Abstract
We have created new genomics tools for chromatin research by genetically engineering the human and mouse major apoptotic nucleases that are responsible for internucleosomal DNA cleavage, DNA fragmentation factor (DFF). Normally, in its inactive form, DFF is a heterodimer composed of a 45-kDa chaperone inhibitor subunit (DFF45 or ICAD), and a 40-kDa latent endonuclease subunit (DFF40 or CAD). Upon caspase-3 cleavage of DFF45, DFF40 forms active endonuclease homo-oligomers. Although Saccharomyces cerevisiae lacks DFF, expression of caspase-3 is lethal in this organism, but expression of the highly sequence-specific tobacco etch virus protease (TEVP) is harmless. Therefore, we inserted TEVP cleavage sites immediately downstream of the two caspase-3 cleavage sites within DFF45, generating a novel form of DFF (DFF-T) whose nuclease activity proved to be exclusively under the control of TEVP. We demonstrate that co-expression of TEVP and DFF-T under galactose control results in nucleosomal DNA laddering and cell death in S. cerevisiae. We also created synthetic DFF genes with optimized codons for high-level expression in Eschericia coli or S. cerevisiae. We further demonstrate the excellence of the synthetic gene products for in vitro mapping of the nucleosome positions and hypersensitive sites in specific genes such as the yeast PHO5.
INTRODUCTION
Eukaryotic genomic DNA is organized into repeating arrays of nucleosomes that are the structural units of chromatin (1). The nucleosome is also known to play a central role in regulating gene transcription from promoters (2). For more than 30 years, micrococcal nuclease (MNase), DNase I and restriction endonucleases have been essential tools for chromatin research (1). MNase in particular has been the gold standard for characterizing nucleosomal structures in chromatin, because of its relative high specificity toward the internucleosomal linker region. However, this enzyme also digests DNA within nucleosome core particles, attacks single-stranded nucleic acids, possesses exonuclease activity and has significant DNA sequence cleavage preferences (3–5).
One of the hallmarks of the terminal stages of programmed cell death or apoptosis is internucleosomal DNA breakdown (6). The endonuclease primarily responsible for mediating apoptotic nucleosomal DNA laddering is DNA fragmentation factor-40 (DFF40), also called caspase-activated deoxyribonuclease (CAD) (7–10). DFF is a heterodimer in its inactive form, composed of a 45-kDa inhibitor and chaperone subunit (DFF45), also called inhibitor of CAD (ICAD), and a 40-kDa latent endonuclease subunit (DFF40/CAD) (7–13). This protein complex possesses nuclear localization sequences and resides within the cell nucleus (9,14–16). Caspase-3 or -7 cleavage of DFF specifically cuts only DFF45, releasing DFF40 from the complex, which in turn forms enzymatically active homo-oligomers (11–13,17,18).
During the course of our studies on characterizing the enzymatic properties of DFF40 on chromatin substrates, we recognized its potential as an excellent reagent for chromatin research because of its exquisite selectivity for linker region cleavage, its exclusive generation of double-stranded breaks and lack of both exonuclease activity and attack within nucleosome core particles (10,13,19–23). Indeed, evolutionary pressure to create ‘bite-size’ pieces of chromatin for phagocytotic clearance of apoptotic products appears to have optimized DFF40 to specifically attack the linker regions in polynucleosomal chains. However, DFF's nuclease dependency on caspase-3, which has numerous other cellular protein substrates (24), in combination with the protease's high cost, limits DFF's potential for chromatin studies. We therefore decided to engineer DFF nuclease activity to be dependent on TEVP cleavage (DFF-T) because this protease has a seven amino acid recognition cleavage sequence instead of the less stringent four amino acid target sequence of caspase-3 (24,25). Importantly, caspase-3 expression in yeast is lethal, but expression of the highly sequence-specific TEVP is harmless (26–28). Thus, TEVP expression per se should not cause any artificial alterations in chromatin structure and gene expression.
We demonstrate here the successful expression of recombinant forms of these proteins in either Eschericia coli or Saccharomyces cerevisiae and that after TEVP cleavage highly active DFF endonuclease is generated. We further demonstrate that DFF-T is an excellent reagent for mapping nucleosome positions and hypersensitive sites in specific genes as revealed by chromatin footprinting of the well-studied PHO5 gene, whose promoter and upstream region displaces four nucleosomes upon transcriptional induction via chromatin remodeling and binding of Pho4p (29–33).
