Abstract
To identify novel sperm alloantigens relevant to immune infertility, sera from infertile men containing antisperm antibodies (ASA) were employed on 2-D immunoblots of human sperm proteins. An immunoreactive protein spot (MW: 44 k Da, pI: 4.5) was microsequenced and the related cDNA was cloned yielding a 309 amino acid sequence corresponding to a gene currently annotated in Genbank as TSGA2 homolog (mouse) to signify ‘testis specific gene A2’. In Genbank the protein deduced from this gene is currently named human meichroacidin, an orthologue of meichroacidin previously identified in mouse spermatocytes. Human TSGA2 mapped to chromosome 21q22.3. Human meichroacidin (hMCA) contained a single potential tyrosine phosphorylation site and five casein kinase phosporylation motifs. The N-terminus contained a Membrane Occupation Recognition Nexus (MORN) motif found in the lipid kinase-phosphatidylinositol 4-phosphate 5- kinase (PIP5K) family and junctophilins. However hMCA lacked the characteristic kinase homology domain of PIP5K. Northern blot analysis revealed 1.5 kb hMCA transcripts in testis and trachea with lower levels in thyroid and spinal cord. A semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis demonstrated occurrence of the mRNA messages in all the ciliated tissues tested with highest levels of messages in testis and trachea. Western blot analysis showed the presence of hMCA protein in brain, thyroid and trachea at the identical mass, 44 k Da, as in human testis. However, this immunoreactive pattern differed from that of sperm in which a 38 k Da form was also evident suggesting that hMCA undergoes proteolytic processing. In human testis, hMCA localized to the tails of developing spermatids and did not localize to the nucleus of either spermatocytes or spermatids. EM immunocytochemistry localized hMCA within the radial spokes of the axonemal complex of the sperm flagellum, and immunofluorescence studies revealed h-meichroacidin in the cilia of epithelial cells in the trachea and ependyma. Bioinformatic identification of orthologues of meichroacidin in several lower organisms including ciliates and flagellates suggest the protein plays a role in flagellar motility across phyla. We propose the term radial spoke protein 44 as an accurate designation, preferable to human meichroacidin because it denotes the restricted localization of the protein to the radial spokes of the axonemes of both sperm and cilia. Further, since the human gene is expressed in brain, thyroid, trachea and lung in addition to testis, we suggest that the gene name be changed from TSGA2 [testis specific gene A2] to radial spoke protein 44 [RSP44].
Keywords: RSP44, TSGA2, MORN, flagellum
1. Introduction
ASA are thought to impair fertility by inhibiting sperm motility (Hass, 1986), sperm penetration of the cervical mucus (Hass, 1987), capacitation (Caron and Sailing, 1991), or the acrosome reaction (Lee et al., 1990); or they may invoke the complement cascade resulting in sperm lysis (D'Cruz et al., 1991; Bronson et al., 1982). Anti-sperm antibodies may also be formed as a result of cross-reactivity between sperm antigens and bacterial antigens (Witkin et al., 1995). A complete understanding of the mechanism behind immunologic infertility, as well as improved diagnosis and treatment, depends on knowledge of the identities of specific sperm antigens capable of eliciting the production of functionally relevant sperm antibodies. Utilizing vectorial labeling and 2-D electrophoresis by isoelectric focusing (IEF) our laboratory identified 6 auto- and isoantigens which were not recognized by fertile sera and possibly relevant to antibody mediated immunoinfertility (Shetty et al., 1999). Further studies led to the identification and characterization of several post-meiotically expressed sperm alloantigens residing in the acrosomal membranes (Hao et al., 2002; Shetty et al., 2003) and equatorial segment (Wolkowicz et al., 2003). In the current study an interesting alloantigen residing in the sperm tail is characterized.
Formation of the sperm tail is one of the final events in spermiogenesis, a process of marked morphological change during differentiation of haploid round spermatids into mature sperm. Mammalian spermatozoa and ciliated cells share a complex structure, the axoneme, which is responsible for motility of both cilia and flagella. The fine structural and molecular components of the axoneme are highly conserved through evolution from lower to higher eukaryotes (Pennarun et al., 2002). Axonemal ultrastructure reveals one central microtubular doublet and nine outer-doublet microtubules with attached inner and outer dynein arms, radial spokes and nexin links.
In addition to the axoneme and its associated proteins, the sperm flagellum also consists of a fibrous sheath (FS) and outer dense fibers (ODF), as well as proteins that interact with both FS and ODF, such as Oppo1 (Nakamura et al., 2002), Shippo 1(Egydio de Carvalho et al., 2002) and Spag 4 (Shao et al., 1999). However there are few reports on proteins associated with the sperm axonemal complex. Using a proteomic approach we report here the cloning and characterization of a human sperm axonemal complex protein, currently annotated in Genbank as h-meichroacidin. Mouse meichroacidin was originally reported to be a male meiotic chromosome associated acidic protein (meichroacidin), specific to pachytene spermatocytes through the round spermatid stages and not expressed in somatic cells (Tsuchida et al., 1998). The human orthologue of the same protein was later reported by Matsuka et al (2005), who demonstrated it to be a component of the sperm flagellum and specifically expressed in the testes. Our study of human meichroacidin indicates that this protein, in addition to be a component of the sperm tail, localizes to the radial spokes of the axoneme, as well as to cilia of epithelial cells suggesting a fundamental role in flagellar and ciliary action. Interestingly, the protein is a human alloantigen. Because this protein's distinct and specific localization in human spermatids and mature sperm differs from that reported for the mouse orthologue, we propose the name radial spoke protein 44 for this protein and gene (RSP44) in humans.
