AtCPSF30 concentration was 300 nM. The percent of full-length RNAs (with the 0 time point set as 100%) remaining after at each time point was plotted as a function of time.
The transient accumulation of RNA breakdown intermediates in the time-course studies suggested that AtCPSF30 might possess endonuclease activity; such intermediates were also observed with RNAs end-labeled at either their 5′ or 3′ ends (data not shown). To further test this, assays were conducted using circular RNAs. As shown in Figure 1C, such RNAs were also susceptible to the nuclease activity of AtCPSF30. Moreover, the kinetics of breakdown of the circular RNAs were similar to those seen with the linear RNA. These results indicate that AtCPSF30 can act as an endonuclease.
One possible function for the endonuclease activity of AtCPSF30 is the processing of the pre-mRNA as a prelude to the polyadenylation. Such a function would require that the nuclease action leaves a 3′-hydroxyl group at the end of the RNA; alternative modes of nuclease action, in contrast, might leave 3′-phosphates or 2′-3′ cyclic phosphates. To examine this, the products of AtCPSF30 endonucleolytic action were end-labeled with RNA ligase + [32P] cytidine 3′,5′-bisphosphate ([32P]-pCp); such a treatment is expected to label RNAs bearing 3′-hydroxyl groups, but not 3′- or 2′,3′-cyclic phosphates. As shown in Figure 2 (lane 1), the end-labeling treatment of the input (unlabeled) RNA yielded a single discrete species. Pre-treatment of the input RNA with 3′-dATP + PAP (Figure 2, lane 2) eliminated this labeling, indicating a requirement for a free 3′-hydroxyl for the labeling with [32P]-pCp. Subsequent treatment of the 3′-blocked input RNA with AtCPSF30 yielded an array of breakdown products that were readily labeled with ([32P]-pCp (Figure 2, lane 3). This result indicates that the nuclease action of AtCPSF30 yields 3′-hydroxyl groups. It should be noted that the RNAs labeled by [32P]-pCp in lane 3 of Figure 2 do not correspond in size to those expected if cleavage was occurring at the three ‘natural’ polyadenylation sites in the substrate RNA (the latter would be expected to yield 5′ cleavage products of 125, 145 and 175 nt) (32,35). Thus, while purified AtCPSF30 processes the substrate at distinct sites, by itself it does not recapitulate the exact 3′-end profile seen in vivo.
