Abstract
Transcription and pre-mRNA splicing are the key nuclear processes in eukaryotic gene expression, and identification of factors common to both processes has suggested that they are functionally coordinated. p100 protein has been shown to function as a transcriptional co-activator for several transcription factors. p100 consists of staphylococcal nuclease (SN)-like and Tudor-SN (TSN) domains of which the SN-like domains have been shown to function in transcription, but the function of TSN domain has remained elusive. Here we identified interaction between p100 and small nuclear ribonucleoproteins (snRNP) that function in pre-mRNA splicing. The TSN domain of p100 specifically interacts with components of the U5 snRNP, but also with the other spliceosomal snRNPs. In vitro splicing assays revealed that the purified p100, and specifically the TSN domain of p100, accelerates the kinetics of the spliceosome assembly, particularly the formation of complex A, and the transition from complex A to B. Consistently, the p100 protein, as well as the separated TSN domain, enhanced the kinetics of the first step of splicing in an in vitro splicing assay in dose-dependent manner. Thus our results suggest that p100 protein is a novel dual function regulator of gene expression that participates via distinct domains in both transcription and splicing.
INTRODUCTION
Transcription and pre-mRNA splicing are the two key nuclear steps regulating gene expression in eukaryotes. Both transcription and splicing are highly ordered processes involving intricate protein–protein interactions. Transcription is regulated by protein complex composed of transcription factors, basal transcriptional machinery and a group of co-activators (1), while pre-mRNA splicing is carried out by spliceosome that consists of five conserved small nuclear ribonucleoprotein particles (snRNPs) U1, U2, U5, base-paired U4/U6 and a large number of less precisely defined non-snRNP proteins (2).
Accumulating evidence supports the concept that transcription and splicing are coordinated (3–5). The key factor linking these processes is the C-terminal domain (CTD) of pol II large subunit which plays essential role both in transcription and RNA processing (6,7). Additionally, CTD associates with general RNA processing machineries, functioning in mRNA capping, pre-mRNA splicing, polyadenylation and alternative splicing (8–10). However, recent evidence supports the idea that CTD is required for efficient co-transcriptional editing of some pre-mRNA, but is not required for efficient splicing of the pre-mRNA (11). Thus, the CTD is most likely a coordinator, rather than a simple enhancer of RNA processing. Additional evidence supporting the coupling between transcription and polyadenylation is emerging from recent proteomic analyses of the human spliceosome which revealed that at least 30 spliceosomal proteins are also participating in other gene expression steps besides splicing (12–14). For example, transcription cofactor such as TAT-SF1 (TAT specific factor 1) (15), CA150 (16) and SKIP (Ski-binding protein) (17) have also been identified in the spliceosome. These results imply that gene expression machineries are extensively coupled via protein–protein interactions. However, although the evidence of CTD recruitment of mRNA processing factors is well documented, the interactions between CTD and other proteins that link transcription and RNA processing have been poorly characterized. Hence, identification of functional protein complexes and the shared protein components between them is an essential step in understanding the physiological mechanisms that couple transcription and RNA processing.
p100 protein was first identified as a co-activator of EBNA2 (Epstein–Barr virus nuclear protein 2) (18), and subsequently discovered as co-regulator of pim-1 (19), and STAT6 transcription factor in IL-4 mediated gene regulation (20,21). p100 functions also as a co-activator of STAT5 in prolactin (PRL) signaling (22), and in mammary epithelial cells p100 protein levels increase in response to PRL during lactation and correlate with induction of β-casein gene expression (23). In addition, p100 has been linked to the pathogenesis of autosomal-dominant polycystic kidney disease (ADPKD) (24), and identified in the RISC (RNA-induced silencing complex) (25). These studies suggest that p100 protein participates in several biological responses and that the protein may play distinct roles in various cellular events.
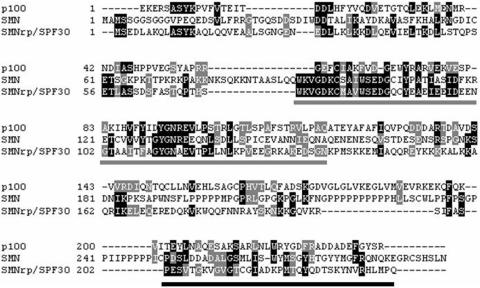
p100 protein consists of four similar domains with homology to the staphylococcal nucleases (SN), followed by a C-terminal Tudor-SN (TSN) domain (26). The SN-like domains of p100 have been implicated in protein interactions, and the SN-like domains of p100 protein recruit CBP and RNA pol II to STAT6 and facilitate the formation of STAT6 enhanceosome (20). The function of p100 TSN domain has remained elusive, but the Tudor domain is 30% homologous to SMN (survival of motor neuron protein) Tudor domain, which functions in the assembly of snRNP complexes and pre-mRNA splicing process (27,28; Figure 6).
Figure 6.
Sequence alignment of the human SPF30, SMN and p100 proteins. Identical and similar amino acids are indicated in black and light shading, respectively. The positions of the Tudor domain in SPF30, SMN and p100 proteins are indicated with gray bars. The positions of Sm-protein-binding domains of SPF30 and SMN are indicated with black bars.
In this report, we demonstrate a novel function for p100 protein as an interaction partner for spliceosomal snRNPs. First, we used an in vitro pull-down assay to demonstrate that TSN domain of p100 protein associates with a subset of U5 snRNP core proteins, and is capable of precipitating all snRNAs. We next showed that exogenously added p100 protein can kinetically enhance both the in vitro spliceosome complex formation and the first step of pre-mRNA splicing. Thus our results suggest that human p100 protein is a novel dual function regulator of both gene transcription and pre-mRNA splicing.