2. Materials and methods
2.1 Preparation of Spermatozoa and Capacitation
All semen samples for this study were obtained under informed consent using forms approved by the University of Virginia Human Investigation committee. Ejaculates with normal semen volume, sperm count, and motility were used in this study. After liquefaction of semen samples at room temperature, fresh sperm were separated from seminal plasma, germ cells, white blood cells, and epithelial cells by Percoll (Pharmacia Biotech, Bromma, Sweden) density gradient centrifugation and washed in Ham's F-10 medium (Gibco BRL, Life Technologies, Gaithersburg, MD) within 2–3 h of ejaculation as described earlier (Shetty et al., 1999). To obtain capacitated spermatozoa, motile sperm were prepared by the swim-up method for 60 min in Biggers, Whitten, and Whittingham (BWW) medium (Irvine Scientific, Santa Ana, CA) containing no human serum albumin. Motile spermatozoa were subsequently collected from the supernatant and incubated in BWW medium supplemented with 3.0% human serum albumin (HSA) (Sigma, St. Louis, MO) at 37°C in 5% CO2 for 6 h to induce capacitation (Bronson and Tung, 1992).
2.2. Serum Samples
Antisperm antibody (ASA) positive serum samples were obtained from infertile men and women with unexplained infertility. The infertile subjects did not have any previously diagnosed hormonal, infective, or physical causes for their infertility. ASA in the serum samples from infertile females were detected by indirect immunobead binding as previously described (Luconi et al., 1996).
2.3 Animals
Adult female Sprague-Dawley virgin rats of 6-9 weeks were used for immunization. All experiments were performed in accordance with the guidelines for the Use and Care of Laboratory Animals (NIH Publication 85-23).
2.4. 2-D gel electrophoresis and Western blotting
Human semen samples were obtained from healthy donors as described earlier (Shetty et al., 1999). Proteins were extracted in Celis reagent (Celis et al., 1992). Two-dimensional electrophoresis was performed according to the method of O'Farrell et al (O'Farrel et al., 1977) with modifications (Shetty et al., 2001). Isoelectric focusing (IEF) was performed in 15 × 0.15-cm acrylamide tube gels. The second dimension SDS-PAGE was carried out in 0.15 cm thick 16 × 16-cm slab gels using linear gradients (9-16%) of acrylamide. Electrotransfer to nitrocellulose membranes and immunobloting was performed as described previously (Shetty et al., 2001).
2.5. One-dimensional SDS-PAGE
Protein electrophoresis was performed on Ready to Load 12.5% Criterion gels (Biorad, Hercules, CA). Gels were stained with Coomassie blue or blotted. Electrotransfer and Western blotting were as described above for 2-D gels.
2.6. Microsequencing of the 2D SDS-PAGE-Resolved Protein Spots
Coomassie-stained protein spots were cored from 1.5-mm-thick 2-D gels. Proteins in the cored spots were destained, minced, and processed for microsequencing by tandem mass spectrometry (W. M. Keck Biomedical Mass Spectrometry Laboratory at the University of Virginia) as described earlier (Shetty et al., 2001). The peptides were interpreted manually. The data were compared with the non-redundant and expressed sequence tag (EST) databases using the Sequest algorithm at GenBank.
2.7. Cloning and Analyses of hMCA
The peptide sequences from mass spectrometry and Edman sequencing matched to a mouse testis EST sequence (Gen Bank accession No. gi1669577). Single gene-specific forward (5′GGG GAC ACA TAT GAA GGA AGC TAT GAG3') and reverse (5′GCG ATA TTC ACC GTG CTG TTC GCA TC3') primers were designed from the EST sequence corresponding to peptides # and # identified by mass spectrometry (Table 1). Primers were obtained from Invitrogen (Carlsbad, CA). The 5′ and 3′ ends of the generated cDNAs were amplified by RACE polymerase chain reaction (PCR) from adaptor-ligated human testicular Marathon ready cDNA (Clontech, Palo Alto, CA) using adaptor primer and reverse or forward gene-specific primers, respectively. PCR was performed for 40 cycles in a PTC 200 DNA Engine (MJ Research, Watertown, MA). PCR reaction products were separated on agarose gels, and bands were isolated and subcloned in the pCR2.1-TOPO vector (Invitrogen). Multiple cDNA clones were sequenced in both directions using vector-derived primers on a Perkin-Elmer Applied Biosystems DNA sequencer (Biomolecular Research Facility, University of Virginia Health System, Charlottesville, VA).
Table 1.
Peptide sequences obtained from an in-gel tryptic digest of the three protein spots determined by tandem mass spectrometry.
| Peptide Number |
Measured Molecular Weight |
Peptide Sequence | Spot identification code |
|---|---|---|---|
| 1. | 1184.6 | _ _EXD---R | MA1 |
| 2. | 1434.2 | HGQGTFIYPDGSR | MA1/MA2/MA3 |
| 3. | 1511.5 | HGQGTYLYAETGSK | MA1/MA3 |
| 4. | 1677.7 | LPNGDTYEGSYEFGK | MA1/MA2/MA3 |
| 5. | 1773.2 | YVFDIGCEQHGEYR | MA1/MA2 |
| 6. | 802.5 | HGQGIYK | MA3 |
| 7. | 899.7 | YIGEYVR | MA3 |
| 8. | 1252.3 | YEGEWANDLR | MA3 |
| 9. | 1773.2 | YVFDVGCEQHGEYR | MA3 |
| 10. | 2523.3 | GTWVN--- | MA3 |
-, a single unknown amino acid; ---, unknown number of unknown amino acids
2.8. Expression of recombinant hMCA and production of antiserum
To express hMCA in E. coli, a DNA fragment encoding the entire open reading frame was amplified by PCR. The amplified fragment was fused in frame with a His-tag at the C-terminus of pET 28b using the Xho I and Pci I sites. The plasmid was then transformed into the host strain E. coli BL21 DE3. The recombinant protein was affinity purified on a Ni-nitriacetic acid (Ni-NTA) column under denaturing conditions. The recombinant protein was further purified on a model 491 Prep Cell (Biorad Laboratories) to a single band, and was verified by SDS-PAGE and used for immunizations.
