Abstract
Oligoribonuclease is the only RNase in Escherichia coli that is able to degrade RNA oligonucleotides five residues and shorter in length. Firmicutes including Bacillus subtilis do not have an Oligoribonuclease (Orn) homologous protein and it is not yet understood which proteins accomplish the equivalent function in these organisms. We had previously identified oligoribonucleases Orn from E. coli and its human homolog Sfn in a screen for proteins that are regulated by 3′-phosphoadenosine 5′-phosphate (pAp). Here, we identify YtqI as a potential functional analog of Orn through its interaction with pAp. YtqI degrades RNA oligonucleotides in vitro with preference for 3-mers. In addition, YtqI has pAp-phosphatase activity in vitro. In agreement with these data, YtqI is able to complement both orn and cysQ mutants in E. coli. An ytqI mutant in B. subtilis shows impairment of growth in the absence of cysteine, a phenotype resembling that of a cysQ mutant in E. coli. Phylogenetic distribution of YtqI, Orn and CysQ supports bifunctionality of YtqI.
INTRODUCTION
Degradation of RNA is an important factor in the regulation of gene expression. Impairment of regulation of mRNA stability was implicated in the pathogenesis of cancer, inflammatory diseases and Alzheimer's disease (1). Enzymes involved in RNA degradation fall into two major classes: endoribonucleases, which cleave RNAs internally and exoribonucleases, which degrade RNAs from the ends. The phylogenetic distribution of endo- and exoribonucleases (2–4) in genomes clearly shows that different species vary considerably with respect to the number and variety of the RNases they harbor. Interestingly, two ribonucleases that are essential in Escherichia coli do not have homologous counterparts in Bacillus subtilis: RNase E (5) and oligoribonuclease (6). Belonging to the degradosome, RNase E is widely believed to be the enzyme initiating mRNA decay (7).
Oligoribonuclease, Orn, is the only known exoribonuclease that is essential in E. coli (6). The essentiality in E. coli is due to its unique ability to degrade RNA oligonucleotides with a length of 5 nt and shorter (8–10), and oligonucleotides of these lengths were shown to accumulate in a conditional orn-mutant (6). We would like to introduce the term ‘nanoRNA’ here to distinguish these extremely short oligonucleotides from the longer microRNAs. We chose the term nano in reference to its roots: Nano originates from the Greek word nanos, which means dwarf. Micro on the other hand descends from the Greek word mikros, which means small. Nano is therefore used in this context simply to articulate ‘smaller than’ micro.
A recent study reveals the structural basis for the constraints preventing RNase II as a member of the RNR exoribonuclease family from degrading oligonucleotides shorter than 5 nt (11–14). Another member of this exoribonuclease family, RNase R was shown to processively degrade RNA in a 3′ to 5′ directed manner until a di- or trimer remains which cannot be degraded further by this enzyme (12). This size limit is therefore likely to be common at least among the members of this important family of exoribonucleases. This highlights the importance of enzymes that have the ability to degrade nanoRNA and thus bring the degradation of RNA to completion. Absence of an oligoribonuclease in Firmicutes is in contrast to its general presence in Gram-negative prokaryotic genomes as well as in eukaryotic genomes (3). This prompted us to question which enzyme could functionally replace oligoribonuclease in these organisms.
We had recently discovered an unexpected link between sulfur- and RNA metabolism: oligoribonuclease binds to 3′-phosphoadenosine 5′-phosphate (pAp) and is sensitive to micromolar amounts of the nucleotide (15). pAp is generated in sulfur assimilation and was implicated in the molecular mechanism of lithium's action in the treatment of bipolar disorder due to strong inhibition of pAp-phosphatase by lithium (16). The interaction between pAp and oligoribonuclease was documented for E. coli oligoribonuclease, Orn and its human homolog, Sfn (15). The purpose of this work was to explore whether the conserved interaction between pAp and oligoribonucleases could be exploited to identify a functional analog of Orn in B. subtilis. Surprisingly, the protein identified by this route, YtqI, points to the existence of an even closer link between sulfur- and RNA-metabolism in this organism: YtqI can degrade both nanoRNA and pAp in vitro. The pAp-degrading activity of YtqI is similar in magnitude to that of CysQ, the pAp-phosphatase from E. coli. Consistent with its in vitro activities, YtqI can replace both Orn and pAp-phosphatase (CysQ) in E. coli.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
Escherichia coli strains were grown in LB or MOPS minimal medium (17) containing 40 μg/ml of amino acids as indicated, K-phosphate at 2 mM, vitamin B1 at 0.0005%, biotin at 0.001% when needed, glycerol at 0.4%, glucose or arabinose as indicated. Bacillus subtilis was grown in minimal medium (18). Ampicillin (100 μg/ml), kanamycin (25 μg/ml) or erythromycin (1 μg/ml) was added for plasmid maintenance or to select for chromosomal marker. Anhydrotetracycline (Atc) was added at 250 ng/ml for induction of PLtetO-1.
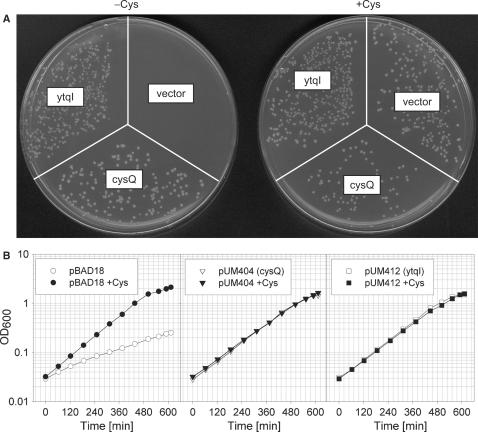
To test growth in the absence of cysteine (Figure 7), overnight cultures grown in MOPS minimal medium containing all amino acids were washed twice with medium lacking cysteine before inoculation into medium containing cysteine (100 μM) or lacking this amino acid.
Figure 7.
Complementation of the cysQ mutant by the expression of YtqI. (A) Transformations of strain UM285 with pBAD18 (vector control), pUM404 (arabinose-inducible cysQ) or pUM412 (arabinose-inducible ytqI) were spread on MOPS-minimal plates containing 0.4% glycerol and 0.2% arabinose in the presence or absence of cysteine. (B) Growth of strain UM285 carrying the plasmids indicated in MOPS-medium containing 0.4% glycerol and 0.02% arabinose in the presence or absence of cysteine (100 μM).