Figure 2.
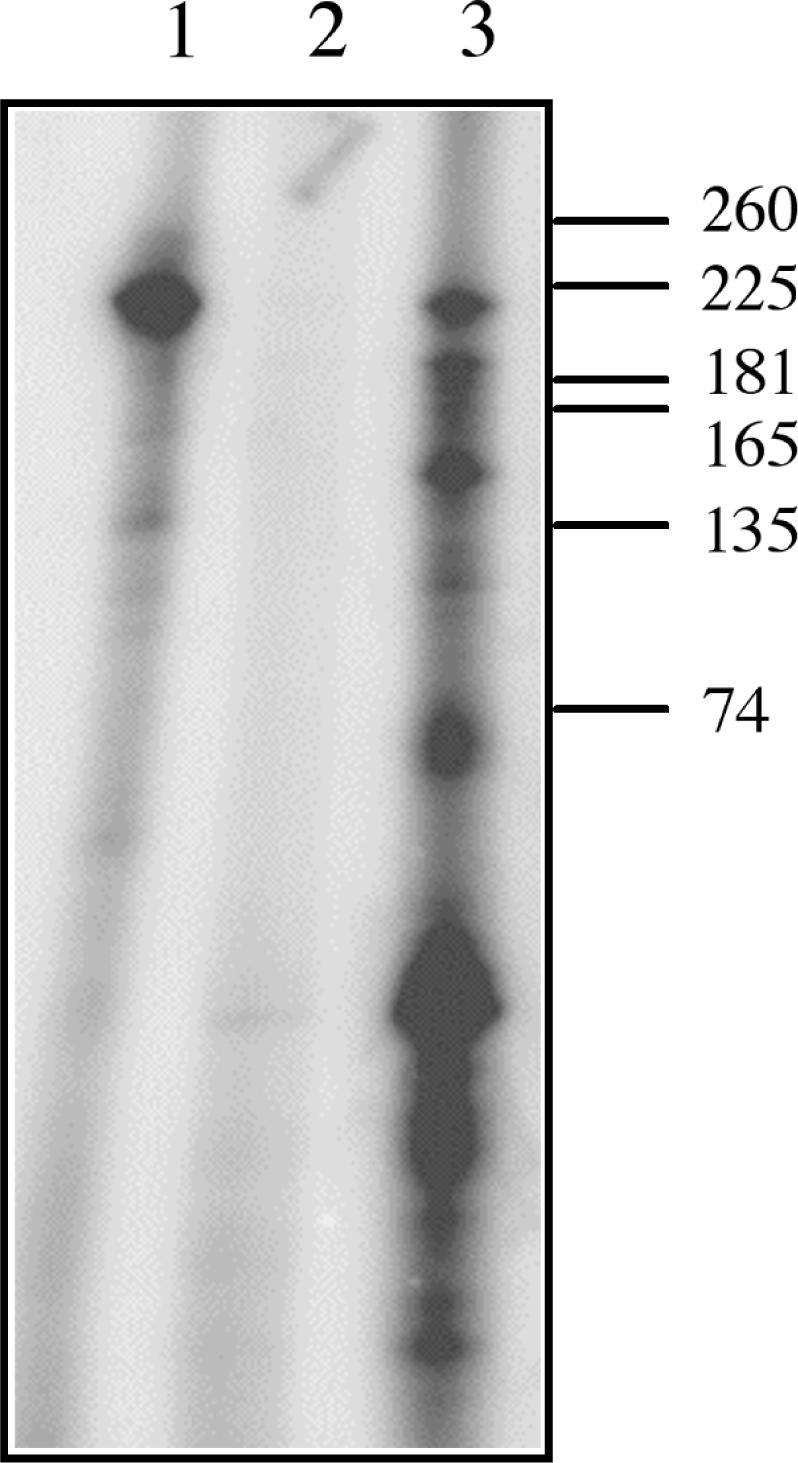
Cleavage by AtCPSF30 generates 3′OH groups. Unlabeled RNA (200 nM) was ligated with [32P]-pCp before pre-treatment with PAP + 3′-dATP (lane 1), after treatment with PAP + 3′-dATP (lane 2) and after treatment with PAP + 3′-dATP and subsequent incubation with AtCPSF30 (300 nM) for 10 min at 30°C (Lane 3). The positions of RNA size standards are indicated to the right.
Absence of sequence specificity in the nuclease activity of AtCPSF30
The distinctive processing pattern seen in Figure 2, while not identical to the pattern expected based on the in vivo handling of this RNA, nonetheless suggests some sequence preference in the action of AtCPSF30. Such preferences may be related to one or more of the polyadenylation-related motifs—FUE, NUE or CE—present in the RNA. To explore this possibility, the action of AtCPSF30 on a battery of other RNA substrates was explored. These RNAs consisted of smaller parts of the E9 polyadenylation signal (Figure 3A). One of these (‘ΔFUE’ in Figure 3A) had a deletion in its FUE region. Another (‘PC’) retained the FUE and two of the NUEs, and ended at the middle of the three sites in this 3′-UTR (this site is the predominant site utilized in vivo) (32,35); this RNA is thus analogous to a pre-cleaved RNA. A third (‘ΔNUE’) had a deletion extending from 60 nt upstream to 40 nt downstream from the middle poly(A) site; this deletion removes all of the NUEs and cleavage sites contained in this signal, but retains the FUE. A fourth (DE) extended from the middle polyadenylation site to 80 nt downstream from this site. As shown in Figure 3B, all of these RNAs could serve as substrates for AtCPSF30 in the nuclease assay. Moreover, no dramatic differences could be seen in the rates with which these RNAs were degraded. An RNA (labeled ‘GFP’ in Figure 3) with no known polyadenylation-related elements was degraded with similar kinetics. The results indicate that AtCPSF30 does not have an obvious preference for polyadenylation-related signals for its nucleolytic activity.