Female virgin Lewis rats (3 rats per each group) weighing 160-200 grams, were injected with purified recombinant hMCA in PBS emulsified with an equal amount of Freund's complete adjuvant. Each rat received 100μg of protein s.c. in 0.3 ml of PBS for the primary injection and for subsequent injections three and six weeks later. Antibody titers were checked by western blot analysis 10 days after each injection and the animals were sacrificed 10 days after the last boost. Specificity of the rat anti-sera for hMCA was tested by Western blotting against rec-hMCA as well as human sperm protein extracts.
2.9. Northern blot analysis
Two multiple tissue northern membranes containing 8 tissues each were purchased from Clontech. The PCR amplified 930 bp cDNA corresponding to the entire open reading frame of hMCA was used as a probe and the analysis was done as described previously (Shetty et al., 2003).
2.10. Semi-quantitative RT-PCR Analysis
Total RNA from several ciliated tissues including brain, spinal cord, thyroid, lung, trachea, liver, testis and fallopian tube were obtained from Stratagene (Stratagene, La Jolla, CA). Reverse transcription was performed using Advantage RT-for-PCR kit from Clontech following the protocol recommended by the manufacturer. RT-PCR exponential phase was determined on 25-35 cycles to allow semi-quantitative comparisons among cDNAs developed from identical reactions. Each PCR regime involved a 3 min initial denaturation step at 94°C followed by 25 amplification cycles of 94 for 30 s, 52 for 30 s, and final extension at 68°C for 2 min. The primers are h-Meichroacidin- forward. primer: 5'- cggggacacctacgaaggga; h-Meichroacidin- reverse primer: 5'-ggtgtccacagggccaattc. For G3PDH, the amplimer set from Clontech was used. The predicted sizes of PCR products were 546 bp (h-meichroacidin) and 983 (G3PDH). Equal volumes of PCR mixtures were resolved on 1.8% agarose gels and visualized by ethidium bromide staining. The intensities of the cDNA bands for each tissue were normalized to G3PDH band intensities. Images of the ethidium bromide (EB)-stained agarose gels were acquired by Alpha Inotech Imaging Systems (Alpha Inotech Corporation, San Leandro, CA) and the bands were analyzed by FluorChem v.20 software.
2.11. Immunofluorescence and Electron Microscopic Analysis
The human spermatozoa were purified by the swim-up method (Mandal et al., 1999) and were air-dried onto poly-L-lysine coated slides (Polysciences, Warrington, PA). Following 3 washes with PBS for 10 min each, slides were blocked in PBS containing 10% normal goat serum for 1 h with or without 0.1% saponin for permeabilization. The sperm were then incubated with a 1:200 dilution of anti-hMCA primary antiserum or preimmune serum for 1 h at 22°C, followed by washing in PBS 3 times for 10 min each at 22°C. Following incubation with the secondary goat-anti-rat IgG conjugated with TRITC at 1:100 dilution for 1 h, the slides were washed in PBS, incubated for 5 min with equilibration buffer followed by Slow Fade (Molecular Probes, Eugene, OR) and mounted with coverslips. Images were captured with an Axioplan fluorescence microscope (Zeiss, Oberkochen, Germany).
Processing of pooled sperm for electron microscopy was performed as described earlier (Hao et al., 2002). To stain sections, they were first blocked in undiluted normal goat serum for 15 min at 22°C. They were then incubated for 16 h at 4°C with either preimmune or rat-anti-hMCA serum at the dilution of 1:50 containing 1% normal goat serum, 1% BSA and 0.1% Tween-20. After washing, the sections were incubated with 1:100 dilution of 5 nm gold-conjugated goat-anti IgG (Goldmark Biologicals, Phillipsburg, NJ) for 1.5 hours at 22°C. The sections were washed in distilled water, stained with uranyl acetate, and observed with a JEOL 100CX electron microscope.
2.12. Localization of hMCA in Testis, Trachea Spinal cord and Liver
Testes were obtained from three patients undergoing elective orchiectomies. Testes were sliced once with a razor blade and immersed in neutral buffered formalin (4%) solution (Sigma) for 1 h. The tissue was then minced and placed into fresh fixative overnight. The tissue was dehydrated in a graded series of ethanols, cleared in xylene, and embedded in paraffin. Sections (2.5 mm thick) were cut, mounted onto slides, deparaffinized, dehydrated, and permeabilized with 100% methanol. Sections were incubated in blocking solution containing 10% normal goat serum (NGS) in PBS, incubated with anti-rec hMCA antiserum or preimmune serum (1:200) in PBS containing 1% NGS (PBS–NGS), washed, incubated with FITC-labeled goat anti-rat IgG (1:400; Jackson Immuno Research, West Grove, PA) in PBS–NGS, washed, and mounted with Slow Fade (Molecular Probes, Eugene, OR) containing DAPI II counterstain (Vysis, Downers Grove, IL). Sections were observed by epifluorescence microscopy using a Zeiss microscope. Individual blue and green fluorescent images were obtained using a digital camera (Hamamatsu) and compiled using Openlab software (Improvision, Inc., Boston, MA).
Paraffin sections of adult human trachea and liver were obtained from BioChain Institute, Inc (Hayward, CA), and paraffin sections of adult human spinal cord were generously provided by Dr. M. Beatriz Lopes, Department of Pathology, University of Virginia. Both tissues were sectioned and processed as above to localize hMCA.