All experiments were performed in accordance with the European regulation requirements concerning the use of Genetically Modified Organisms (level 1 containment, agreement n°2735).
The plasmid for expression of his-tagged YtqI under control of the arabinose-inducible promoter Para (pUM412) was constructed as follows: Primer UM175 and UM176 were used to PCR-amplify ytqI from B. subtilis 168 chromosomal DNA. The EcoRI, XhoI digested fragment was used to replace the EcoRI/XhoI fragment of pUM407 coding for Orn leaving the region coding for the C-terminal his-tag and the ribosomal-binding site intact.
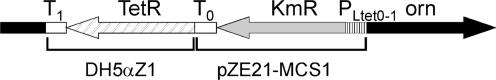
The conditional E. coli orn mutant (strain UM341) uses the anhydrotetracycline (Atc)-inducible promoter PLtetO-1 (19) together with a Tet-repressor (TetR) to ensure tight control in the absence of Atc. This strain was created by introducing the PLtetO-1 promoter in front of orn together with a cassette coding for TetR and a kanamycin selection marker (KmR). Two PCR fragments were amplified: PCR1 amplified PLtetO-1, kmR and the transcription terminator T0 from pZE21-MCS1 (19) using primer UM153 and UM156, PCR2 amplified tetR including its constitutive promoter PN25 and terminator T1 from chromosomal DNA of DH5αZ1 (19) using primer UM155 and UM154. pZE21-MCS1 and DH5αZ1 were kindly provided by Hermann Bujard. The outside primers UM155 and UM156 and equimolar amounts of PCR fragments 1 and 2 were used to perform overlapping PCR. The obtained PCR fragment was then cloned into pGEMT-Easy (Promega) by TA cloning followed by sequencing using primer UM172 and UM173. A verified clone was used as template for PCR amplification using primers UM155 and UM156. The obtained PCR fragment was transformed into CF10230 to create the orn mutant by lambda Red-assisted recombination according to the protocol of Yu et al. (20). CF10230 is a nic+ derivative of DY329 (20) that was kindly supplied by Michael Cashel (Cashel,M., unpublished data). Mutants were verified by confirmation of the 5′ site of integration into the chromosome by means of PCR using primers UM158 and UM159 yielding a 432 bp fragment, as well as the 3′ site of integration using primer UM160 and UM161, yielding a 446 bp fragment.
The cysQ mutant we used here has been described before (15). We will refer to cysQ mutant 1 as UM285 from now on. UM285 has a replacement of the complete coding DNA sequence (CDS) for CysQ by kmR.
The ytqI mutant strain (BSF66) was part of the European/Japanese effort to inactivate the whole gene set of B. subtilis 168 and has an insertion of pMUTIN2MCS after the codon for amino acid 108 (21).
pAp-agarose binding
pAp-agarose-binding experiments were performed as described previously (15). Two hundred milliliter cultures of B. subtilis 168 or a protease-deficient mutant, DB430 (22) were grown in minimal medium containing 1.5 mM MgSO4 at 37°C to an OD600 between 1.6 and 1.8. Cells were harvested and washed once with 50 mM NaPO4 pH 8.0, 300 mM NaCl before freezing. Frozen pellets were resuspended in 2 ml pAp-agarose buffer (50 mM HEPES, pH 7.5, 10 mM CaCl2, 50 mM KCl) containing 100 mM NaCl, 0.4 mM phenylmethylsulfonyl fluoride (PMSF) and 130 μg/ml lysozyme. After incubation for 45 min on ice, the cells were opened using a Fastprep apparatus (Bio101). Blocking with agarose beads, incubation with pAp-agarose, elution, PAA electrophoresis and identification of proteins was done exactly as described before (15).
Purification of his-tagged YtqI and activity assays
YtqI was purified from a 200 ml culture of MG1655 carrying pUM412 according to the his-tag purification protocol described previously (15).
Activity assays determining nanoRNase activity were performed using custom-made RNA oligo 5-mers or 3-mers (5′Cy5-CCCCC3′ or 5′Cy5-CCC3′) as substrates in reactions containing 50 mM HEPES, pH 7.5, 5 mM MnCl2, 1.6–3.4 μM substrate. At intervals, 4.5 μl reaction aliquots were taken and stopped by adding to an equal volume of sample buffer (4× TBE, 100 mM DTT, 16% glycerol, 20 mM EDTA) and frozen at −20°C. For analysis of the reaction products, 1.5 or 2.5 μl of samples were applied to PAA gel electrophoresis on a 22% SDS-PAA gel containing 2× TBE and run in 2× TBE. Fluorescent RNA oligos were visualized using a Molecular Dynamics STORM 860 in 650-nm long-pass filter mode. Quantification of the data was done by calculating the percent of fluorescence of each band at a given time point relative to the total fluorescence of the same time point. Separating reaction products on 22% SDS-PAA gels, we observed a reverse migration phenomenon. This effect can be accounted for by the fact that cyanine dyes have a lower net negative charge than nucleic acids: thus, removing nucleotides will reduce the charge relative to the mass of the oligonucleotide and cause it to shift up instead of down.
Inhibition of YtqI-catalyzed degradation of RNA 5-mers (5′Cy5-CCCCC3′) by pAp was performed in 30 μl reactions containing 3 μM substrate RNA 5-mers, 9 μg YtqI and pAp as indicated. Reactions were incubated for 30 min at 37°C. Relative activities were assigned as conversion of the substrate into monomers. The activity in the reaction without pAp was set to 100%.