MATERIALS AND METHODS
Plasmids and cells
HeLa cells and COS-7 cells were cultured as described previously (21). The HeLa-p100-Flag stable cell lines were cloned as described previously(21). Plasmid encoding AdML pre-mRNA was kindly provided by Dr R. Reed (14). The pSG5 vector expression plasmids containing full-length p100 tagged with Flag sequence, GST-p100-TSN and GST-p100-SN were generated as previously described (21). GST-SMN-Tudor was generated by cloning PCR products corresponding to amino acids 92–156 of SMN and inserted into pGEXT-4T-1 with EcoRI and NotI. pBluescript plasmids containing different domains of U5-220, which could be used for in vitro translation, were constructed by cloning PCR products corresponding to amino acids 1–209 (domain 1), 303–668 (domain 2), 673–907 (domain 3), 914–1170 (domain 4), 1167–1698 (domain 5), 1697–2121 (domain 7), 2040–2332 (domain 7). All PCR products were sequenced.
GST pull-down assays
GST pull-down experiments were performed as described previously(21). GST fusion proteins were bound on glutathione-Sepharose 4B beads (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK) and incubated with HeLa cell nuclear lysates or in vitro-translated 35S-labeled different domains of U5-220 in binding buffer (12.5 mM HEPES pH 7.4, 0.1 mM EDTA, 0.05% NP-40, 1 mM DTT, 2 mg/ml aprotinin, 0.5% bovine serum albumin). The beads were washed with buffer containing 300 mM NaCl, and then separated by SDS–PAGE and visualized by silver staining, Coomassie blue staining or autoradiography.
Mass spectrometry
For mass spectrometric analysis, in order to avoid the contamination of GST fusion protein, the precipitated proteins were dissociated from bead-bound GST-p100-TSN fusion protein by incubating in the elution buffer containing 2 M NaCl, and then concentrated and desalted with Microcon column (cut-off 30 kDa, Millipore Corporation, Bedford, MA, USA) by centrifugation at 13 000 r.p.m. for 30 min. The precipitated proteins were separated by SDS–PAGE and visualized by silver staining or Coomassie blue staining. The bands corresponding to the 220-kDa, 200-kDa and 116-kDa proteins were cut out from Coomassie blue-stained gel and subjected to trypsin digestion as described previously (21). The molecular masses of the peptide mixtures were determined by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. The molecular masses of the tryptic peptides of the 220 kDa, 200 kDa and 116 kDa proteins were used to search the OWL protein sequence database for candidate proteins using the ProFound program.
Immunoprecipitation
The nuclear extracts of p100 stable HeLa cells were immunoprecipitated with anti-Prp8 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Flag M2 agarose (Sigma, St Louis, MO, USA), anti-IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA), or anti-TMG (trimethyl-guanosine) monoclonal antibody cross-linked to agarose beads (Calbiochem, San Diego, USA). The bound proteins were subjected to SDS––PAGE and blotted with anti-Prp8 antibody, anti-Flag M2 antibody or anti-p100 antibody (rabbit polyclonal sera were raised against TSN domain of p100 protein).
Protein purification
COS-7 cells were transfected with pSG5-p100-Flag as previously described (21). Transfected COS-7 cells were suspended in Nonidet P-40 lysis buffer (50 mM Tris–HCl, pH 7.6, 300 mM NaCl, 0.1 mM EDTA, 0.5% Nonidet P-40, 20% glycerol, 0.1 mM sodium orthovanadate, 1 mM sodium butyrate). The total cell lysates were incubated with mouse monoclonal anti-Flag M2 agarose at 4°C for 2 h with head-over-tail rotation. After washing with stringent buffer (containing 700 mM NaCl), the purified p100 proteins (P-p100) were eluted from the agarose with Flag peptide (100 μg/ml) (Sigma, St Louis, MO, USA), and concentrated with Microcon column. The purity of P-p100 was confirmed by SDS–PAGE and silver staining.
GST-p100-TSN and GST-p100-SN fusion proteins were bound on glutathione- Sepharose 4B beads. After washing, the p100-SN or p100-TSN proteins were cleaved from the GST with thrombin (Sigma, St Louis, MO, USA), and concentrated with Microcon column. The GST proteins were detached from the beads with reduced glutathione (10 mM) (Sigma, St Louis, MO, USA). The purity of GST, p100-SN and p100-TSN was detected by SDS–PAGE and silver staining.
In vitro splicing assays and spliceosome analysis
Capped adenovirus splicing substrates that were uniformly labeled with 32P-UTP were produced by in vitro transcription with T7 RNA polymerase (Promega, Madison, WI, USA) using plasmid pAdML linearized with HindIII as template (14). The labeled RNAs were purified in denaturing polyacrylamide gel before use. The splicing reactions, containing 40% (v/v) HeLa nuclear extracts, 2 mM MgCl2, 10 mM DTT, 20 mM creatine phosphate, 2 mM ATP, were supplemented with various recombinant proteins and pre-incubated at 30°C for 10 min, followed by the addition of 10 000 c.p.m. pre-mRNA. After incubating at 30°C for different time points, the reactions were stopped by addition of 150 µl PK buffer (20 mM Tris, 10 mM EDTA, 300 mM NaCl, 4.5 mg/ml proteinase K, 0.5% SDS and 0.14 mg/ml glycogen), and the incubation was continued for 30 min at the same temperature (29). Reactions were extracted with phenol and the RNA was recovered by ethanol precipitation. Subsequently, RNA pellets were resolved in gel loading buffer containing 7.5 M urea, denatured at 95°C for 5 min, chilled on ice before loading on an 8% denatured PAGE with 7 M urea. The gels were visualized by phosphoimager (Fuji FLA-5010).
Native gel analysis on 4% acrylamide was performed as described before (30) and visualized by phosphorimager and autoradiography. Heparin (1 mg/ml final concentration) was added to the splicing reactions prior to loading.