Identification of AtCPSF30 domains involved in nuclease activity and RNA binding
AtCPSF30 consists of a number of domains (30); these include a central region of three CCCH-type zinc finger motifs that are conserved to some extent in all eukaryotic CPSF30 proteins, an N-terminal region with a highly-conserved, plant-specific region enriched in acidic amino acids and a plant-specific C-terminal region. To study the possible contributions of these various domains to the nuclease activities of AtCPSF30, a number of mutant forms of the enzyme (Figure 4A) were prepared and assayed. These includes derivatives lacking either the N-terminal or C-terminal plant-specific domains (m4 and m9) (30) and point mutants in which the last five amino acids of each zinc finger motif (CxxxH) were replaced with the motif STxxY; this replacement eliminates two of the four putative zinc-coordinating amino acid side chains in the motif. As was noted previously (30), purified preparations of the full-sized MBP-AtCPSF30 consisted of two polypeptides (Figure 4B); the smaller of these (noted with an * in Figure 4B) is a breakdown product whose C-terminus lies near that of the m4 mutant (Figure 4B, the ‘m4’ lane on the right of the panel). These variants were assayed for nuclease activity. As shown in Figure 4C, the m9 and ZF1 mutants displayed activities that were similar to that of the wild-type enzyme. The m4 and ZF2 mutants both had activity, but less than that seen with the wild-type enzyme (this is apparent as still-detectable precursor RNA after the 30-min incubation). The ZF3 mutant showed little or no detectable activity under these conditions. These results indicate that several parts of the protein are needed for full activity, but point to the third zinc finger as being most important. Importantly, the effects of the point mutations, and especially the ZF3 mutant, indicate that the nuclease activity under study is a property of the AtCPSF30 polypeptide (as opposed to a co-purifying bacterial contaminant that the stringent washes and ion exchange purification protocols did not separate).
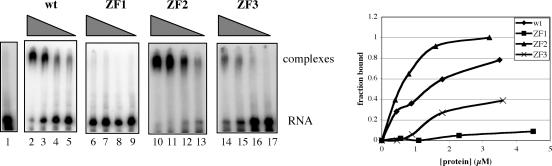
One possible explanation for the absence of nuclease activity with the ZF3 mutant is that it no longer binds RNA as does the wild-type protein. This hypothesis was tested using a gel-shift assay and the rbcS-E9 RNA depicted in Figure 3. As shown in Figure 5, the ZF2 mutant had an RNA-binding activity that was similar to the wild type. The ZF3 mutant was also able to bind RNA, albeit at a somewhat reduced level. The ZF1 mutant displayed no ability to bind RNA. These results indicate that ZF1, which is dispensable for nuclease activity (Figure 4) is primarily responsible for the RNA-binding activity of AtCPSF30. Ablation of ZF3 partially reduces RNA binding but completely eliminates the nuclease activity. Interestingly, alteration of ZF2 had no effect on RNA binding.
Figure 5.
RNA binding by the zinc finger mutants. Uniformly labeled RNA (final concentration of 300 nM) was incubated with 2-fold serial dilutions of the wild-type AtCPSF30 and with the three zinc finger mutants shown in Figure 4A and RNA binding assessed using the electrophoretic mobility shift assay (see the Methods section). RNA with 3 µM MBP is shown in lane 1. Other lanes and their corresponding protein concentrations: Lane 2—3.5 µM wt protein; lane 3—1.8 µM wt protein; lane 4—0.88 µM wt protein; lane 5—0.44 µM wt protein; lane 6—4.5 µM ZF1 mutant; lane 7—2.3 µM ZF1 mutant; lane 8—1.1 µM ZF1 mutant; lane 9—0.56 µM ZF1 mutant; lane 10—3.2 µM ZF2 protein; lane 11—1.6 µM ZF2 protein; lane 12—0.8 µM ZF2 proteins; lane 13—0.4 µM ZF2 protein; lane 14—3.6 µM ZF3 protein; lane 15—1.8 µM ZF3 protein; lane 16—0.9 M ZF3 protein; lane 17—0.45 µM ZF3 protein. The autoradiographs are shown on the left, and the results plotted on the right. In the graph, the fraction of RNA present in the complexes was plotted as a function of protein concentration.