Assays determining degradation activity on a longer substrate were done using a custom-made RNA 24-mer (5′CACACACACACACACACACACACA3′) that was 5′-end labeled with [γ-33P]ATP. This oligonucleotide was labeled using the MirVana Probe and Marker Kit (Ambion) in a 20 μl reaction containing 100 pmol oligo, 6.7 pmol [γ-33P]ATP (20 μCi), 90 pmol ATP and 1 μl T4 Polynucleotide Kinase. Incubation was done for one hour at 37°C. The reaction was stopped by the addition of 2 μl of 10 mM EDTA and incubation at 95°C for 2 min. The reaction mixture was purified from the unincorporated nucleotides using NucAway spin columns (Ambion) according to the instructions of the supplier. Three microliter of the labeled RNA (∼800 000 c.p.m.) were used in a 20 μl reaction containing 5 mM MnCl2, 50 mM HEPES pH 7.5 and 3 μg enzyme; incubation was for 30 min at 37°C after which the reaction was stopped by the addition of 20 μl loading buffer and incubation for 3 min at 95°C. An aliquot of 5 μl of the samples were resolved on a 20% PAA, 7 M Urea gel containing 2× TBE that was ran in 2× TBE. Labeling of the decade-marker was done with [γ-33P]ATP (125 μCi, 1.7 pmol) as suggested by the manufacturer including purification from unincorporated nucleotides. One-fourth of the total volume of labeled decade-marker was used per gel; this amount corresponded to ∼800 000 c.p.m. A previously characterized nuclease (with DNase and RNase activity) from B. subtilis, YhaM (23) served as positive control in experiments on RNA 24-mers. This protein was purified employing a C-terminal his-tag. HEPES was replaced by Tris pH 8.0 in the reaction containing YhaM as this enzyme is less active in HEPES.
pAp degradation was assayed in 20 μl reactions containing 6 mM pAp, 2 mM MnCl2, 50 mM HEPES (pH 7.5) at 37°C. Reactions were started by the addition of 1 μg YtqI. Aliquots of 4.5 μl were taken as indicated and mixed with 0.5 μl 100 mM EDTA before resolving them by polyethyleneimine (PEI) thin-layer chromatography with 0.8 mM LiCl as solvent. Authentic pAp and AMP were used as migration standards. Accumulation of reaction products was estimated after visualization by UV.
Expression of his-tagged proteins was monitored by PAA gel electrophoresis followed by staining with Bio-Safe Coomassie stain (BIO-RAD) or by western blot using Anti-His6 Peroxidase antibodies (Roche) at 1:200 in 1× PBS, 1% skim milk, 0.1% Tween and ECL Plus Western Blotting Detection System (GE Healthcare). For these experiments, different expression levels of the Para-controlled genes were achieved by growing cultures in liquid LB in the presence or absence of arabinose (0.2 or 0.02%).
Phylogenetic analysis
A total of 393 completely sequenced bacterial genomes published before 4 January 2007 (http://www.ebi.ac.uk/genomes/) were analyzed for the presence of YtqI, Orn and CysQ orthologous proteins. Orthologs were defined by searching for bi-directional best hits (BBH) (24) based on the following parameters: ⩾40% amino acid similarity and ⩽20% difference in protein length. The phylogenetic tree presented in Figure S1 was constructed from 141 representative species based on 16S rRNA similarity.
RESULTS
Identification of YtqI among the proteins binding to pAp
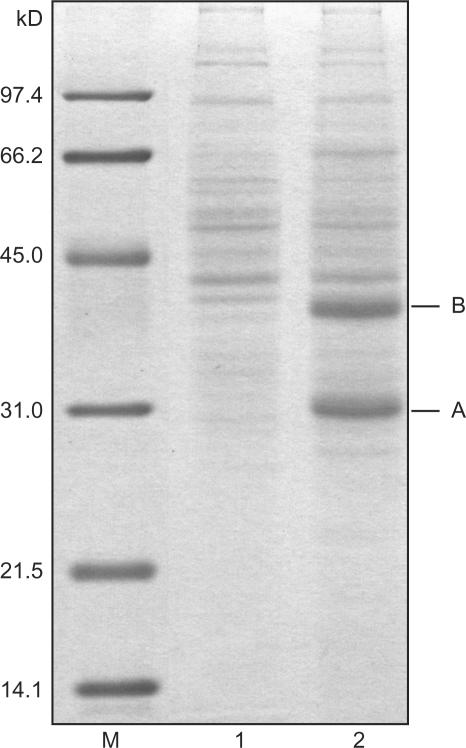
The conserved interaction of pAp and oligoribonucleases between E. coli and human cells encouraged us to ask if we could identify a functional analog of oligoribonuclease in B. subtilis among the pAp-binding proteins from this organism. Extracts of B. subtilis 168 and a protease minus mutant (DB430) (22) were used in pAp-binding experiments. The protein pattern obtained looked similar for both strains; we therefore present only the data acquired for the wild-type strain. Two major protein bands were visible in the pAp-binding fraction (Figure 1). Analysis of band A by liquid chromatography tandem mass spectrometry (LC-MS/MS) revealed HisIE (SwissProt, O34912) with an overall score of 2080 and 5 identified peptides covering 22.5% of the total mass of the protein. HisIE was identified previously as pAp-binding protein in E. coli (15). The second major band gave high scores for two proteins: GuaC (SwissProt O05269), GMP reductase with an overall score of 292 and 4 peptides covering 13% of the total mass, and YtqI (SwissProt O34600) with an overall score of 140 and 4 peptides covering 11.5% of the total mass. YtqI is an unknown protein that belongs to the DHH/DHHA1 family (25). This family of proteins consists of enzymes with phosphoesterase activity, including RecJ. YtqI was therefore our best candidate for a potential functional Orn analog.
Figure 1.
pAp-binding proteins from B. subtilis. Shown is a colloidal Coomassie stained SDS-PAA gel separating the pAp-binding fraction of B. subtilis extract. M: marker, lane 1: fraction binding to blocked agarose beads (control), lane 2: pAp-binding fraction.
YtqI complements an E. coli orn mutant
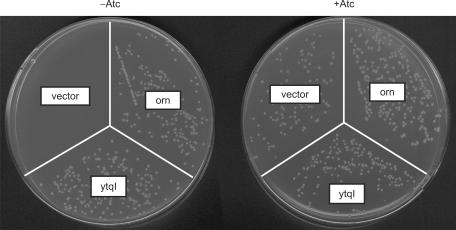
In order to perform complementation experiments, we created a conditional promoter mutant of the essential orn gene in E. coli (Figure 2). This mutant (strain UM341) uses the anhydrotetracycline (Atc)-inducible promoter PLtetO-1 (19) together with a Tet-repressor (TetR) to ensure tight control in the absence of Atc. A growth defect of this mutant was easily observable in cultures lacking Atc grown in LB liquid medium. While growth of the orn mutant carrying a plasmid-borne copy of orn (pUM408) was not affected by the absence of Atc (42 versus 41 min doubling time for cultures minus and plus Atc, respectively), mutants carrying the vector control (pBAD18) had a 1.9-fold longer doubling time when Atc was missing (77 versus 43 min, respectively) and their growth leveled off at an OD600 of ∼0.5. On plates, a similar effect could be observed in the absence of Atc; transformants of strain UM341 with pBAD18 produced pinpoint-sized colonies that stopped growing, while transformants with the orn carrying plasmid were significantly larger after overnight exposure and continued to grow (Figure 3).