MATERIALS AND METHODS
Generation of mouse or human DFF nuclease activities dependent on TEVP cleavage by mutagenesis and expression in E. coli
The TEVP cut site (25), encoding the amino acid sequence ENLYFQS, was inserted between amino acid positions 117–118 and 224–225, immediately downstream of each of the two caspase-3 cleavage sites in the coding sequence of mouse DFF45. We created all three possible combinations of these insertion mutations, termed I1WT, WTI2 and I1I2. Similarly, we created the corresponding I1I2 human DFF45 species, and also substituted the caspase-3 cleavage sites with TEVP cut sites in all combinations in mouse DFF45. Using the two-plasmid E. coli expression system for DFF (34) kindly provided by Gregor Meiss, the individual modified forms of mouse DFF45 along with the corresponding wild-type control were co-expressed with mouse GST-DFF40. The resulting GST-fusion protein-containing complexes were purified on GSH-Sepharose. Human DFF species were cloned into pRSFDuet™ for co-expression in E. coli (Novagen) and purification by nickel chromatography.
Assay of DFF45 cleavage and DFF40 endonuclease activity on plasmid DNA substrate
Recombinant caspase-3 was prepared as previously described (9), and TEVP was purchased from Invitrogen. One microgram of either wild-type, swap or insertion mutated DFF40/DFF45 heterodimer were incubated with caspase-3 or 0.5 unit of TEVP at 37°C in reaction buffer consisting of 10 mM KCl, 100 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol and 20 mM Tris–Cl, pH 7.5. After 20 min of incubation, samples were separated on 12% polyacrylamide–SDS gels, and then proteins were electrophoretically transferred onto nitrocellulose membranes. Membrane-immoblilized proteins were probed with the following commercial antibodies: rabbit anti-human DFF40 polyclonal antibodies and/or rabbit anti-human DFF45 N-terminus polyclonal antibodies (Pharmingen). The antigen–antibody complexes were visualized using enhanced chemiluminescence (ECL) western blotting detection reagents (Amersham Biosciences). For the endonuclease activity assay, 1 μg of naked plasmid DNA was incubated for 30 min at 37°C with DFF nuclease species pre-incubated with appropriate protease, as indicated in the legends of Figure 1. Aliquots of the endonuclease reaction were stopped by gel loading buffer containing 0.6% SDS, 50 mM EDTA, 30% glycerol and samples were then separated on 1.5% agarose gels using TAE as the running buffer. After electrophoresis, DNA was stained with ethidium bromide, and gels were scanned with a FluorImager (Molecular Dynamics Inc., Sunnyvale, CA, USA).
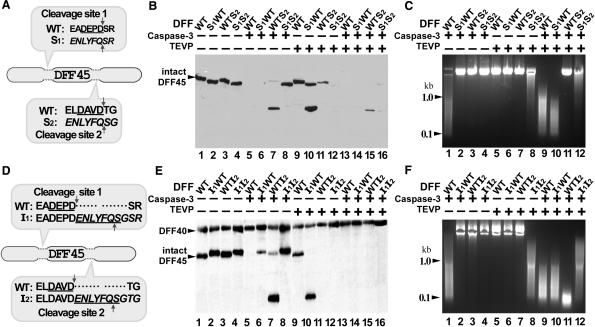
Figure 1.
Inserting TEVP cleavage sites downstream of caspase-3 cleavage sites within DFF45 is effective in generating DFF40 nuclease activity to be dependent on TEVP cleavage. (A and D) Amino acid sequence of mouse DFF45 (fragments containing both protease cleavage sites). The caspase-3 recognition sequences are underlined, the TEVP recognition sequences are in italics. (B and E) The indicated forms of mouse DFF heterodimers, either wild-type (WT), swap (S) or insertion (I) mutants, were expressed in E. coli, purified and treated either with caspase-3, TEVP, both or neither. Western blot of a SDS–PAGE gel probed with anti-DFF45 antibodies (B) or with a mixture of anti-DFF40 and anti-DFF45 antibodies (E); arrowheads mark the positions of intact DFF45 and GST-DFF40. (C and F) Agarose gel of plasmid DNA substrate incubated with the indicated proteinase-treated DFF species, either wild-type (WT), swap (S) or insertion (I) mutants.