3. Results
3.1. hMCA is an alloantigen
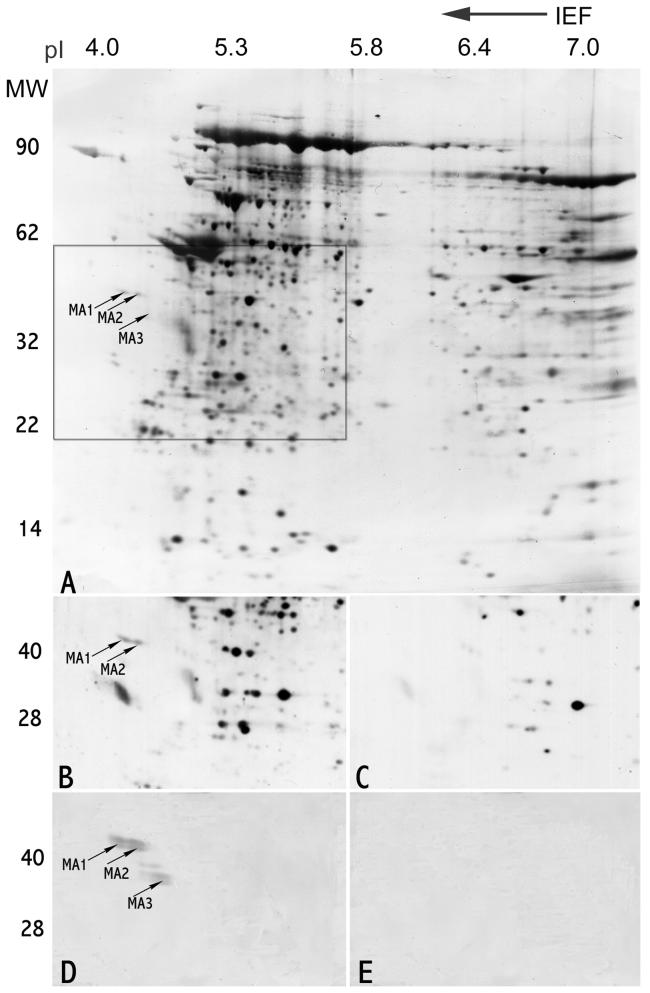
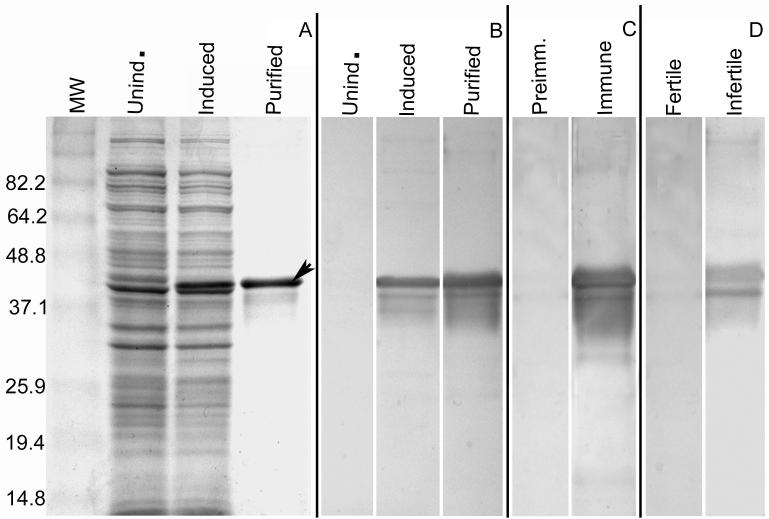
2D immunoblotting of infertile sera that were known to contain antisperm antibodies (ASA) demonstrated a unique reactivity to a pair of acidic spots (pI 4.2, 4.3; MW 42 kDa) that were not recognized by ASA negative fertile subjects in separate 2D western blot experiments (Fig. 1 B and C). Two of 14 ASA positive sera from infertile male patients reacted with the identical spots, identified (see below by microsequencing) in Genbank as h-meichroacidin, demonstrating that this human sperm protein is alloantigenic.
Fig. 1.
Identification of alloantigenic RSP44 (h-meichroacidin) spot by 2-D gel electrophoresis. A. Silver stained 2-D gel of Celis (Celis et al., 1992) extract of human sperm proteins showing the locations of h-meichroacidin (MA1, MA2 and MA3). The boxed area denotes the region of the gel shown in B-E. B: Sequential probing of a blot of sperm proteins using sera from 5 antisperm antibody positive infertile patients recognized 2 spots corresponding to h-meichroacidin, MA1 and MA2. C: Immunoblotting with 5 antisperm antibody negative, fertile subjects' sera did not recognize these spots. D: 2-D blot of human sperm proteins probed with rat serum against purified rec RSP44. The region corresponding to the originally microsequenced spots bound the antibody. E: Preimmune control for D.
3.2. Mass Spectrometry and Cloning of hMCA
The alloantigenic spots were cored for mass spectrometry analysis along with several spots in the neighboring area of the gel. Overlapping of tryptic peptides contributed by different spots and data base analysis of the same (Table 1) revealed that the cored spots MA1, MA2 and MA3 (Fig.1A) belonged to the same protein and matched to a mouse testis EST (Genbank accession No.gi1669577). The mouse EST sequence and the deduced peptide sequences were used to design primers to PCR amplify the cDNA. In the first step of PCR amplification a truncated fragment of human cDNA was obtained. The missing 5' and 3' ends of the DNA were generated by 5' and 3' Race PCR. Fig. 2 A shows the nucleotide and amino acid sequence for the h-MCA protein. The protein of 309 aa has a predicted pI of 4.56 and molecular weight of 35.12 kDa. During analysis of the cloned sequence we found a mouse orthologue of the same protein that had been designated meichroacidin based on its association with meiotic chromosomes (Tsuchida et al., 1998).
Fig. 2.