Figure 2.
Design of the conditional PLtetO-1 orn mutant. The PLtetO-1 promoter together with a cassette coding for the kanamycin resistance marker (KmR) and the Tet-repressor (TetR) were inserted directly in front of orn. The arrows correspond to the direction of transcription. DH5aZ1 (19) was the source of TetR and terminator T1, and plasmid pZE21-MCS1 (19) was the source of the PLtetO-1, kmR and terminator T0.
Figure 3.
Complementation of the conditional orn mutant by expression of YtqI. Transformants of strain UM341 with pBAD18 (vector control), pUM408 (arabinose-inducible orn) or pUM412 (arabinose-inducible ytqI) were spread on LB plates containing 0.2% arabinose in the presence or absence of anhydrotetracycline (Atc).
C-terminally his-tagged YtqI was expressed under control of the arabinose-inducible Para promoter (plasmid pUM412) for complementation experiments. As seen in Figure 3, ytqI expression completely rescued the growth defect of the orn mutant on plates lacking Atc. Expression was induced by the addition of 0.2% arabinose. Expression levels of YtqI and Orn were similar under these conditions as judged from Coomassie-stained gels (data not shown). Complementation could be seen even in the absence of arabinose. As opposed to the expression level of YtqI in the presence of 0.2% arabinose, in the absence of arabinose expression was not visible on a Coomassie-stained protein gel and was below the amount that could be detected by western blotting using Anti-His6 antibodies (data not shown). We concluded therefore that even low levels of YtqI expression were sufficient for Orn complementation.
YtqI degrades nanoRNA in vitro, with 3-mers being a preferred substrate
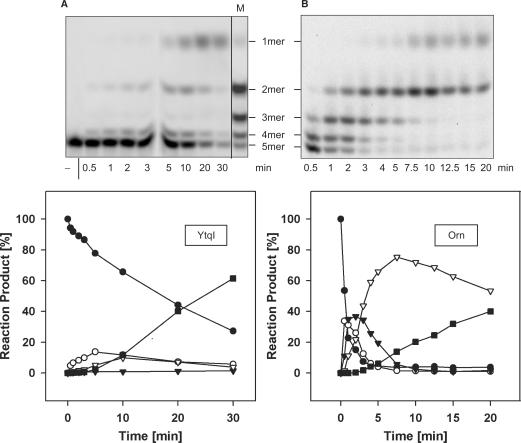
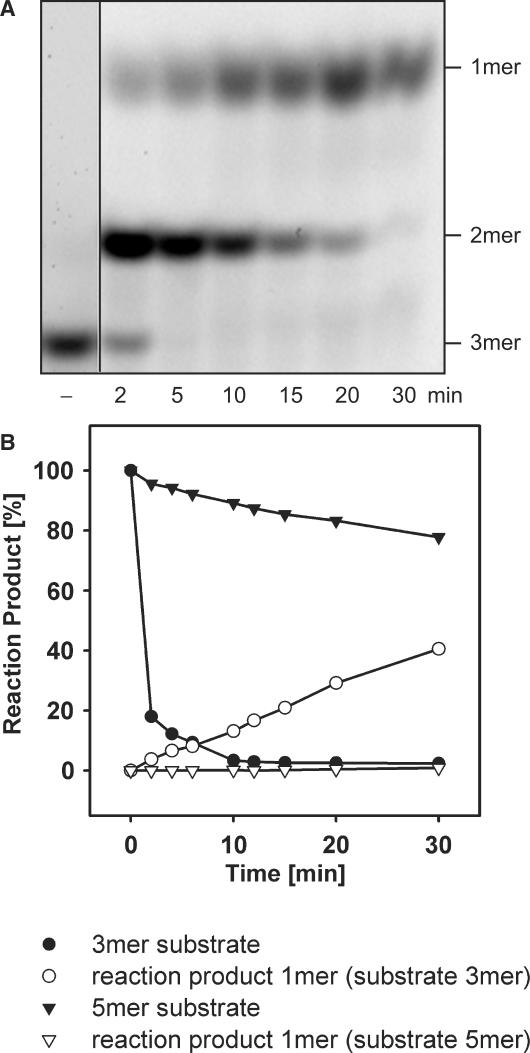
Purified recombinant YtqI was tested for nanoRNase activity. In the presence of manganese, YtqI was able to degrade nanoRNA 5-mers (Figure 4). The activity in the presence of other ions tested (magnesium, zinc and calcium) was negligible (data not shown). Comparing YtqI- and Orn-catalyzed degradation of nanoRNA 5-mers, we noticed significant differences: The amount of YtqI required for appreciable activity was two orders of magnitude higher than that necessary for Orn-catalyzed activity. In addition, the pattern of degradation products as well as the kinetics of this reaction looked very different. Here, 3-mers were virtually missing and other intermediates (2-mers and 4-mers) accumulated less than in Orn-catalyzed hydrolysis (Figure 4). Therefore, we hypothesized that 3-mers might be a preferred substrate for YtqI and as such they might be hydrolyzed so fast that accumulation could not be observed. We tested this hypothesis by comparing degradation of 3-mers and 5-mers (Figures 4 and 5). We used three times more enzyme in the reaction with 5-mers as substrate in order to obtain appreciable conversion into monomers (Figure 4A) as compared to the reaction on 3-mers (Figure 5A). Turnover numbers for 3-mers were one order of magnitude higher than for 5-mers (1.5 versus 0.14 pmol/μg/min). In Figure 5B, we compare the kinetics of the disappearance of different substrates (3-mers or 5-mers) and the appearance of the final reaction product monomers in reactions with equal amounts of YtqI (1.5 μg). These results clearly document that 3-mers were a much better substrate for YtqI than 5-mers. Moreover, it seems that degradation of 3-mers to 2-mer was the fastest step in catalysis as the 2-mers formed here disappeared considerably slower.