Expression of mouse DFF-T and TEVP in S. cerevisiae
The TEVP gene's coding region with SV40 nuclear localization signals added to its N- and C-termini, under the control of the GAL1 promoter, and cloned into a TRP1 integration vector (28), which was kindly provided by Kim Nasmyth and used to stably transform the yeast P2 strain (35). DFF40 and wild-type DFF45 or the other three insertion mutants, I1WT, WTI2 and I1I2, were cloned into pESC-His dual yeast expression vector (Novagen) containing two divergently orientated galactose inducible GAL1 and GAL10 promoters. The pESC-DFF vectors were introduced into yeast P2 strains containing or lacking integrated TEVP via standard lithium acetate transformation and selection on synthetic complete medium lacking histidine and tryptophan, supplemented with 2% dextrose. To assay the effects of TEVP and DFF expression on cell viability, exponentially growing cultures of individual transformants were serially diluted 10-fold in 10 mM Tris–HCl (pH 8.0), 1 mM EDTA and 5 μl aliquots were spotted onto selective media supplemented with 2% raffinose and 2% galactose. The number of viable cells forming colonies at 30°C was determined. For in vivo endonuclease activity assay, yeast genomic DNA was isolated and analyzed. Briefly, 10 ml cultures of yeast cells transformed as indicated in Figure 2A, were harvested by centrifugation following galactose induction for 6 h. The cells were washed with 1 ml of distilled H2O, re-suspended in 200 μl of 2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris–Cl, pH 8.0, 1 mM EDTA, pH 8.0, vortexed at highest speed for 3 min with 200 μl glass beads and 200 μl phenol/chloroform/isoamyl alcohol, followed by the addition of 200 μl TE buffer and centrifugation for 5 min. The genomic DNA in the aqueous layer was ethanol precipitated and re-suspended and treated with 100 μg/ml RNase for 20 min at 37°C. The DNA was phenol extracted, ethanol precipitated and re-suspended in 50 μl TE buffer. The integrity of 5 μg DNA samples was assessed by agarose gel electrophoresis.
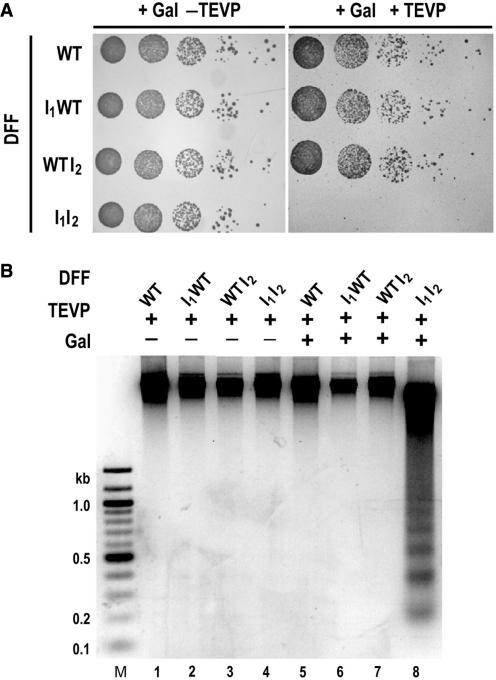
Figure 2.
Expression of DFF-T in yeast. (A) Co-expression of TEVP, GST-DFF40 and the TEVP-cleavable DFF45 I1I2, is lethal in yeast cells. Suspended yeast cells with or without an integrated TEVP gene and harboring the indicated forms of mouse DFF in a yeast expression plasmid were serially diluted 10-fold, and 5 μl aliquots were spotted onto galactose plates. Plates were photographed after 3 days of growth at 30°C. (B) Nucleosomal DNA laddering in vivo in yeast requires the combined expression of TEVP, GST-DFF40 and the DFF45 I1I2 mutant. Agarose gel of genomic DNA purified from yeast harboring the indicated forms of mouse DFF in a yeast expression plasmid after 6 h of galactose induction.
Generation of synthetic genes for high-level expression of DFF in E. coli
We took advantage of the ‘Center for Invention’ resource at UT Southwestern to generate a computer-assisted prediction for the design of synthetic genes and their robotic synthesis from pairs of overlapping 40-mer oligonucleotides (36). We have created, through this core, synthetic genes for both mouse and human forms of DFF40 and DFF45. After ligation of oligonucleotides, we amplified the products with high-fidelity PCR in preparation for cloning and sequencing. Any mutations that occurred were cured by additional high-fidelity PCR reactions with appropriate primers and repeated subcloning and sequencing. We have designed these synthetic genes to possess hexa-his tags at the C-terminal ends of DFF40 species, to either lack or possess the TEVP-cut sites as described below (sequences will be made available upon request), as well as the appropriate restriction sites for cloning into pRSFDuet™ E. coli expression vectors (Novagen). Eschericia coli BL21(DE3) cells transformed with this plasmid were induced with 0.5 mM IPTG for 3 h when the optical density at a wavelength of 600 nm (OD600) was 0.5. Cells were lysed by thawing in wash buffer solution (WB; 50 mM NaH2PO4, 300 mM NaCl containing lysozyme (1 mg/ml), RNase A (10 μg/ml) and DNaseI (5 μg/ml). Lysates were clarified by centrifugation and allowed to bind to nickel affinity resin (Ni-NTA superflow, Qiagen) for 1 h at 4°C with gentle mixing. After washing with WB, the protein was eluted with elution buffer solution (10 mM NaH2PO4, 100 mM NaCl, pH 8.0, 250 mM imidazole).