RNA extractions and northern blotting
The nuclear extracts of HeLa cells that displayed stable expression of p100 were immunoprecipitated with anti-IgG or anti-TMG monoclonal antibody cross-linked to agarose beads. After washing, the beads were incubated with 300 µl PK buffer at 65°C for 60 min. After extraction with phenol/chloroform, RNA was precipitated in ethanol. The bead-bound GST fusion proteins were incubated with HeLa cell nuclear lysates (100 mM or 300 mM NaCl), the precipitated RNA was extracted as described above. The RNA was separated on denatured 6% PAGE gels, transferred to a nylon filter with semi-dry blotter (Owl Scientific, Woburn, MA, USA) in 0.5X Tris–EDTA buffer using a constant 3 mA/cm2 current for 1.5–2 h, followed by cross-linking with stratalinker (Stratagene, La Jolla, CA, USA).
Hybridization condition for snRNA blots has been described previously (31). Radiolabeled probes of U1 snRNA, U2 snRNA, U4 snRNA, U5 snRNA and U6 snRNA, were made by in vitro transcription of the linearized snRNA plasmids. U7 and 7SK probes were made by PCR and labeled with 32P-UTP as described before (31).
RESULTS
Identification of U5 snRNP specific proteins as interacting proteins of p100
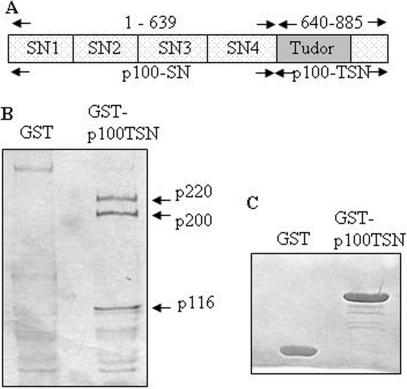
Human p100 protein is composed of four repeats of SN-like domains followed by a C-terminal TSN domain (Figure 1A). The SN consists of two subdomains. The first subdomain belongs to the large oligonucleotide/oligosaccharidebinding (OB)-fold superfamily (26), and the second subdomain consists of two independently folded α-helices. The TSN domain is a hybrid SN domain, in which the OB-domain is divided by a domain found in Drosophila melanogaster Tudor protein. According to the crystal structure of p100 TSN domain (Shaw,N. et al., submitted for publication), the Tudor domain is flanked by two segments of SN, thus we refer it as TSN domain instead, to differentiate it from the bona fide Tudor domains.
Figure 1.
Interaction between p100 protein and U5 snRNP specific proteins. (A) The schematic structure of p100 protein (B) Pulldown of U5 specific proteins using TSN domain of p100. Aliquots of nuclear lysates from HeLa cells were incubated with either GST alone or with GST-p100-TSN. The bound proteins were subjected to SDS–PAGE and visualized by Coomassie blue or silver staining and identified by mass spectrometry. (C) The loading control of GST and GST-p100-TSN proteins.
In order to identify p100 TSN domain interacting proteins, a pull-down assay with GST-p100-TSN fusion protein was carried out. Equal amounts of GST or GST-p100-TSN fusion proteins were bound to glutathione coupled beads and incubated with nuclear extracts of HeLa cells. The precipitated proteins were prepared as described above. Due to the relatively large cut-off (30 kDa) of the Microcon columns and the high salt concentration used in the elution, many of the larger complexes were expected to be dissociated, and the smaller protein components of such complexes were most likely lost during the centrifugation step. As shown in Figure 1B, several proteins were found to specifically interact with TSN domain of p100 protein. The bands corresponding to 220 kDa, 200 kDa and 116 kDa proteins were recovered and subjected to in-gel trypsin digestion. The molecular masses of the digested peptides were analyzed by MALDI-TOF mass spectrometry. The program ProFound was used to compare the mass maps obtained against theoretical tryptic peptide mass maps in the OWL protein sequence database. The comparison resulted in the identification of a group of U5 snRNP specific proteins: p220 is human U5-220 (splicing factor Prp8 in yeast), p200 is human U5-200 (DEXH box RNA helicase, Brr2 in yeast), p116 is human U5-116 (a putative GTPase homologous to the ribosomal elongation factor EF-2, Snu114 in yeast).
p100 protein can interact with snRNPs complexes in vivo and in vitro
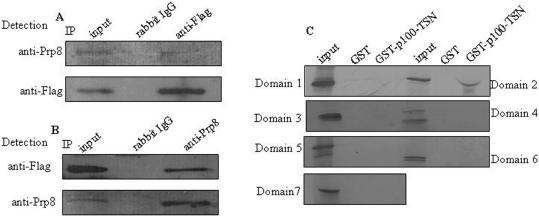
To analyze the possible in vivo association of p100 protein and U5-220, co-immunoprecipitation experiments were carried out in HeLa cells which stably express Flag-tagged human p100 protein (HeLa-p100). The nuclear extracts of these cells were immunoprecipitated with anti-Flag agarose, or rabbit anti-IgG antibody as a control. The precipitated proteins were separated by SDS–PAGE and blotted with anti-Flag or anti-Prp8 antibodies. As shown in Figure 2A, anti-Flag antibodies, but not IgG control, precipitated the endogenous U5-220.
Figure 2.
p100 protein associates with U5-220 in vivo. (A) The nuclear extracts of HeLa-p100 cells were immunoprecipitated with anti-Flag agarose or anti-IgG as control. The bound proteins were subjected to SDS–PAGE and blotted with anti-Prp8 antibody (upper panel) or anti-Flag antibody (lower panel). (B) Alternately, the nuclear extract of HeLa-p100 cells were immunoprecipitated with anti-Prp8 or anti-IgG as control. The bound proteins were subjected to SDS–PAGE and blotted with anti-Flag (upper panel) or anti-Prp8 antibody (lower panel). The input is 15% of the lysate for IP reaction. (C) TSN domain of p100 protein interacts with U5-220 in vitro. Different domains of U5-220 were 35S-labeled by in vitro translation and incubated with beads loaded with GST-p100-TSN or GST. The bound proteins were subjected to 8% SDS–PAGE and visualized by autoradiography. Twenty percent of the in vitro-translated protein was included as a control. The domain 2 has similar molecular weight as the GST-p100-TSN; due to large amount of GST-p100-TSN protein present in the lane, the migration of the domain 2 is somewhat aberrant.