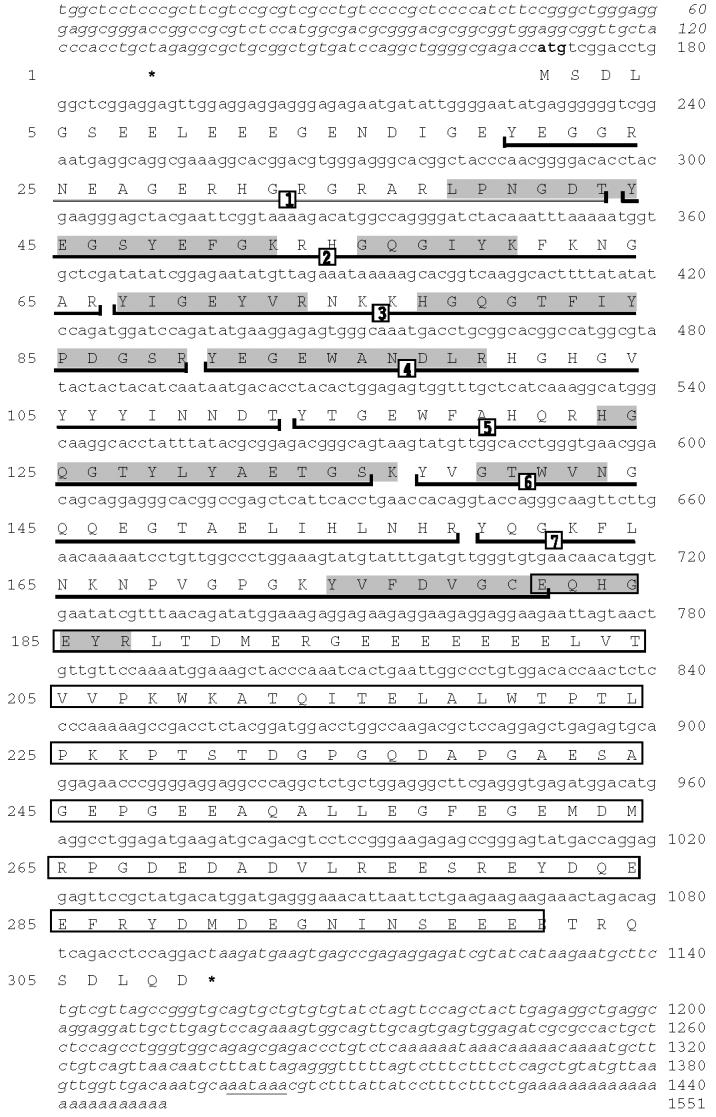
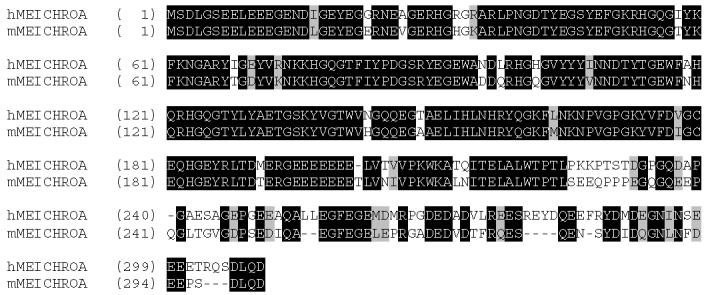
Fig 2: AThe nucleotide and deduced amino acid sequences of RSP44 cDNA. The deduced amino acid sequence is shown below the cDNA sequence. The numbers on the left refer to the amino acid sequence; numbers on the right refer to the nucleotide sequence. The consensus ATG of the ORF is shown in bold. The termination codon (TAA) is marked with an asterisk. The partial 5' and 3' untranslated regions are shown in italics. The polyadenylation signal is underlined with a thin line. The calculated molecular mass and pI of the predicted protein were 35.1 and 4.5 respectively. The sequences shaded in gray indicate the tryptic peptides obtained by microsequencing the gel spots noted in Fig. 1. The 7 MORN repeat sequences are underlined with a dark line and are numbered. The glutamic acid rich region is boxed.
Fig. 2 B: Amino acid sequence alignment of RSP44 and mouse MCA. The identical amino acids are shaded in dark background with white letters and the similar amino acids are shaded in gray background with dark letters.
Human meichroacidin showed a MORN motif repeat structure. This motif is found in some members of the lipid kinase-phosphatidylinositol 4-phosphate 5-kinase (PIP5K) and junctophilin families. The c-terminal region of hMCA (aa 181 to 300) was particularly rich in the acidic amino acid glutamic acid. In particular, a continuous stretch of 7 glutamic acid residues was present at position 195-201. hMCA contained several potential phosphorylation sites including 1 tyrosine phosphorylation site (residues 37-44), 1 cAMP phsophorylation site (residues 226-229), and 5 CK2 phosphorylation sites (residues 297-300, 88-91, 189-192, 229-232 and 243-246). A single potential ASN-glycosylation site was present at position 110-113 and 3 myristylation sites at positions 139-144, 240-245, and 293-298. Although the N-terminal region of hMCA showed homology to several members of the lipid kinase phosphatidyl-inositol 4-phosphate 5-kinase (PIP5K) family in having the MORN repeats, hMCA lacked the characteristic kinase homology domain of the PIP5 kinases. hMCA showed identity to a wide range of sequences in the animal kingdom, including unicellular organisms. The sequence identity between mouse and human MCA is more than 72% (Fig.2B), while the similarity is 80 %. Database searches revealed that meichroacidin orthologues are conserved across various chordates including primates (Pan troglodytes), carnivores (Canis familiaris), ruminants (Bos Taurus), chicken (Gallus gallus), amphibians (Xenopus laevis), teleosts (Cyprinus carpio, Tetraodon nigroviridis), as well as being present in other phyla, including echinoderms (Strongylocent-rotus purpuratus), insects (Tribolium castaneum, Drosophila melanogaster), trematodes (Schistosoma japonicum), and several unicellular organisms (Leishmania major, Tetrahymena thermophila, Trypanosoma cruzi, Plasmodium berghei, Chlamydomonas reinhardtii, Paramecium tetraurelia) (Table 2). Consistent in meichroacidin-like proteins from all species is the presence of 5-14 MORN domains. hMCA also showed similarity to several uncharacterized proteins with MORN domains from the plant kingdom (eg. giǀ18403768 from Arabidopsis thaliana). These findings indicate that meichroacidin is an essential molecule conserved in many organisms during evolution. The human genomic sequence for meichroacidin was localized to 21q22.3.