Mapping nucleosome positions on the inactive and induced PHO5 gene's upstream and coding regions
Saccharomyces cerevisiae strain W303 was grown over night at 30°C in yeast nitrogen base phosphate-free medium. Cells were centrifuged, suspended in fresh medium, either phosphate-free or supplemented with 14 mM KH2PO4 and grown for an additional 9 h. Cells were centrifuged and washed with water. One g of wet cell pellet was suspended in 2 ml of yeast lysis buffer (1 M sorbitol, 5 mM β-mecaptoethanol supplemented with 2 mg/ml of Zymolase 100T, ICN) and incubated at room temperature for 3–4 min with occasional agitation in 2-ml round-bottom microcentrifuge tubes. Cells were centrifuged for 15 s, washed in 1 M sorbitol, then suspended in 4 ml of digestion buffer (1 M sorbitol, 50 mM NaCl, 10 mM Tris–Cl pH 7.6, 5 mM MgCl2, 1 mM CaCl2, 1 mM β-mecaptoethanol, 0.5 mM spermidine, 0.075% NP-40) supplemented with 2 μg/ml of RNaseA and incubated for 15 min at room temperature (DFF is inhibited by RNA) (37). Aliquots measuring 400 μl of the resulting suspension were mixed with 100 μl of MNase (Worthington, 1 or 2 U) in digestion buffer. Alternatively, 400 μl aliquots of the resulting suspension were diluted with 400 μl of digestion buffer and mixed with 500 μl solution of TEVP-activated human DFF-T (100 or 200 pmol) in digestion buffer (100 pmol of DFF-T was pre-incubated with 50 U of AcTEV protease for 15 min at room temperature). Digestion mixtures were incubated for 10 min at 33°C, and then reactions were stopped by adding 1/5 vol of stop solution (2% SDS, 100 mM EDTA and 2 mg/ml proteinase K). Mixtures were incubated for 6 h at 50°C and for additional 4 h at 65°C, then phenol/chlorophorm extracted and isopropanol precipitated. DNA was purified by routine RNaseA treatment, phenol/chlorophorm extractions and isopropanol precipitation. DNA concentration was assessed by fluorometry; usually ∼25 μg of DNA was recovered from 0.1 g of wet yeast cell pellet. Ten micrograms of purified DNA was digested with ApaI restriction enzyme (50 U for 15 h at 28°C), electrophorectically separated on a 1.6% agarose gel and transferred onto Zeta-probe (Bio-Rad) nylon membrane using alkali (0.4 M NaOH, 1.5 M NaCl). The membrane was pre-hybridized for 4 h at 64°C with 0.5 M sodium phosphate (pH 7.2), 7% SDS, 1 mM EDTA and then hybridized over night in the same buffer supplemented with 0.1 mg/ml of denatured calf thymus DNA and 10 ng/ml of 32P-labeled PCR-amplified PHO5 probe, which was a 200-bp fragment adjacent to the ApaI site generated with the following primers 5′-GTC TTC AGC GTC AAC TTT AG-3′ and 5′-TAA CGA TGA TGG TCC CTT AA-3′ (32).
RESULTS AND DISCUSSION
Generation of TEVP-cleavage-dependent DFF nuclease activity
In an attempt to convert mouse DFF's nuclease activity to be dependent on TEVP cleavage, we first swapped one or both of the caspase-3 cleavage sites within DFF45 with a TEVP recognition sequence (Figure 1A). After co-expression of the different recombinant forms of DFF in E. coli and purification of the heterodimers, we separately assayed the cleavage pattern of DFF45 mutant forms by either caspase-3 or TEVP (or both) via western blotting, and the activity of DFF40 nuclease by digestion of a plasmid DNA substrate. As shown in Figure 1B, the various forms of DFF45 were cleaved as expected by the appropriate proteases. However, in spite of the fact that the double swap mutant was effectively cleaved by TEVP (Figure 1B, lane 12), DFF40 nuclease activity was only weakly generated compared with the wild-type control after cleavage by caspase-3 ± TEVP (Figure 1C, compare lanes 8 and 12 with 9). Interestingly, the S1WT but not the WTS2 mutant was also fully activated after cleavage by both proteases (Figure 1C, compare lanes 10 and 11). It therefore appears as if maintenance of the second caspase-3 cleavage site is most important for maintaining DFF45 chaperone function. We conclude that the swapped amino acid sequence at caspase-3 cleavage sites within DFF45 apparently plays a chaperone role in folding DFF40 into a potential nuclease capable of activation.