The N-terminal domain of an Arabidopsis Fip1 ortholog inhibits the AtCPSF30-associated nuclease activity
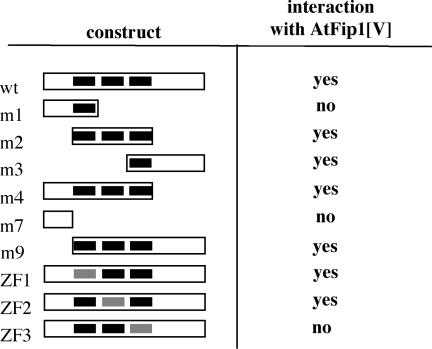
The third zinc finger of AtCPSF30 corresponds to the motif of Yth1 that is involved in its interaction with Fip1, an interaction that affects the binding of Yth1 to RNA (14). Since this is also the motif that is needed for endonuclease activity (Figure 4), it seemed possible that plant Fip1 orthologs might have effects on the nuclease activity of AtCPSF30. Previously, it was reported that an Arabidopsis Fip1 ortholog, encoded by At5g58040, was able to interact with AtCPSF30, and that the N-terminal 137 amino acids of the 1196 amino acid Fip1 ortholog [AtFip1(V)] contained the domain responsible for this interaction (24). This interaction was further dissected using a standard yeast two-hybrid assay. For this, the battery of AtCPSF30 mutants illustrated in Figure 4A, along with others described previously (30), were tested for interactions with the N-terminus of AtFip1(V). As summarized in Figure 6, all of the variants that possessed the third zinc finger retained the ability to interact with AtFip1(V); importantly, alteration of just the third zinc finger eliminated the interaction, indicating that this interaction requires just the third zinc finger motif of AtCPSF30.
Figure 6.
Two-hybrid analysis of the interaction between AtCPSF30 and AtFip1(V). The battery of mutants tested in the assay is shown on the left; as in Figure 4A, the three zinc fingers are represented as black bars, and mutated zinc fingers depicted as gray bars. The extents of each protein present in the two-hybrid construct are shown for each variant; the representation is not drawn to scale. The results of the tests are summarized on the right; ‘yes’ indicates that different independently isolated dual transformants were able to grow on the selective media (and therefore denotes an interaction in yeast cells), whereas ‘no’ indicates an inability to grow on such media.
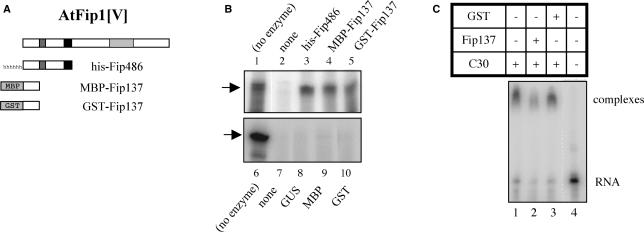
Accordingly, the effects of the N-terminal portion of AtFip1(V) on the nuclease activity of AtCPSF30 were tested, using three different forms of the N-terminus of the Arabidopsis protein. One form consisted of a histidine-tagged segment containing the first 483 amino acids of AtFip1(V); this segment includes the CPSF30-binding region as well as the conserved Fip1 domain that defines this group of proteins (this is illustrated in Figure 7A). Two other forms consisted of the N-terminal 137 amino acids fused to either GST or MBP. These three forms were purified and added in 2-fold molar excesses to nuclease reactions. The results, shown in the upper panel of Figure 7B, indicate that all three forms inhibit the nuclease activity of AtCPSF30. Purified histidine-tagged GUS (Figure 7B, lane 8), MBP (Figure 7B, lane 9) or GST (Figure 7B, lane 10) had no effects on the nuclease activity. Moreover, the purified control proteins themselves possessed no noticeable nuclease activity (data not shown). These results indicate that the interaction of AtCPSF30 with the N-terminus of AtFip1(V) inhibits the nuclease activity of this protein.