Figure 4.
Comparison of YtqI and Orn-catalyzed degradation of nanoRNA 5-mers. Shown are the separation of reaction products on 22% PAA gels (upper panel) and the corresponding quantification (lower panel). Reactions contained 12 μg YtqI (A) or 0.14 μg Orn (B) and 1.5 μM or 2.7 μM RNA 5-mer (5′Cy5-CCCCC3′), respectively. The minus indicates a control lacking enzyme. M specifies a size marker obtained by Orn-catalyzed reaction. Closed circle: 5-mers, open circle: 4-mers, closed triangle: 3-mers, open triangle: 2-mers, square: 1-mers.
Figure 5.
Substrate preference of YtqI for RNA 3-mers. (A) YtqI-catalyzed degradation of RNA 3-mers. A 30 μl reaction contained 3 μg YtqI and 3.5 μM RNA 3-mers (5′Cy5-CCC3′). The minus indicates a control-lacking enzyme. (B) Comparison of YtqI-catalyzed degradation of RNA 5-mers or 3-mers. Shown are the amounts of substrate and reaction product monomer present at times indicated. Numbers indicate fractions of the total amount of fluorescent present in substrate, intermediate and complete reaction products. Reactions contained 2.7 μM substrate RNA 5-mers or 3-mers and 1.5 μg YtqI.
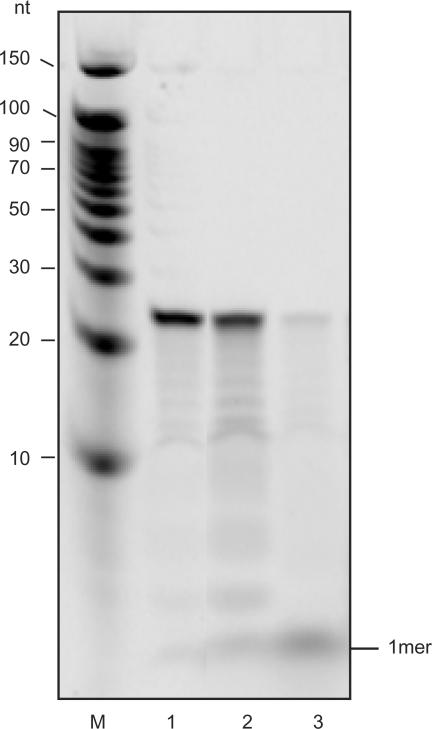
In order to ask whether YtqI degrades specifically nanoRNA or is active on longer substrates as well, we tested degradation of a RNA 24-mer 5′-end labeled with 33P. Figure 6 shows that activity of YtqI on this substrate was insignificant. The YtqI-catalyzed turnover of 24-mers into monomers could be roughly estimated from this experiment as 0.01 pmol/μg/min.
Figure 6.
Activity of YtqI on RNA 24-mers. Reactions containing 5′ 33P-labeled RNA 24-mers (5′CACACACACACACACACACACACA3′) were incubated for 30 min at 37°C. M, decade marker; 1, no enzyme control; 2, YtqI; 3, positive control (YhaM).
Sensitivity of YtqI to pAp
Binding of YtqI to pAp could point to the following possibilities: (i) activity of YtqI is affected by pAp or (ii) pAp can be a substrate for YtqI. We had reported before that Orn-catalyzed degradation of nanoRNA is highly sensitive to pAp (15). Therefore, we decided to test the possibility (i) first. Unlike what we observed with Orn, the addition of small amounts of pAp to the YtqI-catalyzed reaction (10, 20 and 50 μM) did not produce an easily observable effect on degradation of nanoRNA. At 100, 200 or 500 μM pAp the activity of YtqI based on the conversion of 5-mer into monomers in 30 min dropped to 28, 4 and 1%, respectively (data not shown). The effect produced by 500 μM pAp was comparable to the effect seen in the presence of 20 μM pAp in an Orn-catalyzed reaction with 0.07 μg Orn and 3 μM substrate (15).
YtqI is also a pAp-phosphatase
The observed effect of pAp on YtqI-catalyzed degradation of nanoRNA did not exclude the possibility of pAp being a substrate for YtqI. We therefore tested the ability of YtqI to degrade pAp in vitro. Remarkably, YtqI was able to degrade pAp to AMP (data not shown). YtqI converted 6 nmol of pAp/μg/minute. The pAp-degrading activity of YtqI was similar in magnitude to that of CysQ (33 nmol/μg/min), the pAp-phosphatase from E. coli (15). Unlike CysQ activity, pAp-degrading activity of YtqI was not affected by either LiCl or CaCl2 at concentrations of 5 mM (data not shown).
To test if this in vitro activity of YtqI has physiological relevance, we asked whether the expression of ytqI could complement the cysQ mutant phenotype, i.e. the growth impairment of CysQ-lacking cells in the absence of cysteine. Figure 7 shows that complementation could indeed be achieved. Transformants of UM285 (ΔcysQ) with the vector control formed very small colonies when plated on medium lacking cysteine (Figure 7A). In liquid medium, growth of the vector control strain was severely affected in the absence of cysteine (Figure 7B). Transformants of UM285 with a plasmid expressing YtqI (pUM412) or CysQ (pUM404) however formed normal size colonies (Figure 7A). In liquid medium, UM285 strains transformed with plasmids expressing YtqI or CysQ were not affected in their growth when omitting cysteine (Figure 7B). A comparison of expression levels of YtqI and CysQ in the presence of 0.02% arabinose showed that CysQ was expressed at a somewhat higher level than YtqI (data not shown).
The ability of ytqI to complement a cysQ mutant in E. coli, prompted us to investigate the phenotype of an ytqI mutant (BFS66) in B. subtilis. Growth rates of the B. subtilis wild type and BFS66 were compared either in the absence or in the presence of cysteine. Doubling times were similar in the presence of cysteine with 42 and 44 min for wild type and the ytqI mutant, respectively, but varied considerably in the absence of cysteine with 43 versus 68 min. This phenotype resembled that of a cysQ mutant in E. coli. The latter seemed however more pronounced as withdrawal of cysteine affected growth more severely (88 versus 203 min) (Figure 7B).