In vivo complex formation was confirmed by a reciprocal experiment in which the nuclear extracts of HeLa-p100 cells were immunoprecipitated with anti-Prp8 antibody, or rabbit anti-IgG antibody as a control, and precipitated proteins were detected by blotting with anti-Prp8 antibody or anti-Flag antibody. As shown in Figure 2B, U5-220 but not IgG control, precipitated the p100-Flag. These results demonstrate that p100 protein and U5-220 are associated, either directly or indirectly, in vivo. We did not have access to antibodies specific to Brr2 or Snu114, and therefore, we were not able to confirm the in vivo association of p100 protein and the other two U5 snRNP specific proteins identified in our mass spectrometry analysis.
In order to further verify the interaction of p100 protein and U5-220, GST pull-down assay was performed by incubating the in vitro-translated different domains of U5-220 with beads-bound GST-p100-TSN fusion protein, or GST as control. As shown in Figure 2C, domain 2 (aa 303–668) of U5-220 interacted with TSN domain of p100, but not with GST control. Meanwhile, other domains of U5-220 did not associate with either GST alone, or GST-p100-TSN fusion protein. These results demonstrated that p100 protein interacts with U5-220 protein both in vivo and in vitro.
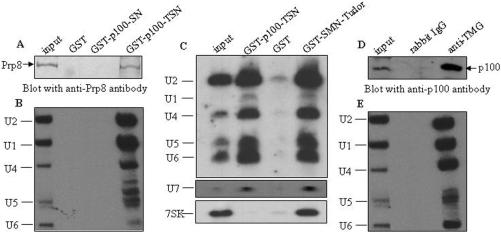
To ask if the other snRNPs are the targets of the TSN domain of p100, GST pull-down assays were performed with GST-p100-TSN and HeLa nuclear lysates. After pulldown, the beads were washed in the buffer containing 300 mM NaCl, followed by both western blotting analysis (with anti-Prp8) and northern blot analysis using snRNA-probes specific to the major U2-dependent spliceosome (U1, U2, U4, U5 and U6 snRNAs), U7 snRNA functioning in histone mRNA processing, or 7SK snRNA, a component of the transcription elongation factor P-TEFb.
Consistent with the original pulldown and proteomic analysis, the western blotting results showed that the GST-p100-TSN precipitated U5-220, while GST-p100-SN or GST protein alone did not (Figure 3A). Additionally, northern analysis of the pull-down reactions using spliceosomal snRNA-specific probes and U7-specific probe revealed, similarly to the control SMN pull-down, an efficient pull-down with the GST-p100-TSN fusion protein (Figure 3C). In contrast, no interaction was observed with the 7SK, which is another nuclear snRNP. This suggests that the common features of spliceosomal U-snRNPs and U7 snRNP, Sm-proteins and trimethylated cap structure may be recognized by p100-TSN domain, because both features are missing from 7SK RNP. U6 snRNP lacks these features as well, but its precipitation is promoted by base-pairing with U4.
Figure 3.
p100 protein associates with snRNP via TSN domain. (A) TSN domain of p100 protein interacts with a complex containing U5-220. Nuclear lysate of HeLa cells were incubated with either GST alone, GST-p100-SN or GST-p100-TSN fusion proteins. The bound proteins were resolved in SDS–PAGE and immunoblotted with anti-Prp8 antibody. The input is 10% of lysate. (B) The precipitated RNAs were analyzed by northern blotting with U1, U2, U4, U5 and U6 snRNA probes, and then visualized by autoradiography. The positions of different snRNAs are indicated on the left. The input is 5% of extracted snRNA. (C) In a parallel experiment, the GST-p100-TSN and GST-SMN-Tudor (as positive control) protein precipitated RNAs were analyzed by northern blotting with both U2-spliceosome and non-spliceosome (U7 and 7SK) snRNA probes. The positions of different snRNAs are indicated on the left. The input is 5% of extracted snRNA. (D) p100 protein is precipitated with anti-TMG antibody in vivo. The nuclear extracts of HeLa-p100 cells were immunoprecipitated with anti-TMG antibody or anti-IgG as control. The immunoprecipitates were separated by SDS–PAGE, immunoblotted with anti-p100 antibody. The input is 10% of lysate. (E) The precipitated RNAs were analyzed as in panel B. The input is 5% of extracted snRNA.
To further validate the association of p100 protein with snRNPs, the nuclear extracts of HeLa-p100 cells were immunoprecipitated with anti-trimethylguanosine (TMG) cap antibody (32), or rabbit anti-IgG antibody as control using the same salt concentration as above (300 mM NaCl). The TMG-cap is present in U1, U2, U4 and U5 snRNAs (33), while the U6 snRNA contains a monomethyl cap instead, but is base-paired in vivo with the U4 snRNA which contains a TMG-cap. Northern analysis revealed that U1, U2, U4, U5 and U6 snRNAs were all present in the anti-TMG immunoprecipitates, but not in the anti-IgG control sample (Figure 3E). Additionally, western blot analysis of the anti-TMG-cap immunoprecipitates indicated that the p100 protein was co-immunoprecipitated with anti-TMG-cap antibody, but not with the anti-IgG antibody (Figure 3D). The efficient co-immunoprecipitation of p100 with TMG antibody indicates that the TMG-cap structure itself is not a target for p100. Together, these results suggest that the p100 protein can interact with both U5-specific proteins and with Sm-proteins.