Figure 7.
Inhibition of AtCPSF30 by the N-terminal domain of AtFip1(V) (A) Illustration of the parts of AtFip1(V) that were tagged and used for these assays. At the top is a depiction of the full-sized AtFip1(V) protein, showing an N-terminal acidic domain (gray bar), Fip motif (black bar) and C-terminal RNA-binding domain [light gray bar (24)]. Beneath this are shown the parts of the protein present in the ‘Fip137’ and ‘Fip486’ constructs. The Fip137 segment was fused to MBP and GST, and the Fip486 segment to a histidine tag; the tags are not drawn to scale in the illustration. (B) Nuclease activities of AtCPSF30 in the presence of the Fip1 preparations. AtCPSF30 (300 nM) was assayed in the absence (lanes 2 and 7) of additional protein or in the presence of histidine-tagged Fip486 (lane 3), MBP-Fip137 (lane 4), GST-Fip137 (lane 5), histidine-tagged GUS (lane 8), purified MBP (lane 9) or purified GST (lane 10). For the reactions in lanes 3–5 and 8–10, the concentrations of the respective additional protein was 600 nM. RNA incubated without enzyme is shown in lanes 1 and 6. In addition to lane numbers, the proteins used to supplement AtCPSF30 nuclease reactions are shown above or beneath their respective lanes; (no enzyme) denotes samples in which RNA was incubated with buffer and no protein. In all reactions, the RNA concentration was 200 nM. (C) Effects of GST-Fip137 on RNA binding by AtCPSF30. RNA (300 nM) and AtCPSF30 (3.5 µM) were incubated and analyzed as described in the Methods section. AtCPSF30 was assayed by itself (lane 1) or with 4 µM GST-Fip137 (lane 2) or 4 µM GST (lane 3). The sample in lane 4 had no protein. GST-Fip137 and GST have no RNA-binding activity [(24); data not shown]. These component combinations are summarized in the table above the autoradiogram.
This result suggests that AtFip1(V) regulates the nuclease activity of AtCPSF30. In contrast to this, the GST-Fip137 protein had little apparent effect on RNA binding by AtCPSF30 (Figure 7C). Thus, RNA binding by AtCPSF30 (Figure 7C, lane 1) was still apparent in the presence of added GST-Fip137 (Figure 7C, lane 2) or GST (Figure 7C, lane 3). A slight reduction in binding in the presence of GST-Fip137 was apparent; this is consistent with the reduced RNA binding of the ZF3 mutant (Figure 3). However, the extent of diminution of RNA binding by GST-Fip137 was far less than the inhibition of nuclease activity seen with the same preparation.
DISCUSSION
AtCPSF30 as a processing endonuclease
The nature of the events associated with processing of the pre-mRNA remains somewhat unclear. Bai and Tolias (26,27) reported that the Drosophila CPSF30 ortholog, the so-called clipper protein, possesses an inherent nuclease activity. Based on this observation, it was suggested that CPSF30 is the processing endonuclease (8). Such a suggestion would seem to be consistent with the observation that Yth1 associates with the processing site in vitro (11). However, others have suggested that CPSF73 is the processing endonuclease. These arguments are based on a number of observations: the AAUAAA-dependent crosslinking of CPSF73 to the cleavage site, the similarity of CPSF73 to Zn-dependent hydrolytic enzymes related to metallo-beta-lactamases, the requirement of amino acids residues in CPSF73 that are required for hydrolytic function in metal-beta-lactamases and the observation that recombinant CPSF73 possesses endonucleolytic activity (6,7). Based on the results presented in this study, AtCPSF30 would seem to be an excellent candidate for an endonuclease involved in processing pre-mRNAs prior to polyadenylation in Arabidopsis. It possesses a distinctive endonuclease activity (Figure 1) and leaves a 3′-terminus that is suitable for subsequent poly(A) tail addition by PAP (Figure 2). Poly(A) site choice in vivo is altered in a mutant deficient in AtCPSF30 (Zhang et al., submitted for publication), indicating that normal poly(A) site selection in Arabidopsis requires the presence of AtCPSF30. However, if AtCPSF30 is in fact a processing endonuclease, it cannot be the only one active in plants. This follows from the realization that this protein is not essential for plant growth and development (30), and that plants deficient in this protein produce bulk poly(A) that is indistinguishable in length and quantity from wild-type plants (Zhang et al., submitted for publication). Wickens and Gonzalez (36) have speculated that the cleavage step in polyadenylation may be performed by any of a multiplicity of processing endonucleases, and has likened the 3′-processing machinery to the ‘package’ of nucleases that comprise the exosome. It may be that such a situation is in fact in force in plants, and that AtCPSF30 and AtCPSF73 are both processing endonucleases.