Because the caspase-3 sites within DFF45 are important for its chaperone activity, we created another series of DFF45 mutants in which the TEVP cleavage sequence was inserted immediately downstream of the caspase-3 cleavage sites (Figure 1D). Surprisingly, even though the caspase-3 recognition sequences DEPD and DAVD in mouse DFF45 were not altered by the adjacent insertions of the TEVP sites, the corresponding caspase-3 sites are no longer cleavable by that enzyme, as demonstrated by the maintenance of intact DFF45 I1I2 mutant protein after caspase-3 treatment, as revealed by western blotting (Figure 1E, lane 8), and by the inability of caspase-3 alone to activate the nuclease (Figure 1F, lanes 2–4). We conclude that the ability of caspase-3 to cleave at these sites is dependent on the amino acid sequence context and that the additional seven amino acids may affect the spatial properties of the caspase-3 cleavage sites. As shown in Figure 1F (lane 8), only the heterodimer containing the double-TEVP-site-insertion mutant I1I2 could be activated by TEVP treatment alone to a specific activity nearly equal to that of the caspase-3 ± TEVP treated wild-type enzyme (Figure 1F, lanes 1 and 9). We conclude that a novel modified form of mouse DFF nuclease (DFF-T) has been successfully generated, expressed and purified, whose activation is specifically under the control of TEVP.
Co-expression of TEVP and DFF-T in S. cerevisiae
We intended to develop a highly reproducible system in yeast for the regulated expression and activation of DFF-T for the purposes of in vivo footprinting of the positions of nucleosomes and transcription factors on specific genes. Previously, Simpson and Wang (38) were successful in developing a DNase I expression system in yeast for in vivo footprinting experiments by putting the DNase I gene into a high copy shuttle expression vector under the control of the GAL1 promoter. Here we have taken advantage of a system developed by Nasmyth and co-workers (28), who engineered the TEVP gene's coding region by adding SV40 nuclear localization signals to its N- and C-termini, which was placed under the control of the GAL1 promoter, and integrated into the trp1 locus. We therefore created a yeast strain with a galactose-inducible TEVP gene. We next inserted various engineered forms of mouse DFF45 and mouse DFF40 genes into the bicistronic, high copy number shuttle expression vector pESC-HIS (Stratagene), which divergently expresses introduced sequences from GAL1-10 promoters, and transformed yeast strains that either lacked or possessed the TEVP gene under GAL control with these expression vectors, and optimized expression in this system by titration with different concentrations 3-amino-1,2,4-triazole. As shown by the plating assay in Figure 2A, expression of TEVP together with various engineered forms of mouse DFF results in cell death only when the DFF45 I1I2 mutant and DFF40 are co-expressed together with TEVP. It is significant that this in vivo result is in agreement with the in vitro activation data for the corresponding recombinant proteins expressed in E. coli (Figure 1F above). Furthermore, yeast DNA undergoes nucleosomal laddering in vivo in a galactose-dependent fashion, only in the strain harboring the genes encoding the DFF45 I1I2 mutant, DFF40 and TEVP (Figure 2B, lane 8). We conclude that this form of DFF expression could be useful as a genomics tool to study yeast chromatin structure in vivo. It should be noted, however, just as in the case of the DNase I yeast expression system developed by Simpson and Wang (38), there is ca. 6-h time lag after GAL induction before significant DNA breakdown occurs, and it would be most desirable to be able to much more rapidly activate the nuclease to obtain a snapshot of the in vivo chromatin structure. This might be achieved by the high-level constitutive expression of codon-optimized DFF-T prior to GAL induction of TEVP. In addition, it may be possible to engineer TEVP for rapid regulated transport from the cytoplasm to the nucleus. A further limitation of DFF, particularly in the yeast system, is that these cells have a very high RNA/DNA ratio and RNA is an inhibitor of the enzyme (37).