p100 protein accelerates the spliceosome assembly and pre-mRNA splicing process in vitro
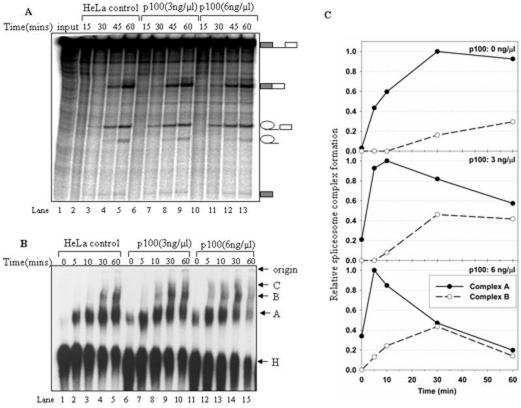
In order to investigate the functional significance of p100-snRNP interaction, we tested the effect of the p100 on in vitro splicing using splicing substrate (pre-mRNA) derived from the adenovirus major late transcription unit (AdML). Splicing reactions were either pre-incubated with purified p100 protein or mock-treated, followed by the addition of radiolabeled splicing substrate and subsequent time-course analysis of the splicing reactions. The results in Figure 4A show that the appearance of the ligated mRNA and exon-lariat intermediate is faster and respond in a dose-dependent manner (lanes 6–8, 10–12) as compared to the reactions containing no exogenously added p100 protein (lanes 2–4). In particular, the first detectable signal from ligated mRNA and exon-lariat intermediate were consistently observed at earlier time point in splicing reactions containing p100. For example, ligated mRNA appeared at 15 min time point with 6-ng/μl p100 protein (lane 10), and at 30 min with 3-ng/μl p100 protein (lane 7), while it was hardly detectable at 30 min in the control (lane 3). Similar trend is seen with ligated exons, which have reached their maximal intensity already at 45 min time point while the control reaction has not (lanes 8 and 12 compare with lane 4). Significantly, there were no major differences in the levels of splicing intermediates and products at the terminal 60-min time point (lanes 5, 9 and 13) or after (data not shown), suggesting that the added p100 does not increase the overall level of in vitro splicing, but rather accelerates the kinetics of the splicing reaction. As the second step is not markedly influenced, that is, the amount of excised intron was quantitatively similar in the splicing reactions with or without addition of p100 protein (compare lanes 4, 8 and 12, lanes 5, 9 and 13), the results suggest that either first step, or the assembly steps prior to catalysis were affected. Control reactions with added BSA did not show any stimulatory effect on splicing, thus ruling out possible effects of overall protein concentration (data not shown).
Figure 4.
Effect of p100 protein on the spliceosome assembly and splicing activity. In vitro splicing or spliceosome complex assembly reactions were carried out with AdML splicing substrate in condition indicated above each lane. (A) The kinetics of the splicing reaction following the addition of purified p100 protein. HeLa cell nuclear extracts were first incubated for 20 min on ice in the presence protein p100 storage buffer (lanes 2–5), or different concentrations (3 ng/μl, 6 ng/μl) of p100 protein (lanes 6–13), followed by an addition of ATP and creatine phosphate and a further incubation at 30°C for 10 min. Subsequently, α-32P-labeled AdML pre-mRNA was added, and the incubation continued at 30°C for the time indicated above each lane. RNA products were separated by electrophoresis in denaturing PAGE gels, and visualized by autoradiography. Splicing intermediates and products are indicated on the left. (B) The effect of p100 protein on spliceosomal complex formation in vitro. Native gel analysis of spliceosome complexes formation in a mock-treated splicing reaction (lanes 1–5), or reactions pre-incubated with increasing concentrations (3 ng/μl, 6 ng/μl) of purified p100 protein (lanes 6–15). The gel was visualized by autoradiography. The bands corresponding to the H, A, B and C complexes as well as the gel origin are indicated on the right. (C) Quantitative analysis of spliceosome formation. Native gels were quantified by phosphoimager and normalized by setting the highest A complex value observed within each time series to 1.0.
To examine the possibility that the assembly steps prior to the catalysis were affected by p100, we analyzed the formation of pre-spliceosomal complex A, fully assembled spliceosomal complex B, and the catalytically active late spliceosomal complex C using native gel electrophoresis with AdML substrate. Figure 4B shows a time-course analysis of spliceosomal complexes using different amounts of p100 protein. The reactions were either kept on ice (0-min time point), or incubated at 30°C up to 60 min prior to gel analysis. In the control reaction (Figure 4B, lanes 1–5) the non-specific complex H is first detected at 0 min time point, complex A at 5 min time point and the later B and C complexes after 30 min of incubation. In the presence of p100 protein, we reproducibly observed relatively strong complex A formation in reactions on ice (lanes 1, 6 and 11) in a p100 protein dose-dependent manner. Additionally, in those reactions the complex A band has reached its maximal intensity already at 5 min time point. Furthermore, the kinetic appearance of B and C complexes was also accelerated. The complex B could be detected already at 5 min time point in the reaction with 6-ng/μl p100 protein (lane 12), and at 10 min (lane 8) with 3-ng/μl p100 protein, while at 30 min (lane 4) in the control reaction. Finally, there appears to be a faster turnover of the splicing reaction particularly with the higher concentration of the p100 as the level of complexes detected in the native gel drops significantly after 60 min of incubation (compare lanes 5, 10 and 15). Furthermore, the stimulation of the complex formation appears to be dose-dependent as the complex formation is faster in the reaction containing a large amount of the p100 protein (compare reactions 6–10 and 11–15), which is consistent with the results observed in splicing reactions (Figure 4A). To validate this point further, we determined the intensities of A and B complex from Figure 4B using a phosphoimager and normalized each set by setting the highest value of complex A to 1. As shown in Figure 4C, the addition of p100 protein accelerated the kinetics of complex A and B formation, the transition of complex A to B (at 30 min in control, 10 min with 3 ng/μl and 5 min with 6 ng/μl p100 protein), and also the decay at later time points.