Table 2.
Suspected Orthologues of Meichroacidin in other Species
| Species | Name of the Protein | Accession No. |
Length (aa) |
Identity to hMeichroacidin |
Morn domains |
|---|---|---|---|---|---|
| Homo sapiens | meichroacidin | giI21542279 | 309 | 100% | 7 |
| Pan troglodytes | Similar to Meichroacidin | gi|55657511 | 346 | 251/274 (91%) | 7 |
| Canis familiaris | Similar to testis-specific gene A2 |
gi|74001485 | 322 | 208/277 (75%) | 7 |
| Mus musculus | TSGA2 | gi|73402277 | 301 | 196/273 (71%) | 6 |
| Rattus norvegicus |
TSGA2 | gi|54038556 | 300 | 194/273 (71%) | 6 |
| Bos taurus | Unknown (protein for MGC:134494 |
gi|8340543 | 270 | 204/263 (77%) | 5 |
| Gallus gallus | Similar to TSGA2 | gi|50730001 | 338 | 128/199 (64%) | 6 |
| Xenopus laevis | LOC496054 protein | gi|56269556 | 300 | 126/191 (65%) | 7 |
| Ciona intestinalis | similar to TSGA2 | gi|23588198 | 306 | 117/185 (63%) | 7 |
| Cyprinus carpio | meichroacidin-like sperm specific protein |
gi|37625514 | 218 | 117/182 (64%) | 6 |
|
Tetraodon nigroviridis* |
Unnamed protein product |
gi|47215789 | 343 | 97/150 (64%) | 14 |
| Strongylocent- rotus purpuratus |
Similar to TSGA2 | gi|72171594 | 287 | 102/167 (61%) | 5 |
| Schistosoma japonicum |
SJCHGC09106 protein | gi|56758472 | 159 | 78/143 (54%) | 5 |
| Tribolium castaneum |
Similar to TSGA2 | gi|91080421 | 307 | 77/150 (51%) | 6 |
| Drosophila melanogaster |
CG5458-PA | gi|19921216 | 344 | 64/150 (42%) | 5 |
| Leishmania major |
Hypothetical protein | gi|68128010 | 358 | 65/184 (35%) | 14 |
| Tetrahymena thermophila |
Hypothetical protein | gi|89296645 | 410 | 62/148 (41%) | 12 |
| Trypanosoma cruzi |
hypothetical protein | gi|71410032 | 201 | 64/180 (35% | 8 |
| Plasmodium berghei |
Hypothetical protein | gi|68009960 | 196 | 55/154 (35% | 7 |
| Chlamydomonas reinhardtii |
radial spoke protein 1 | gi|83284723 | 814 | 55/136 (40%) | 6 |
| Paramecium tetraurelia |
MORN repeat protein | gi|50057566 | 401 | 54/150 (36%) | 8 |
note: partial sequence
3.3. Expression of hMCA in various tissues
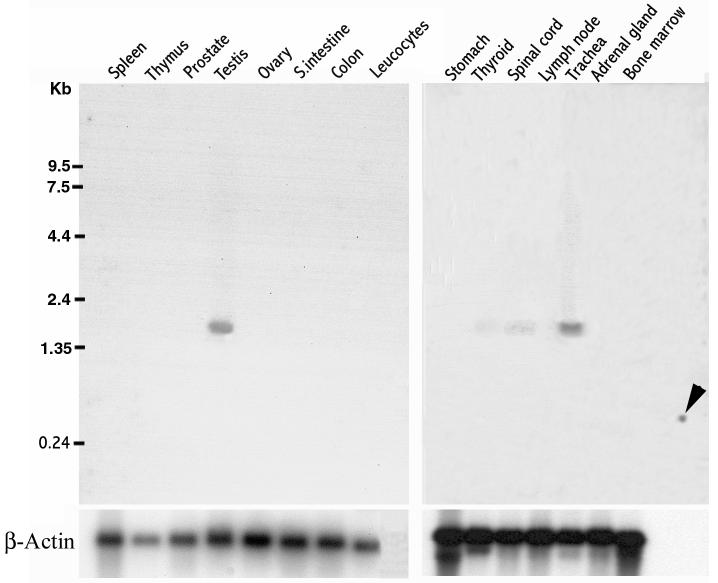
A northern blot analysis to determine the expression of the mRNA in various tissues showed that hMCA is expressed in trachea at levels comparable to that of testis (Fig.3). Lower levels of message were also present in the spinal cord and thyroid gland. A data base analysis of hMCA ESTs also revealed transcripts in trachea, thyroid and brain.
Fig. 3.
Northern blot analysis of RSP44. Two northern blots (Clontech) containing poly (A)+ mRNA from human tissues (2 μg per lane) were hybridized with radiolabeled RSP44 cDNA and exposed for 96 h. RNA markers are indicated on the left. RSP44 transcripts of ∼1.8 kb were clearly detected in testis and trachea. A low amount of mRNA expression was also detected in thyroid and spinal cord. The arrow indicates the reactivity to a testis mRNA that was spotted as a positive control.