Phylogenetic distribution of YtqI
Analysis of the phylogenetic distribution of Orn and YtqI (Supplementary Figure S1), clearly demonstrated that the majority of bacterial species possess only one of the two proteins. YtqI was present in Firmicutes, Bacteroidetes, Chlorobi and in the delta subdivision of Proteobacteria. Orn, however, was present in beta and gamma-Proteobacteria and in Actinobacteria. This distribution points to some anti-correlation: the presence of one of the genes seemed to exclude the presence of the second one (26). The two proteins of different origin might therefore exert the same function. Some Actinobacteria were exceptional in that they had both orn and ytqI. Cyanobacteria and alpha-proteobacteria had neither Orn nor YtqI.
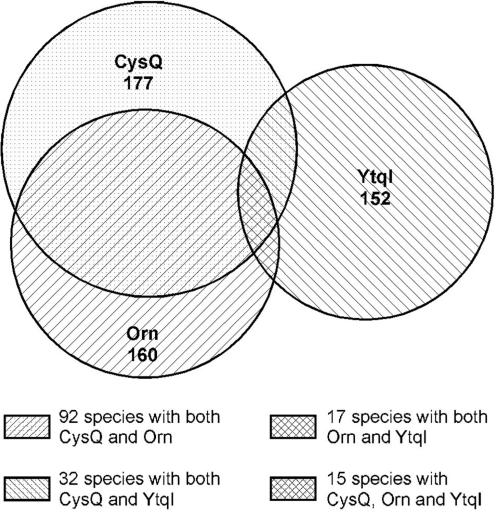
Figure 8 shows the distribution of YtqI, Orn and CysQ in 393 completely sequenced genomes. This figure shows that most organisms that had YtqI, did not have CysQ or Orn. Whereas the overlap between organisms carrying both Orn and CysQ was considerable (58% of the species having Orn have also CysQ), only 21% of the species having YtqI had also CysQ, and only 11% of species having YtqI carried also Orn. This distribution supports our hypothesis that YtqI might fulfill the function of two proteins, Orn and CysQ.
Figure 8.
Distribution of CysQ, Orn and YtqI in bacterial genomes.
Interestingly, while Orn was absent in all sequenced archeal genomes (27), YtqI was represented in 42% of them.
A complete list of genomes investigated and details concerning presence of YtqI, Orn or CysQ orthologs is shown in Table S1.
DISCUSSION
This study was conducted in order to search in the model organism B. subtilis for a functional analog of E. coli oligoribonuclease, Orn. Encouraged by the observation that the pAp-oligoribonuclease interaction is conserved between E. coli and humans, we identified YtqI as potential functional Orn analog through its binding to pAp. The other B. subtilis proteins interacting with pAp, HisIE and GuaC, are of known function and were not the focus of this study. It is however noteworthy that the interaction between HisIE and pAp was observed previously using E. coli extracts (15), which points to biological relevance of this interaction.
YtqI belongs to the DHH family of phosphoesterases, more specifically to the DHHA1 subfamily (25), some members of which are involved in nucleic acid metabolism. It was therefore a good candidate for a functional Orn analog. YtqI can complement a conditional orn mutant in E. coli when expressed at similar levels as Orn. This complementation does not require high amounts of YtqI, as expression levels that are below the detection limit of Anti-His6 antibodies are sufficient. Recombinant YtqI is able to degrade nanoRNA 5-mers in vitro in the presence of manganese. Whereas Orn is essential in E. coli, YtqI is not essential in Bacillus. This points to the existence of at least one more enzyme with the ability to degrade nanoRNA.
The pattern of degradation products on the PAA gel as well as the kinetics of their appearance make it clear that Orn and YtqI employ different mechanisms for the degradation of nanoRNA. 5-mers are not a good substrate for YtqI, they might be degraded in a distributive rather than a processive way. Another obvious difference was the absence of 3-mers from the degradation pattern. One possible explanation for this could be a preferred degradation of 3-mers into 2-mers. When used as substrate, 3-mers are degraded much faster than 5-mers, requiring approximately 10 times less enzyme than 5-mers for complete degradation. We therefore concluded that 3-mers are much better substrates than 5-mers. This in vitro result could reflect the intriguing possibility that YtqI acts preferentially on 3-mers in vivo and cannot efficiently degrade 5-mers. In this case, the fact that YtqI can complement Orn in E. coli could suggest that the accumulation of 3-mers and not 5-mers is the main cause of growth deficiency in E. coli lacking Orn. According to the literature (28,29), 90% of RNA degradation in E. coli is done hydrolytically, implying a more significant contribution of RNase II and RNase R as compared to PNPase. The relative contribution of RNase II and RNase R is under dispute; RNase II was considered to be the main contributor to mRNA degradation (30), but this result was questioned by a genome-wide analysis of mRNA levels in a strain deleted for RNase II (31). The end products of degradation catalyzed by RNase II and RNase R differ slightly in size; for RNase II experimental data indicate 3–5-mers (12) or 4–6-mers (11,32) as final product and 4-mers according to the structural model (13), and RNase R leaves 2–3-mers (12,14) or 1–2-mers (11). The size range of fragments produced by RNase R seems to be more suitable for degradation by YtqI than that of oligonucleotides produced by RNase II. The importance of RNase R is increasingly recognized. RNase R has the ability to degrade stable RNA (12,33) and contributes to quality control of rRNA (33). More recently this enzyme was shown to be involved in the degradation of mRNA substrates with extensive secondary structure (27). In addition, RNase R was shown to increase dramatically under different stress conditions (34–36). Bacillus subtilis harbors only one member of the RNR family of exoribonucleases, RNase R, which seems to be equally important for the degradation of highly structured RNA as its counterpart in E. coli (37). Another requirement for oligonucleotide degradation might also come from systems expressed under the control of cyclic dGMP. Indeed this regulatory molecule is degraded by a phosphodiesterase, which should result in formation of pGpG, a dinucleotide that needs to be further degraded (38).