Creation of synthetic DFF genes and their high-level expression in E. coli
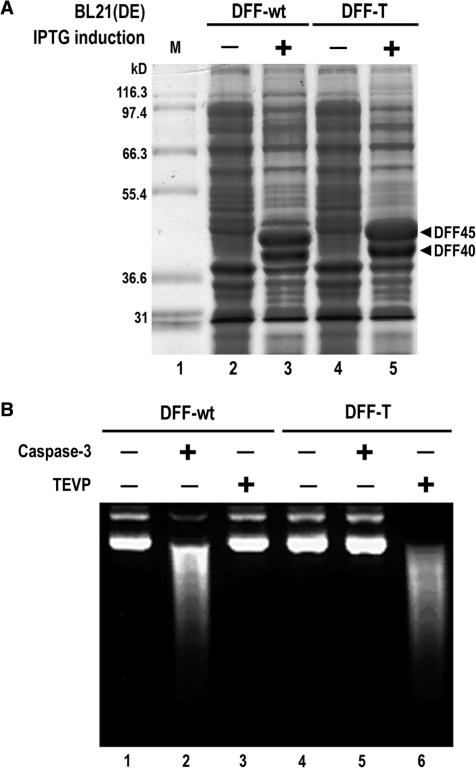
Many codons in DFF cDNAs are rare with respect to their cognate tRNA abundances in E. coli, and only a few micrograms of recombinant proteins are routinely obtained upon expression of recombinant forms of DFF in this organism per liter of culture. To provide a more robust source of DFF proteins to the scientific community we have created codon-optimized synthetic genes for wild-type and I1I2 mutants of the mouse and human DFF proteins through their robotic synthesis from pairs of overlapping 40-mer oligonucleotides (36). After expression of the proteins encoded by these synthetic genes in the pETDuet™ E. coli vector (Novagen), more than 20% of the total E. coli protein corresponds to DFF bands in Coomassie blue-stained SDS–PAGE gels (Figure 3A). This level of expression is highly significant because our previous expression systems using the corresponding wild-type cDNA coding sequences gave no visible new bands after induction in such gels of total E. coli cell protein (data not shown). Furthermore, DFF nuclease activities produced from these synthetic genes were fully dependent on cleavage by the appropriate protease, and both wild-type and insertion mutant proteins have similar nuclease activities (Figure 3B).
Figure 3.
High-level expression from synthetic human DFF genes in E. coli. (A) Coomassie blue-stained SDS–PAGE gel of total cell protein from uninduced or induced E. coli BL21 (DE3) transformants, both carrying DFF40 synthetic genes, and either DFF45 wild-type (DFF-wt) or the DFF45I1I2 mutant (DFF-T) synthetic genes, in the pETDuet™ expression plasmid. (B) Agarose gel of plasmid DNA substrate incubated with the indicated proteinase-treated DFF nucleases.
Mapping hypersensitive sites and nucleosome positions on the inactive and active yeast PHO5 gene with DFF-T produced from synthetic genes
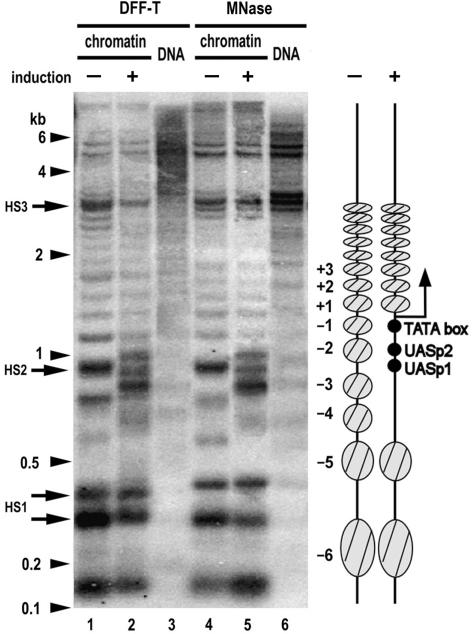
We have demonstrated here that DFF-T could be fully and specifically activated by TEVP both in vitro and in vivo. To further investigate the utility of DFF-T we mapped nucleosome positions and hypersensitive sites within the promoter region of PHO5 gene, one of the most thoroughly studied yeast genes with respect to chromatin structure (29–32). Upon induction of PHO5 gene transcription by phosphate starvation, four nucleosomes are displaced from the upstream promoter region through the binding of Pho4p to UASp1 and UASp2 and the recruitment of chromatin remodeling complexes (Figure 4, right diagram). To evaluate the effectiveness of DFF-T to detect hypersensitive sites, and both nucleosome positioning and displacement, we made a direct comparison on the same chromatin samples with MNase cutting patterns by indirect end-labeling analysis. Here we utilized human DFF-T produced in high-yield from codon-optimized synthetic genes in an E. coli expression system (Figure 3). This comparison reveals a striking degree of similarity with respect to: (i) the positions of hypersensitive sites HS2 and HS3 on the uninduced PHO5 gene, which correspond to the accessible UASp1 between nucleosomes −2 and −3, and the 3′ end of the PHO5 gene, respectively (29,30); (ii) the positions of nucleosome footprints (Figure 4, compare lanes 1 and 4) and (iii) in the displacement of nucleosomes −1 to −4 after gene induction (Figure 4, compare lanes 2 and 5). The partial occupancy by nucleosomes at the −1 position of the activated PHO5 promoter may be attributed to a steady state of disassembly and reassembly of nucleosomes as previously described (39). There is some subtle difference, however, in the hypersensitive sites in the HS1 doublet (Figure 4, compare lanes 1 and 2 with lanes 4 and 5), which corresponds to the upstream region of the YBR094W gene. In summary, this comparison allows us to conclude that DFF-T is an excellent reagent to map the chromatin structures associated with specific inactive or active genes. We speculate that this enzyme may detect chromatin structures missed by other conventional enzymes.