3.4. Western blot analysis
A western blot analysis with anti-hMCA sera raised in rats recognized the identical spots on a 2-D immunoblot (Fig. 1D) as those that were originally microsequenced after recognition by ASA+ sera. 1-D western blot analysis of ejaculated sperm proteins with anti-hMCA sera (Fig. 4) detected 2 bands at ∼ 38 kDa and ∼44 k Da in both capacitated and non-capacitated sperm. However only the ∼ 44 k Da band was detected in testicular extracts indicating that the protein undergoes post-translational modification, possibly proteolytic processing. A 44 k Da band was also detected in brain, spinal cord, thyroid, trachea and lung, the intensity being less in all these tissues than in testis, except for the brain.
Fig. 4.
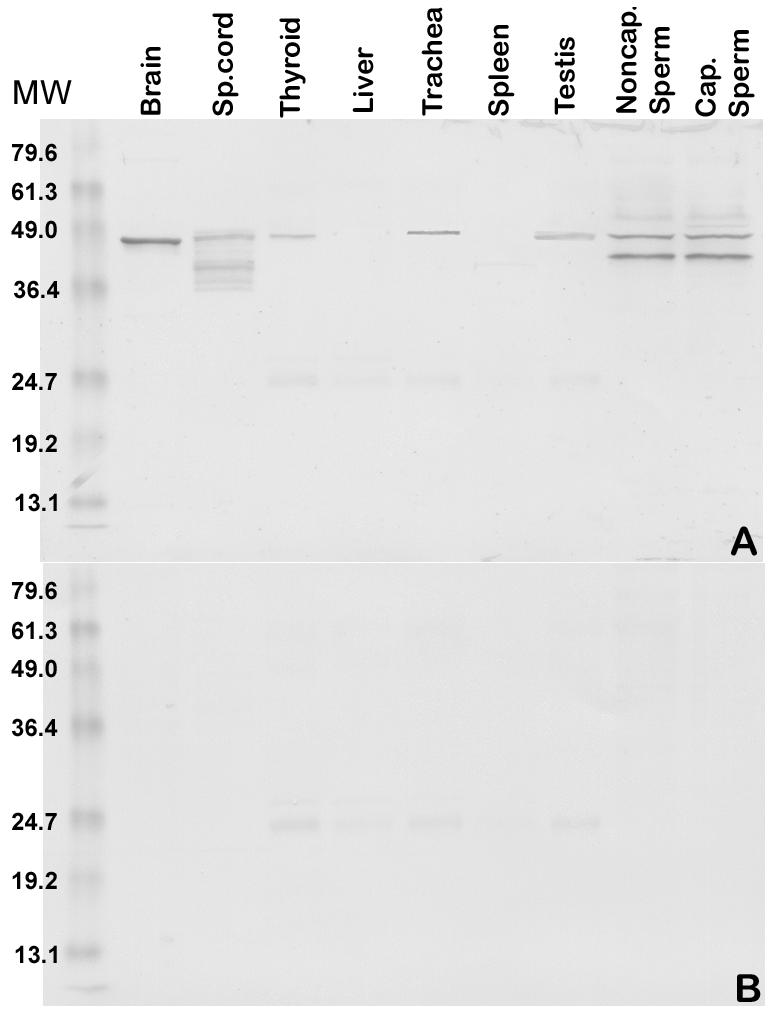
A multiple tissue Western blot analysis of RSP44. A: Protein samples were extracted from adult human tissues with NP40 and examined by Western blot analysis using rat anti-rec RSP44 polyclonal antibody. Protein samples (∼30μg/lane) from the brain, spinal cord, thyroid, liver, trachea, spleen, testis, uncapacitated sperm, and capacitated sperm were loaded. Expression of the protein in sperm was characterized by the appearance of 44 and 38 k Da bands in both un-capacitated and capacitated sperm. Brain and testis showed only the 44 k Da form of RSP44 with an intensity similar to that of sperm MCA. Thyroid and trachea also showed a lower level of expression of the higher molecular weight form of MCA. Spinal cord showed a pattern of expression similar to that of sperm, although the lower molecular weight form appeared to be of lower mass than that of sperm. Liver and spleen did not show any signal indicating the absence of protein in these tissues. B. Pre-immune control for A showed no reactivity at the expected molecular weight range for RSP44, although a low intensity background signal was observed at ∼25 k Da in both A and B.
3.5. hMCA localized to the axoneme of the entire sperm flagellum
Immunofluorescent staining of swim-up ejaculated human sperm revealed hMCA localized to the entire length of the sperm tail including the middle piece, principal piece and end piece (Fig. 5 A). An increase in intensity of staining was observed when the sperm were permeabilized using 0.1% saponin indicating that the protein is localized internally.
Fig. 5.
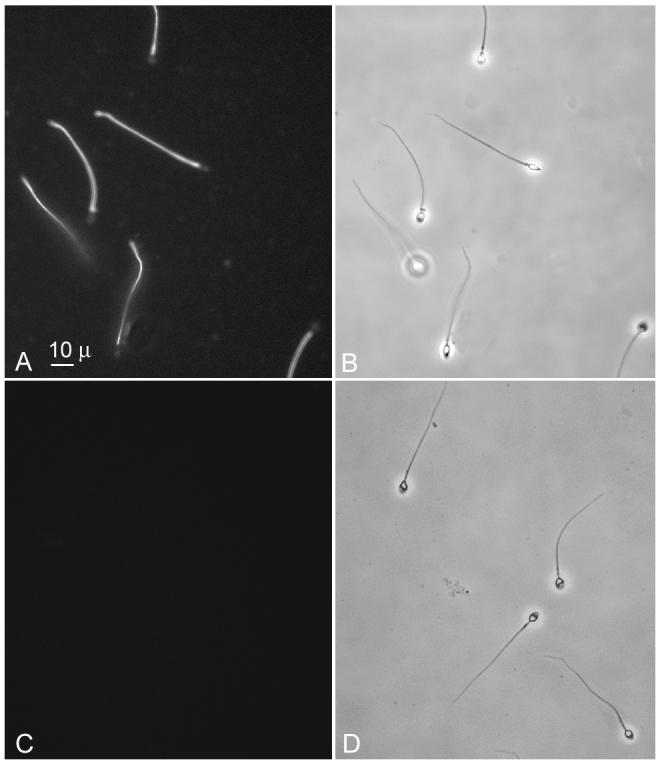
Indirect immunofluorescence of swim-up, permeabilized human sperm, including immunofluorescence staining for meichroacidin (A and C) and phase contrast pictures (B and D). The sperm were permeabilized with 0.1% saponin before probing with serum. The immune serum (A) stained the entire sperm tail including middle piece, principal piece and end piece. No immunostaining was observed with preimmune serum (C).