We previously demonstrated that Orn and Sfn bind pAp, but cannot degrade it, instead pAp is a strong inhibitor of these enzymes (15). YtqI however can degrade pAp in vitro; it also complements a cysQ mutant in E. coli. Both results clearly indicate that YtqI is a pAp-phosphatase. The phenotype of an ytqI mutant in B. subtilis resembles that of an E. coli cysQ deletion: growth is impaired in the absence of cysteine. Withdrawal of cysteine causes doubling times to increase 1.6-or 2.3-fold in an ytqI mutant in B. subtilis or an E. coli cysQ mutant, respectively. The effect of withdrawal of cysteine seems therefore slightly more moderate in B. subtilis lacking YtqI than in E. coli lacking CysQ. In fact, the difference between B. subtilis and E. coli in this respect could be somewhat larger, considering the fact that E. coli strain MG1655 used in our experiments has a rather leaky cysQ mutant phenotype as compared to other strains of E. coli (39). One possible explanation for this interspecies difference could be bispecificity of B. subtilis protein CysH1. This enzyme has the ability to reduce both PAPS and APS (40) in vitro. In addition, expression of B. subtilis cysH1 can complement an E. coli mutant defective for APS kinase encoded by cysC (41). This raises the possibility that APS could be reduced directly in B. subtilis, which would bypass the requirement for PAPS synthesis and thus pAp accumulation could be unnecessary. The direct reduction of APS is commonly used in plants (42) and was documented for some bacteria including Mycobacterium tuberculosis (41), Pseudomonas aeruginosa (43) and Rhizobium meliloti (44).
The existence of a second enzyme able to hydrolyze pAp could be an alternative explanation for the only partial growth defect of the ytqI mutant in the absence of cysteine.
Table 1.
Bacterial strains, plasmids and primers
| Description | Reference | |
|---|---|---|
| Strains | ||
| CF10230 | nic+ derivative of DY329 (20) | Michael Cashel, unpublished data |
| UM285 | As CF10230 but ΔcysQ, kmR | (15) |
| UM341 | As CF10230 but orn under control of P LtetO-1, TetR, kmR | This study |
| DH5αZ1 | tetR under control of PN25, lacI under control of Placiq, SpR | (19) |
| BSF66 | B. subtilis 168, ytqI::pMUTIN2MCS, EmR | (21) |
| DB430 | B. subtilis 168, deficient in multiple proteases | (22) |
| Plasmids | ||
| pBAD18 | Vector, Para, pBR replicon, ApR | (45) |
| pGEM-T Easy | Vector for TA cloning | Promega |
| pZE21-MCS1 | Vector, P LtetO-1, ColE1 replicon, KmR | (19) |
| pUM404 | As pBAD18, CysQ with C-terminal his-tag | (15) |
| pUM408 | As pBAD18, Orn with C-terminal his-tag | (15) |
| pUM412 | As pBAD18, YtqI with C-terminal his-tag | This study |
| Primers | ||
| UM153 | 5′GATTGCGGCCGCGCTTGGATTCTCACCAATAAA3′ | |
| UM154 | 5′GCGTTTTTTATTGGTGAGAATCCAAGCGCGGCCGCAATCATAAAAAA TTTATTTGCTTTCAGGAAAATTT3′ | |
| UM155 | 5′GATACACGCAATTGTGGATCAATATAGTCACTGTGAATGGGTGGAAA ATAGCGTCGACGGCGGATTTGTCCTACTCAGG3′ | |
| UM156 | 5′CCGGTCATCTCAAGATCGATCCAAATCAGGTTGTTTTCATTGGCACTCA TGGTACCTTTCTCCTCTTTAATGAATTCGG3′ | |
| UM158 | 5′TTCCACATCAGCGTGCATAC3′ | |
| UM159 | 5′GCAACGAACAGGTCACTATC3′ | |
| UM160 | 5′CCTGCGCCATCAGATCCTTG3′ | |
| UM161 | 5′TCAGCGCCAGCTGTTCATCA3′ | |
| UM172 | 5′GTGCTGCAAGGCGATTAAGT3′ | |
| UM173 | 5′CGCCAAGCTATTTAGGTGAC3′ | |
| UM175 | 5′GGGGAATTCACCATGAAAACAGAATTGATCAGAACCATATC3′ | |
| UM176 | 5′GGGGCTCGAGCTCGTGTTCTTTACATAATGTTTCCAGATC3′ |
The phylogenetic distribution of YtqI, Orn and CysQ and in particular the anti-correlation is in agreement with the hypothesis that YtqI fulfills the functions of two proteins in E. coli, Orn and CysQ.
Until now, RNase R was the only exoribonuclease known in the small genomes of Mycoplasma species (3). Noteworthy is therefore the presence of YtqI homologs among the small set of proteins of unknown function in the genomes of Mycoplasma genitalium and Mycoplasma pneumoniae, where proteins MG371 and MPN140 respectively, are likely to perform the essential function of nanoRNA degradation.