Figure 4.
Comparison between DFF-T and MNase in revealing specific chromatin structures of the yeast PHO5 gene. Yeast were grown in the absence or presence of inorganic phosphate to induce or repress the PHO5 gene, respectively, spheroplasts were isolated upon Zymolase treatment and then chromatin or purified naked DNA were digested with TEVP-treated human DFF-T or MNase as indicated. DNA was purified, digested with ApaI, electrophorectically separated on a 1.6% agarose gel, and the resulting Southern blot was hybridized with a 32P-labeled probe. Arrowheads and arrows indicate the positions of DNA length markers and nuclease hypersensitive sites HS1–3, respectively. The diagram on the right depicts as gray ovals the locations of positioned nucleosomes in the upstream and coding regions of the PHO5 gene, as closed circles the positions of the TATA-box and the Pho4-binding sites at UASp1 and UASp2, and as a bent arrow the transcription initiation site.
Other potential uses for DFF in functional genomics
In this study, we have shown that DFF-T is an excellent reagent for chromatin structure investigations. The enzyme should also prove to be valuable in other types of footprinting experiments. From our previous and current studies, we also know DFF is extremely suitable for fragmenting chromatin to prepare nucleosomes without them possessing internal DNA nicks. We also propose that DFF nuclease is ideally suited to serve as a substitute for sonication to shear chromatin to nucleosome-sized fragments for the chromatin immunoprecipitation (ChIP) technique, because it only cuts between and not within nucleosomes and thus it cannot over-digest the chromatin DNA as other nucleases can. This nuclease should also be ideal for the purpose of generating genomic libraries for gene cloning/subcloning and shotgun sequencing, as it creates nearly exclusively double-stranded blunt DNA ends, and leaves 5′-phosphate and 3′-hydroxyl groups that are ideal for blunt-end cloning by ligation with T4 DNA ligase. Controlled digestion of DNA to yield fragments of various sizes should be easy to achieve with this enzyme. Thus, DFF-T should be a valuable tool for several lines of investigation.
ACKNOWLEDGEMENTS
This work was supported in part by Grant N30105831/1763 from the Polish Ministry of Scientific Research (to P.W.), Grant RO1-GM29935 from the National Institutes of Health and Grant I-0823 from the Robert A. Welch Foundation (to W.T.G.). Funding to pay the Open Access publication charges for this article was provided by Grant RO1-AI067906 from the National Institutes of Health (to W.T.G.).
Conflict of interest statement. None declared.
REFERENCES
- 1.van Holde KE. Chromatin. Berlin: Springer; 1988. [Google Scholar]
- 2.Mellor J. Dynamic nucleosomes and gene transcription. Trends Genet. 2006;22:320–329. doi: 10.1016/j.tig.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Dingwall C, Lomonossoff GP, Laskey RA. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981;9:2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horz W, Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9:2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson PP, Albright SC, Garrard WT. Nucleosome arrangement with regard to DNA base composition. J. Biol. Chem. 1979;254:9194–9199. [PubMed] [Google Scholar]
- 6.Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 7.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 8.Halenbeck R, MacDonald H, Roulston A, Chen TT, Conroy L, Williams LT. CPAN, a human nuclease regulated by the caspase-sensitive inhibitor DFF45. Curr. Biol. 1998;8:537–540. doi: 10.1016/s0960-9822(98)79298-x. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Li P, Widlak P, Zou H, Luo X, Garrard WT, Wang X. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl Acad. Sci. USA. 1998;95:8461–8466. doi: 10.1073/pnas.95.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J. Cell. Biochem. 2005;94:1078–1087. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 12.Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 13.Widlak P, Lanuszewska J, Cary RB, Garrard WT. Subunit structures and stoichiometries of human DNA fragmentation factor proteins before and after induction of apoptosis. J. Biol. Chem. 2003;278:26915–26922. doi: 10.1074/jbc.M303807200. [DOI] [PubMed] [Google Scholar]
- 14.Samejima K, Earnshaw WC. ICAD/DFF regulator of apoptotic nuclease is nuclear. Exp. Cell Res. 1998;243:453–459. doi: 10.1006/excr.1998.4212. [DOI] [PubMed] [Google Scholar]