Another possibility merits mention. While the obvious role for an endonuclease in the polyadenylation reaction is the generation of the 3′ end for PAP, alternative functions for such an activity may exist as well. CPSF has been implicated in small RNA 3′ end maturation (37,38), 3′ end formation of cell cycle-regulated histone mRNAs (39,40) and cytoplasmic polyadenylation (41,42). While specific roles for CPSF30 in these processes have not been reported, it is nevertheless possible that the endonuclease activity documented here may reflect roles for AtCPSF30 in these, or other as yet unidentified, activities.
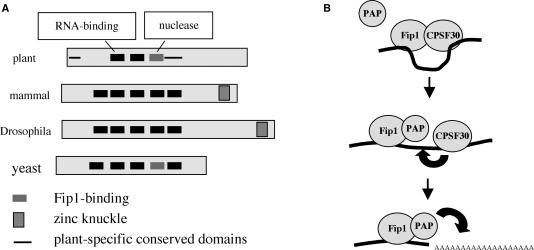
Implications of the structure–function map of AtCPSF30 for mRNA 3′ end formation in plants
The part of AtCPSF30 that is associated with endonucleolytic activity (the third zinc finger) is present in all eukaryotic CPSF30 orthologs (Figure 8A). The corresponding zinc finger motif in Yth1 is required for RNA binding (14). Moreover, with AtCPSF30 and Yth1, this motif is needed for interactions with Fip1 proteins, and interactions with Fip1 inhibit the associated RNA-related activities (nuclease, in the case of AtCPSF30, and RNA binding, in the case of Yth1) of these proteins (14). Tacahashi et al. (14) have proposed that this interplay between Fip1 and Yth1 may reflect a progression through the polyadenylation reaction, with Fip1 displacing Yth1 from the pre-mRNA after the processing step, thereby bringing PAP to the 3′ end left by processing. The characteristics of AtCPSF30 do not lend themselves easily to an analogous model. In particular, the zinc finger motifs of AtCPSF30 implicated in RNA binding and nucleolytic activity are different, and AtFip1 interacts only with the motif involved in nuclease action. Moreover, AtFip1 inhibits the nuclease activity of AtCPSF30 (Figure 7B) but has a much more modest effect on RNA binding by AtCPSF30 (Figure 7C). Therefore, it seems unlikely that AtFip1 would displace AtCPSF30 from a processed pre-mRNA. One plausible alternative to this is a scenario where AtCPSF30 is initially associated with AtFip1, such that the endonuclease activity of AtCPSF30 is repressed (Figure 8B). The endonuclease would become activated with the dissociation of AtFip1(V), perhaps as a consequence of binding of other subunits to AtFip1(V). Along these lines, it is of interest to recall the Arabidopsis PAP isoforms bind to the same 137 amino acid portion of AtFip1(V) that inhibits the nuclease activity of AtCPSF30 (24; B. Addepalli and A. G. Hunt, unpublished data). This remodeling around AtFip1(V) would lead to subsequent processing and polyadenylation.
Figure 8.