Different species seem to have found different solutions to the same cellular problem, the problem being the degradation of nanoRNA or pAp. Yet another solution to the problem of nanoRNA degradation awaits to be discovered, as cyanobacteria and the alpha division of proteobacteria have neither YtqI nor Orn orthologs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We wish to thank Dr Hermann Bujard for the gift of the anydrotetracycline tightly regulated system, Dr Michael Cashel for the gift of strain CF10230 Drs Isabelle Martin-Verstraete and Anne-Marie Gilles for helpful discussions. This work was supported by European Union's 6th Framework Program: BioSapiens-Network of Excellence, Grant LSHG CT-2003-503265, section 66010 and by the French Ministry of Research ACI IMPBIO (program Blastsets). Funding to pay the Open Access publication charges for this article was provided by the Institut Pasteur, Unité Génétique des Génomes Bactériens.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hollams EM, Giles KM, Thomson AM, Leedman PJ. MRNA stability and the control of gene expression: implications for human disease. Neurochem. Res. 2002;27:957–980. doi: 10.1023/a:1020992418511. [DOI] [PubMed] [Google Scholar]
- 2.Condon C, Putzer H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002;30:5339–5346. doi: 10.1093/nar/gkf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condon C. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 2003;67:157–174. doi: 10.1128/MMBR.67.2.157-174.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apirion D, Lassar AB. A conditional lethal mutant of Escherichia coli which affects the processing of ribosomal RNA. J. Biol. Chem. 1978;253:1738–1742. [PubMed] [Google Scholar]
- 6.Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl Acad. Sci. USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kushner SR. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. discussion 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta AK, Niyogi K. A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J. Biol. Chem. 1975;250:7313–7319. [PubMed] [Google Scholar]
- 9.Niyogi SK, Datta AK. A novel oligoribonuclease of Escherichia coli. I. Isolation and properties. J. Biol. Chem. 1975;250:7307–7312. [PubMed] [Google Scholar]
- 10.Zhang X, Zhu L, Deutscher MP. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J. Bacteriol. 1998;180:2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amblar M, Barbas A, Gomez-Puertas P, Arraiano CM. The role of the S1 domain in exoribonucleolytic activity: Substrate specificity and multimerization. RNA. 2007;13:317–327. doi: 10.1261/rna.220407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 13.Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 14.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 15.Mechold U, Ogryzko V, Ngo S, Danchin A. Oligoribonuclease is a common downstream target of lithium-induced pAp accumulation in Escherichia coli and human cells. Nucleic Acids Res. 2006;34:2364–2373. doi: 10.1093/nar/gkl247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.York JD, Ponder JW, Majerus PW. Definition of a metal-dependent/Li(+)-inhibited phosphomonoesterase protein family based upon a conserved three-dimensional core structure. Proc. Natl Acad. Sci. USA. 1995;92:5149–5153. doi: 10.1073/pnas.92.11.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hullo MF, Auger S, Soutourina O, Barzu O, Yvon M, Danchin A, Martin-Verstraete I. Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J. Bacteriol. 2007;189:187–197. doi: 10.1128/JB.01273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, et al. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He XS, Shyu YT, Nathoo S, Wong SL, Doi RH. Construction and use of a Bacillus subtilis mutant deficient in multiple protease genes for the expression of eukaryotic genes. Ann. NY Acad. Sci. 1991;646:69–77. doi: 10.1111/j.1749-6632.1991.tb18565.x. [DOI] [PubMed] [Google Scholar]
- 23.Oussenko IA, Sanchez R, Bechhofer DH. Bacillus subtilis YhaM, a member of a new family of 3′-to-5′ exonucleases in gram-positive bacteria. J. Bacteriol. 2002;184:6250–6259. doi: 10.1128/JB.184.22.6250-6259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 25.Aravind L, Koonin EV. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci. 1998;23:17–19. doi: 10.1016/s0968-0004(97)01162-6. [DOI] [PubMed] [Google Scholar]
- 26.Morett E, Korbel JO, Rajan E, Saab-Rincon G, Olvera L, Olvera M, Schmidt S, Snel B, Bork P. Systematic discovery of analogous enzymes in thiamin biosynthesis. Nat. Biotechnol. 2003;21:790–795. doi: 10.1038/nbt834. [DOI] [PubMed] [Google Scholar]
- 27.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol. Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 28.Chaney SG, Boyer PD. Incorporation of water oxygens into intracellular nucleotides and RNA. II. Predominantly hydrolytic RNA turnover in Escherichia coli. J. Mol. Biol. 1972;64:581–591. doi: 10.1016/0022-2836(72)90084-8. [DOI] [PubMed] [Google Scholar]
- 29.Deutscher MP, Reuven NB. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc. Natl Acad. Sci. USA. 1991;88:3277–3280. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donovan WP, Kushner SR. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl Acad. Sci. USA. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- 32.Amblar M, Barbas A, Fialho AM, Arraiano CM. Characterization of the functional domains of Escherichia coli RNase II. J. Mol. Biol. 2006;360:921–933. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 33.Cheng ZF, Deutscher MP. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl Acad. Sci. USA. 2003;100:6388–6393. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrade JM, Cairrao F, Arraiano CM. RNase R affects gene expression in stationary phase: regulation of ompA. Mol. Microbiol. 2006;60:219–228. doi: 10.1111/j.1365-2958.2006.05092.x. [DOI] [PubMed] [Google Scholar]
- 35.Cairrao F, Cruz A, Mori H, Arraiano CM. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 2003;50:1349–1360. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Deutscher MP. Elevation of RNase R in response to multiple stress conditions. J. Biol. Chem. 2005;280:34393–34396. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- 37.Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH. Participation of 3'-to-5' exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 39.Neuwald AF, Krishnan BR, Brikun I, Kulakauskas S, Suziedelis K, Tomcsanyi T, Leyh TS, Berg DE. cysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J. Bacteriol. 1992;174:415–425. doi: 10.1128/jb.174.2.415-425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berndt C, Lillig CH, Wollenberg M, Bill E, Mansilla MC, de Mendoza D, Seidler A, Schwenn JD. Characterization and reconstitution of a 4Fe-4S adenylyl sulfate/phosphoadenylyl sulfate reductase from Bacillus subtilis. J. Biol. Chem. 2004;279:7850–7855. doi: 10.1074/jbc.M309332200. [DOI] [PubMed] [Google Scholar]
- 41.Williams SJ, Senaratne RH, Mougous JD, Riley LW, Bertozzi CR. 5′-adenosinephosphosulfate lies at a metabolic branch point in mycobacteria. J. Biol. Chem. 2002;277:32606–32615. doi: 10.1074/jbc.M204613200. [DOI] [PubMed] [Google Scholar]
- 42.Bick JA, Leustek T. Plant sulfur metabolism – the reduction of sulfate to sulfite. Curr. Opin. Plant Biol. 1998;1:240–244. doi: 10.1016/s1369-5266(98)80111-8. [DOI] [PubMed] [Google Scholar]
- 43.Bick JA, Dennis JJ, Zylstra GJ, Nowack J, Leustek T. Identification of a new class of 5'-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria. J. Bacteriol. 2000;182:135–142. doi: 10.1128/jb.182.1.135-142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abola AP, Willits MG, Wang RC, Long SR. Reduction of adenosine-5′-phosphosulfate instead of 3′-phosphoadenosine-5′-phosphosulfate in cysteine biosynthesis by Rhizobium meliloti and other members of the family Rhizobiaceae. J. Bacteriol. 1999;181:5280–5287. doi: 10.1128/jb.181.17.5280-5287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.