- 15.Samejima K, Earnshaw WC. Differential localization of ICAD-L and ICAD-S in cells due to removal of a C-terminal NLS from ICAD-L by alternative splicing. Exp. Cell Res. 2000;255:314–320. doi: 10.1006/excr.2000.4801. [DOI] [PubMed] [Google Scholar]
- 16.Lechardeur D, Drzymala L, Sharma M, Zylka D, Kinach R, Pacia J, Hicks C, Usmani N, Rommens JM, et al. Determinants of the nuclear localization of the heterodimeric DNA fragmentation factor (ICAD/CAD) J. Cell Biol. 2000;150:321–334. doi: 10.1083/jcb.150.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Zou H, Widlak P, Garrard W, Wang X. Activation of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease). Oligomerization and direct interaction with histone H1. J. Biol. Chem. 1999;274:13836–13840. doi: 10.1074/jbc.274.20.13836. [DOI] [PubMed] [Google Scholar]
- 18.Woo EJ, Kim YG, Kim MS, Han WD, Shin S, Robinson H, Park SY, Oh BH. Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway. Mol. Cell. 2004;14:531–539. doi: 10.1016/s1097-2765(04)00258-8. [DOI] [PubMed] [Google Scholar]
- 19.Widlak P, Li P, Wang X, Garrard WT. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J. Biol. Chem. 2000;275:8226–8232. doi: 10.1074/jbc.275.11.8226. [DOI] [PubMed] [Google Scholar]
- 20.Widlak P, Garrard WT. Ionic and cofactor requirements for the activity of the apoptotic endonuclease DFF40/CAD. Mol. Cell. Biochem. 2001;218:125–130. doi: 10.1023/a:1007231822086. [DOI] [PubMed] [Google Scholar]
- 21.Widlak P, Li LY, Wang X, Garrard WT. Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates: cooperation with exonuclease and DNase I. J. Biol. Chem. 2001;276:48404–48409. doi: 10.1074/jbc.M108461200. [DOI] [PubMed] [Google Scholar]
- 22.Widlak P, Palyvoda O, Kumala S, Garrard WT. Modeling apoptotic chromatin condensation in normal cell nuclei. Requirement for intranuclear mobility and actin involvement. J. Biol. Chem. 2002;277:21683–21690. doi: 10.1074/jbc.M201027200. [DOI] [PubMed] [Google Scholar]
- 23.Widlak P, Garrard WT. Unique features of the apoptotic endonuclease DFF40/CAD relative to micrococcal nuclease as a structural probe for chromatin. Biochem. Cell. Biol. 2006;84:405–410. doi: 10.1139/o06-063. [DOI] [PubMed] [Google Scholar]
- 24.Lee AY, Park BC, Jang M, Cho S, Lee DH, Lee SC, Myung PK, Park SG. Identification of caspase-3 degradome by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2004;4:3429–3436. doi: 10.1002/pmic.200400979. [DOI] [PubMed] [Google Scholar]
- 25.Dougherty WG, Cary SM, Parks TD. Molecular genetic analysis of a plant virus polyprotein cleavage site: a model. Virology. 1989;171:356–364. doi: 10.1016/0042-6822(89)90603-x. [DOI] [PubMed] [Google Scholar]
- 26.Smith TA, Kohorn BD. Direct selection for sequences encoding proteases of known specificity. Proc. Natl Acad. Sci. USA. 1991;88:5159–5162. doi: 10.1073/pnas.88.12.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang JJ, Schaber MD, Srinivasula SM, Alnemri ES, Litwack G, Hall DJ, Bjornsti MA. Cascades of mammalian caspase activation in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:3189–3198. doi: 10.1074/jbc.274.5.3189. [DOI] [PubMed] [Google Scholar]
- 28.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 29.Almer A, Horz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Jessen WJ, Hoose SA, Kilgore JA, Kladde MP. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat. Struct. Mol. Biol. 2006;13:256–263. doi: 10.1038/nsmb1062. [DOI] [PubMed] [Google Scholar]
- 34.Korn C, Scholz SR, Gimadutdinow O, Pingoud A, Meiss G. Involvement of conserved histidine, lysine and tyrosine residues in the mechanism of DNA cleavage by the caspase-3 activated DNase CAD. Nucleic Acids Res. 2002;30:1325–1332. doi: 10.1093/nar/30.6.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee MS, Garrard WT. Transcription-induced nucleosome ‘splitting’: an underlying structure for DNase I sensitive chromatin. EMBO J. 1991;10:607–615. doi: 10.1002/j.1460-2075.1991.tb07988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers RS, Johnston SA. High-level generation of polyclonal antibodies by genetic immunization. Nat. Biotechnol. 2003;21:1088–1092. doi: 10.1038/nbt858. [DOI] [PubMed] [Google Scholar]
- 37.Widlak P, Garrard WT. The apoptotic endonuclease DFF40/CAD is inhibited by RNA, heparin and other polyanions. Apoptosis. 2006;11:1331–1337. doi: 10.1007/s10495-006-6983-0. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Simpson RT. Chromatin structure mapping in Saccharomyces cerevisiae in vivo with DNase I. Nucleic Acids Res. 2001;29:1943–1950. doi: 10.1093/nar/29.9.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]