To rule out possible stimulatory effects of increased protein concentration, similar native gel analyses were carried out using equivalent amounts of control proteins, such as GST (Figure 5) and BSA, neither of which changed the kinetics of the complex formation (data not shown).
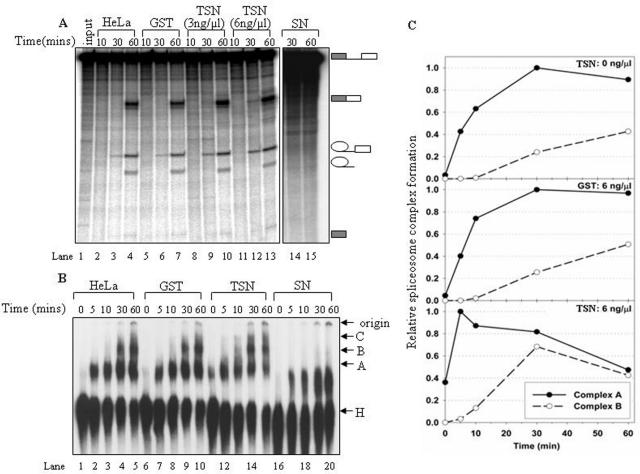
Figure 5.
Effect of p100 TSN domain on the spliceosome assembly and splicing activity. Nuclear extract pre-treatment and the splicing reactions were done essentially as in Figure 4. (A) The kinetics of the AdML in vitro splicing in the presence of p100 storage buffer (lanes 2–4), 3 ng/μl of GST (lanes 5–7), 3 ng/μl p100-TSN (lanes 8–10), 6 ng/μl p100-TSN (lanes 11–13) or 3 ng/μl of p100-SN protein (lanes 14 and 15). Splicing intermediates and products are indicated on the left. (B) Native gel analysis of spliceosome complexes. The nuclear extracts were mock-treated (lanes 1–5), or pre-incubated with 3 ng/μl of GST (lanes 6–10), p100-TSN (lanes 11–15) or p100-SN (lanes 16–20), respectively. The bands corresponding to the H, A, B and C complexes as well as the gel origin are indicated on the right. (C) Quantitative analysis of spliceosome formation similarly as in Figure 4C.
TSN domain of p100 protein participates in the pre-mRNA processing
To identify the domains of p100 protein responsible for the observed stimulation of splicing kinetics we performed in vitro splicing and spliceosome complex analysis using the separated TSN and the SN domains of the p100 protein. As shown in Figure 5A, addition of the recombinant TSN domain resulted in clear accumulation of final product and lariat intermediate of RNA already after 30 min reaction (lanes 9 and 10, 12 and 13). Particularly the lariat intermediate band is 2 to 3.5-fold (in lanes 9 and 12, respectively) more intense than the corresponding bands in lanes 3 and 6. The GST control (lanes 5–7) did not display any differences compared to the reaction without any additional protein (lanes 2–4). Additionally, the isolated TSN-domain displays similar dose-dependent stimulation of the splicing kinetics as the full-length p100 protein (compare lanes 8–10 and 11–13). At 30 min time point (lanes 3, 6, 9 and 12), the amount of ligated mRNA, especially the exon-lariat intermediates were increased with the addition of TSN protein. Surprisingly, the addition of recombinant SN domain of p100 protein led to a total block of the splicing reaction (lanes 14 and 15). Taken together, the splicing analysis indicates that the TSN domain alone is sufficient to accelerate the kinetics of the splicing reaction.
These results were confirmed with the native gel analysis. Consistent with the splicing result, the native gel analysis demonstrated that TSN domain alone has a similar function in splicing as the full-length p100 protein, that is, it accelerated the kinetics of the spliceosomal complex formation (Figure 5B lanes 11–15, Figure 5C), while the GST control had no effect on the pattern of spliceosome assembly (Figure 5B, lanes 6–10). In particular, in reactions containing 6-ng/μl GST-TSN domain the A-complex reached it's maximal intensity after 5 min incubation (Figure 5B, lane 12; 5C bottom panel) while in control reactions, the maximal intensity appeared after 30 min incubation (Figure 5B, lanes 4 and 9; Figure 5C, top and middle panels). In contrast, addition of the recombinant SN domain blocked the formation of all the specific complexes, leading to a formation of an unknown complex with an intermediate mobility between H and A complexes (lanes 16–20). This result is consistent with the observation above (Figure 5A), which indicated that addition of the SN-domain alone caused a complete inhibition of splicing, but at present we do not know if it has any biological significance.
DISCUSSION
The p100 protein is an evolutionarily conserved protein present in the genomes of various eukaryotes from fungi to vertebrates (for example, in Histoplasma capsulatum, Saccharomyces pombe, Mus musculus, Bos taurus and humans) (34,35). Human p100 protein is composed of four repeats of SN-like domains and a TSN domain (Figure 1A). The SN-domains consist of two subdomains of which the first subdomain belongs to the large OB-fold superfamily, and the second subdomain consists of two independently folded-helices (26). The TSN domain of p100 is a hybrid of SN-like domain and a domain that is found in multiple copies in the Drosophila melanogaster Tudor protein.