Models for the structure and function of AtCPSF30. (A) Illustration of the domain organization of AtCPSF30 (30), showing the correspondence with orthologs from mammals, Drosophila and yeast (4,14,26,27). Domains of the plant protein that are associated with particular activities are indicated above the depiction of the plant protein. Symbols are as indicated beneath the representations of the proteins. (B) Model for the action of CPSF30 in the course of pre-mRNA 3′ end processing in plants. Initially, AtCPSF30 is associated with AtFip1(V); in this complex nuclease activity is repressed, the complex may bind RNA via the AtFip1(V) and AtCPSF30 RNA-binding sites. PAP may displace AtCPSF30 from AtFip1(V) as shown in the middle panel; this would activate the endonucleolytic activity of AtCPSF30 (thick black arrow). Subsequent to this processing, PAP would polyadenylate the processed RNA as shown in the bottom panel.
More generally, the mutational analysis of AtCPSF30 presented in this study reveals a protein of interesting and subtle complexity. Two RNA-associated activities of the protein, binding and endonucleolytic activity, are associated with different zinc finger motifs. The third motif and its evolutionarily conserved association with AtFip1 is mentioned above. The first zinc finger motif is the most highly conserved of the motifs when compared with other eukaryotic CPSF30 proteins (30) and is needed for RNA binding by AtCPSF30. The corresponding motif in the bovine and yeast counterparts is also needed for RNA binding (4,11,14). Interestingly, the corresponding motif in Yth1 is needed for the functioning of this protein in the cleavage step of polyadenylation in vitro (14); this is consistent with the model proposed in the preceding paragraph, in which positioning prior to cleavage by the nuclease domain is accomplished by the RNA-binding domain of AtCPSF30.
In Arabidopsis (and likely other plants), CPSF30 is encoded by a complex gene whose transcripts are alternatively processed to yield two mRNAs (30). The smaller of these encodes AtCPSF30, whereas the larger specifies a polypeptide that consists of virtually the entire CSPF30 sequence fused to another domain (the so-called YT521B domain) that is found in splicing-associated proteins in mammals (31) and in Arabidopsis proteins that are associated with so-called calcineurin B-like interacting protein kinases [CIPKs; (43)]. The function of the larger polypeptide is not known, but the present study suggests that it should possess endonuclease activity. The possible roles that the larger polypeptide might play in gene expression are not clear, but the conceptual linkages between mRNA 3′ end formation and both calcium-mediated signaling and pre-mRNA splicing are intriguing.
To summarize, the results presented here constitute strong evidence that AtCPSF30 is a processing endonuclease in the polyadenylation reaction in plants. In addition, they suggest that this function is regulated by AtFip1(V), thereby suggesting an ordered series of events that occurs during the process of mRNA 3′ end formation in plants. With other studies, these data suggest that there is more than one 3′-end processing endonuclease, and possibly an unexpected redundancy in this function, in plants.
ACKNOWLEDGEMENTS
The authors are grateful to Carol Von Lanken for technical assistance and to Kim Delaney and Dr Kevin Forbes for some materials used in this study. They are especially appreciative of the advice of Dr Dan Schoenberg. This work was supported by NSF grant MCB-0313472. Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation (MCB-0313472).
Conflict of interest statement. None declared.
REFERENCES
- 1.Keller W, Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr. Opin. Cell. Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 2.Wahle E, Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 5.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23:616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zarudnaya MI, Kolomiets IM, Hovorun DM. What nuclease cleaves pre-mRNA in the process of polyadenylation? IUBMB Life. 2002;54:27–31. doi: 10.1080/15216540213821. [DOI] [PubMed] [Google Scholar]