We have previously shown that the SN domains of human p100 recruit STAT6 and CBP to RNA pol II, resulting in enhanced STAT6-mediated transcriptional activation (20,21). Here we provide evidence that the TSN domain of p100 protein is associated with U5 snRNP via specific interactions with U5-220 both in vivo and in vitro. Under high salt (700 mM NaCl) concentrations we detected association with the U5 snRNP specific proteins including U5-220, U5-200 and U5-116, which is consistent with previous reports showing that these proteins form a stable, RNA-free subcomplex (36–39). Additionally, under moderate salt concentrations we observed interactions with U1, U2, U4, U5, U6 and U7 snRNPs, but not with 7SK snRNP. The appearance of U7, but not 7SK in p100-TSN precipitation indicates that p100 protein is likely to interact with snRNP via recognition of TMG-cap or Sm-protein both of which are present in spliceosomal snRNPs and in U7 snRNP. The efficient co-immunoprecipitation of p100 along with the spliceosomal snRNPs using TMG-cap-specific antibody, argues that TMG-cap may not be the recognition structure of p100, and suggests two modes for the interaction of p100 and snRNPs, one is via the recruitment of U5 snRNP, the other one is via the association of Sm-protein. Finally, we provide evidence that addition of p100 protein or the TSN-domain to an in vitro splicing reaction accelerates the kinetics of splicing intermediate and product formation by enhancing the complex A formation and the transition from A to B complex
Comparison of the Tudor-domains of p100, splicing factor SMNrp/SPF30 and SMN demonstrates sequence similarity (Figure 6) between the Tudor domains. This is further supported by our recent structural analysis of the isolated TSN-domain of p100 using X-ray crystallography (Shaw,N. et al., submitted for publication) demonstrating that the TSN domain has a hydrophobic core, composed of aromatic residues F715, Y721, Y738 and Y741, which is highly similar to the Tudor domain of SMN. The hydrophobic core mediates interactions between SMN and the Sm-proteins, by binding with the dimethylarginine modifications of the Sm-proteins (28,40,41). Consistent with this hypothesis, mutations of the conserved tyrosine residues (Y738A, Y741A) of the aromatic cage (Shaw,N. et al., submitted for publication) abolished the interactions between TSN domain and snRNAs.
The association of p100 protein and snRNPs resemble the previous reports of SMNrp/SPF30 which interacts with the U4/U6-90 kDa protein present in tri-snRNP. Similarly to our study, all snRNPs are co-immunoprecipitated with SMNrp/SPF30 antibodies, and conversely the SMNrp/SPF30 is present in anti-TMG IP (42,43). Interestingly, SMNrp/SPF30 has been shown to be essential for the recruitment of the tri-snRNP to the spliceosome (42,43), indicating that it functions during the same spliceosome assembly step as p100 protein. It is possible that the p100 utilizes similar interaction mechanisms as the SMNrp/SPF30 during the spliceosome assembly, although the specific molecular interactions are most likely different.
We investigated the functional significance of the p100: snRNP interaction using an in vitro splicing and spliceosome complex-formation assays using a generic splicing substrate (AdML), and found that p100 accelerated the kinetics of the mature spliceosome formation before the first catalysis. Addition of either a purified full-length p100 protein or the plain TSN domain enhanced spliceosome formation, particularly the formation of the complex A and the transition from A to B complex. In contrast, p100 fragment encompassing the SN domains of the p100 severely inhibited the in vitro spliceosome assembly, but we do not know if it has any biological significance. Because p100 only accelerated the kinetics of the spliceosome assembly, but did not affect the overall level of splicing, we hypothesize that p100 may function by recruiting snRNPs, particularly the U4/U6.U5 tri-snRNP, onto spliceosome as supported by the specific interactions between snRNPs and p100.
An important question related to the function of p100 in spliceosome assembly in vivo is that why has p100 passed undetected in the extensive proteomic analyses of spliceosomes (12–14) and why it can enhance the splicing of a generic splicing substrate. A possible explanation is that compared to the previously identified factors functioning during spliceosome assembly, such as SMN or SMNrp/SPF30, the p100 is a large modular protein composed of multiple SN-domains in addition to the TSN domain and has thus other functions besides RNA processing. Our previous studies indicated distinct functions for the different domains, and the SN-like domain alone was sufficient to enhance STAT6-mediated transcription activity in response to IL-4 stimulation (21), while expression of the TSN alone did not affect the transcriptional response. Here we show that the isolated TSN stimulated the kinetics of spliceosome assembly, while the isolated SN-domain was highly inhibitory in the in vitro splicing assay. The stimulation of splicing of a generic splicing substrate may reflect the conditions of the in vitro reaction in which the p100 protein is free in the solution, while in vivo it may be associated with specific genes via promoter regions. Taken together, the results from this and previous studies suggest that the human p100 protein is dual function regulator of gene expression, capable of interacting with different protein complexes through distinct domains. Thus, it is possible that p100 protein may cooperate with RNA pol II and selectively coordinate gene transcription and pre-mRNA splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Paula Kosonen, and Hao Zhang for technical assistance, Dr Robin Reed for providing AdML plasmids, Dr Samuel I. Gunderson for helpful discussions. This work was supported by grants from national natural science foundation of China (30670441, 30300070), Program for New Century Excellent Talents in University (NCET-04-0245), Specialized Fund for the Doctoral Program of Higher Education (20040062003), Tianjin Municipal Science and Technology Commission (07JCZDJC07300, 05YFGHHZ01300), Medical Research Council of Academy of Finland (to O.S. and M.J.F.), Medical Research Fund of Tampere University Hospital, Finnish Foundation for Cancer Research, Tampere Tuberculosis Foundation, Finnish Cultural Foundation and the Sigrid Juselius Foundation. Funding to pay the Open Access publication charges for this article was provided by National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- 2.Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 3.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell. Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Zorio DA, Bentley DL. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 2004;296:91–97. doi: 10.1016/j.yexcr.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 5.Kornblihtt AR, de la MM, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millhouse S, Manley JL. The C-terminal domain of RNA polymerase II functions as a phosphorylation-dependent splicing activator in a heterologous protein. Mol. Cell. Biol. 2005;25:533–544. doi: 10.1128/MCB.25.2.533-544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corden JL, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem. Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 8.Hicks MJ, Yang CR, Kotlajich MV, Hertel KJ. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4:e147. doi: 10.1371/journal.pbio.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 10.de la MM, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Laurencikiene J, Kallman AM, Fong N, Bentley DL, Ohman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmuth K, Vornlocher HP, Luhrmann R. Tobramycin affinity tag purification of spliceosomes. Methods Mol. Biol. 2004;257:47–64. doi: 10.1385/1-59259-750-5:047. [DOI] [PubMed] [Google Scholar]
- 13.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 15.Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith MJ, Kulkarni S, Pawson T. FF domains of CA150 bind transcription and splicing factors through multiple weak interactions. Mol. Cell Biol. 2004;24:9274–9285. doi: 10.1128/MCB.24.21.9274-9285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, Stein GS, MacDonald PN. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 2003;278:35325–35336. doi: 10.1074/jbc.M305191200. [DOI] [PubMed] [Google Scholar]
- 18.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leverson JD, Koskinen PJ, Orrico FC, Rainio EM, Jalkanen KJ, Dash AB, Eisenman RN, Ness SA. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell. 1998;2:417–425. doi: 10.1016/s1097-2765(00)80141-0. [DOI] [PubMed] [Google Scholar]
- 20.Valineva T, Yang J, Palovuori R, Silvennoinen O. The transcriptional co-activator protein p100 recruits histone acetyltransferase activity to STAT6 and mediates interaction between the CREB-binding protein and STAT6. J. Biol. Chem. 2005;280:14989–14996. doi: 10.1074/jbc.M410465200. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Aittomaki S, Pesu M, Carter K, Saarinen J, Kalkkinen N, Kieff E, Silvennoinen O. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 2002;21:4950–4958. doi: 10.1093/emboj/cdf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol. Endocrinol. 2003;17:1805–1814. doi: 10.1210/me.2002-0256. [DOI] [PubMed] [Google Scholar]
- 23.Broadhurst MK, Lee RS, Hawkins S, Wheeler TT. The p100 EBNA-2 coactivator: a highly conserved protein found in a range of exocrine and endocrine cells and tissues in cattle. Biochim. Biophys. Acta. 2005;1681:126–133. doi: 10.1016/j.bbaexp.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Low SH, Vasanth S, Larson CH, Mukherjee S, Sharma N, Kinter MT, Kane ME, Obara T, Weimbs T. Polycystin-1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Dev. Cell. 2006;10:57–69. doi: 10.1016/j.devcel.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 26.Callebaut I, Mornon JP. The human EBNA-2 coactivator p100: multidomain organization and relationship to the staphylococcal nuclease fold and to the tudor protein involved in Drosophila melanogaster development. Biochem. J. 1997;321(Pt 1):125–132. doi: 10.1042/bj3210125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, et al. The SMN Complex: An Assembly Machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- 28.Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 29.Frilander MJ, Steitz JA. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev. 1999;13:851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frilander MJ, Meng X. Proximity of the U12 snRNA with both the 5′ splice site and the branch point during early stages of spliceosome assembly. Mol. Cell. Biol. 2005;25:4813–4825. doi: 10.1128/MCB.25.12.4813-4825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pessa HK, Ruokolainen A, Frilander MJ. The abundance of the spliceosomal snRNPs is not limiting the splicing of U12-type introns. RNA. 2006;12:1883–1892. doi: 10.1261/rna.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bringmann P, Rinke J, Appel B, Reuter R, Luhrmann R. Purification of snRNPs U1, U2, U4, U5 and U6 with 2,2,7-trimethylguanosine-specific antibody and definition of their constituent proteins reacting with anti-Sm and anti-(U1)RNP antisera. EMBO J. 1983;2:1129–1135. doi: 10.1002/j.1460-2075.1983.tb01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersen J, Zieve GW. Assembly and intracellular transport of snRNP particles. Bioessays. 1991;13:57–64. doi: 10.1002/bies.950130203. [DOI] [PubMed] [Google Scholar]
- 34.Porta A, Colonna-Romano S, Callebaut I, Franco A, Marzullo L, Kobayashi GS, Maresca B. An homologue of the human 100-kDa protein (p100) is differentially expressed by Histoplasma capsulatum during infection of murine macrophages. Biochem. Biophys. Res. Commun. 1999;254:605–613. doi: 10.1006/bbrc.1998.9894. [DOI] [PubMed] [Google Scholar]
- 35.Abe S, Sakai M, Yagi K, Hagino T, Ochi K, Shibata K, Davies E. A Tudor protein with multiple SNc domains from pea seedlings: cellular localization, partial characterization, sequence analysis, and phylogenetic relationships. J. Exp. Bot. 2003;54:971–983. doi: 10.1093/jxb/erg096. [DOI] [PubMed] [Google Scholar]
- 36.Laggerbauer B, Achsel T, Luhrmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl Acad. Sci. USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman AJ. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 1997;16:5797–5800. doi: 10.1093/emboj/16.19.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner IA, Norman CM, Churcher MJ, Newman AJ. Roles of the U5 snRNP in spliceosome dynamics and catalysis. Biochem. Soc. Trans. 2004;32:928–931. doi: 10.1042/BST0320928. [DOI] [PubMed] [Google Scholar]
- 39.Achsel T, Ahrens K, Brahms H, Teigelkamp S, Luhrmann R. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 1998;18:6756–6766. doi: 10.1128/mcb.18.11.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 42.Rappsilber J, Ajuh P, Lamond AI, Mann M. SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J. Biol. Chem. 2001;276:31142–31150. doi: 10.1074/jbc.M103620200. [DOI] [PubMed] [Google Scholar]
- 43.Meister G, Hannus S, Plottner O, Baars T, Hartmann E, Fakan S, Laggerbauer B, Fischer U. SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J. 2001;20:2304–2314. doi: 10.1093/emboj/20.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.