- 9.Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- 10.Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 11.Barabino SM, Ohnacker M, Keller W. Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 2000;19:3778–3787. doi: 10.1093/emboj/19.14.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross S, Moore CL. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 2001;21:8045–8055. doi: 10.1128/MCB.21.23.8045-8055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tacahashi Y, Helmling S, Moore CL. Functional dissection of the zinc finger and flanking domains of the Yth1 cleavage/polyadenylation factor. Nucleic Acids Res. 2003;31:1744–1752. doi: 10.1093/nar/gkg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preker PJ, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 16.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002;21:1800–1810. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minvielle-Sebastia L, Preker PJ, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl Acad. Sci. USA. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belostotsky DA, Rose AB. Plant gene expression in the age of systems biology: integrating transcriptional and post-transcriptional events. Trends Plant Sci. 2005;10:347–353. doi: 10.1016/j.tplants.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Hunt AG. Messenger RNA 3′ end formation in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1994;45:47–60. [Google Scholar]
- 21.Li Q, Hunt AG. The polyadenylation of RNA in plants. Plant Physiol. 1997;115:321–325. doi: 10.1104/pp.115.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothnie HM. Plant mRNA 3′-end formation. Plant Mol. Biol. 1996;32:43–61. doi: 10.1007/BF00039376. [DOI] [PubMed] [Google Scholar]
- 23.Herr AJ, Molnar A, Jones A, Baulcombe DC. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl Acad. Sci. USA. 2006;103:14994–15001. doi: 10.1073/pnas.0606536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes KP, Addepalli B, Hunt AG. An Arabidopsis Fip1 homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 2006;281:176–186. doi: 10.1074/jbc.M510964200. [DOI] [PubMed] [Google Scholar]
- 25.Xu R, Zhao H, Dinkins RD, Cheng X, Carberry G, Li QQ. The 73 kD subunit of the cleavage and polyadenylation specificity factor (CPSF) complex affects reproductive development in Arabidopsis. Plant Mol. Biol. 2006;61:799–815. doi: 10.1007/s11103-006-0051-6. [DOI] [PubMed] [Google Scholar]
- 26.Bai C, Tolias PP. Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol. Cell. Biol. 1996;16:6661–6667. doi: 10.1128/mcb.16.12.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai C, Tolias PP. Drosophila clipper/CPSF 30K is a post-transcriptionally regulated nuclear protein that binds RNA containing GC clusters. Nucleic Acids Res. 1998;26:1597–1604. doi: 10.1093/nar/26.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 29.Noah DL, Twu KY, Krug RM. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology. 2003;307:386–395. doi: 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 30.Delaney KJ, Xu R, Zhang J, Li QQ, Yun KY, Falcone DL, Hunt AG. Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 2006;140:1507–1521. doi: 10.1104/pp.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoilov P, Rafalska I, Stamm S. YTH: a new domain in nuclear proteins. Trends Biochem. Sci. 2002;27:495–497. doi: 10.1016/s0968-0004(02)02189-8. [DOI] [PubMed] [Google Scholar]
- 32.Mogen BD, MacDonald MH, Leggewie G, Hunt AG. Several distinct types of sequence elements are required for efficient mRNA 3′ end formation in a pea rbcS gene. Mol. Cell. Biol. 1992;12:5406–5414. doi: 10.1128/mcb.12.12.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl Acad. Sci. USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mogen BD, MacDonald MH, Graybosch R, Hunt AG. Upstream sequences other than AAUAAA are required for efficient messenger RNA 3′-end formation in plants. Plant Cell. 1990;2:1261–1272. doi: 10.1105/tpc.2.12.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickens M, Gonzalez TN. Molecular biology. Knives, accomplices, and RNA. Science. 2004;306:1299–1300. doi: 10.1126/science.1100137. [DOI] [PubMed] [Google Scholar]
- 37.Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, et al. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 38.Morlando M, Ballarino M, Greco P, Caffarelli E, Dichtl B, Bozzoni I. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J. 2004;23:2392–2401. doi: 10.1038/sj.emboj.7600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dickson KS, Bilger A, Ballantyne S, Wickens MP. The cleavage and polyadenylation specificity factor in Xenopus laevis oocytes is a cytoplasmic factor involved in regulated polyadenylation. Mol. Cell. Biol. 1999;19:5707–5717. doi: 10.1128/mcb.19.8.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bilger A, Fox CA, Wahle E, Wickens M. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes Dev. 1994;8:1106–1116. doi: 10.1101/gad.8.9.1106. [DOI] [PubMed] [Google Scholar]
- 43.Ok SH, Jeong HJ, Bae JM, Shin JS, Luan S, Kim KN. Novel CIPK1-associated proteins in Arabidopsis contain an evolutionarily conserved C-terminal region that mediates nuclear localization. Plant Physiol. 2005;139:138–150. doi: 10.1104/pp.105.065649. [DOI] [PMC free article] [PubMed] [Google Scholar]