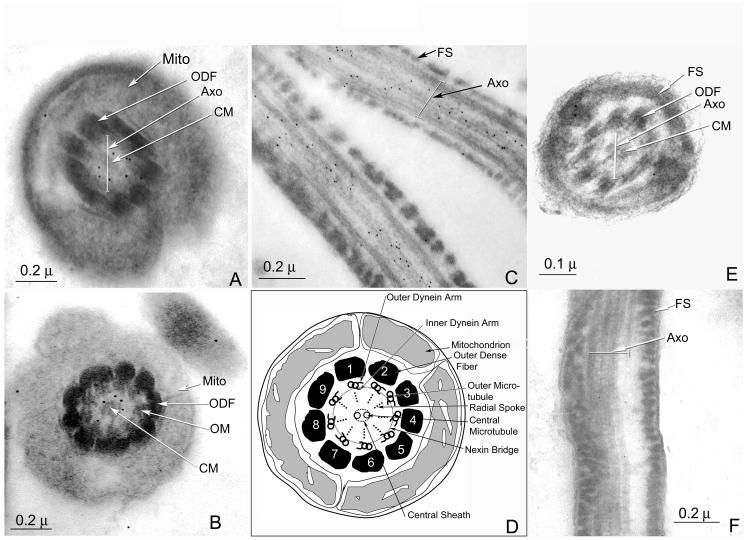
Electron microscopic immunostaining of hMCA using secondary antibody conjugated to 10 nm gold particles localized the protein to the axonemal complex [inside the outer dense fibers along the entire sperm tail] as seen in longitudinal sections (Fig. 6C). Further examination of cross sections of the axonemal complex (Fig. 6A and B) localized the protein to the interval between the central pair and outer doublet microtubules, which corresponds to the location of the radial spokes (Fig. 6D).
Fig. 6.
Immunoelectron microscopy of human spermatozoa: A and B: cross sections of the mid-piece of a human sperm tail, stained with anti-RSP44 antibody. Inside the layer of outer dense fibers, note the gold particles in the axoneme between the central microtules and the outer doublet of microtubules corresponding to the region occupied by the radial spokes. C: A longitudinal section of the principal piece of a human spermatozoon stained with anti-RSP44 serum. Note the gold particles associated with axonemal structure of the sperm tail. D: A schematic diagram of a cross section of the mid-piece of a human sperm illustrating the ultrastructural details of the axoneme (drawn based on Fawcett, 1975). E and F: A cross section (E) and a longitudinal section (F) of the principal piece of the human spermatozoon stained with pre-immune serum reveals absence of gold particles. FS: Fibrous sheath, ODF: outer dense fiber, Axo: Axoneme, Mito: Mitochondrion.
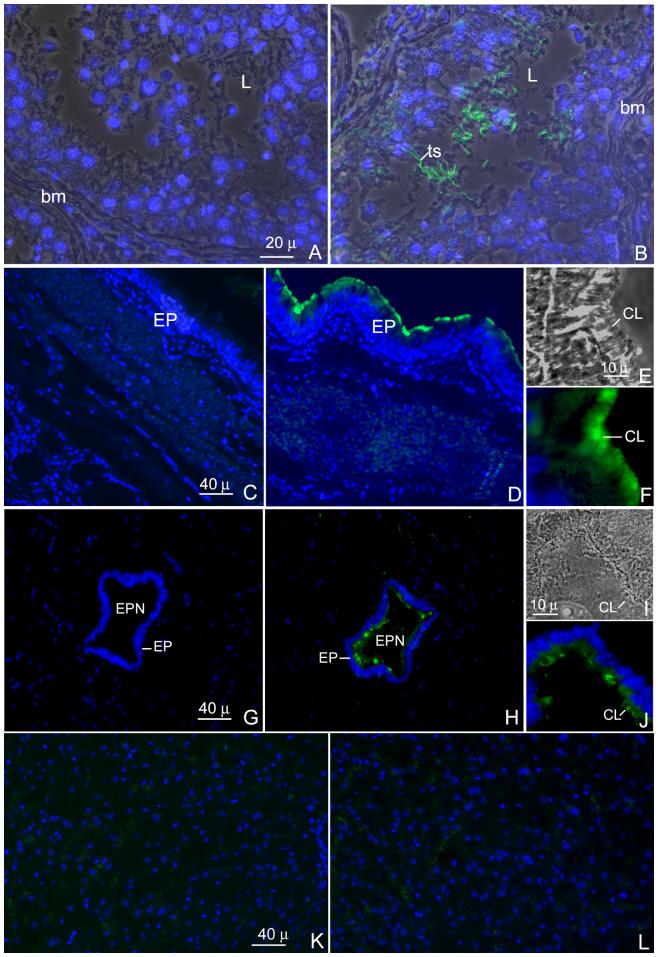
3.6. Localization of hMCA in the seminiferous epithelium, tracheal epithelium and central canal of spinal cord
Fig. 7 (A-L) shows the immunofluorescent localization of h-meichroacdin on paraffin sections of human testis, trachea, spinal cord and liver using the antibody generated to recombinant hMCA. The antibody stained the tails of the spermatids and testicular spermatozoa within the lumen of the seminiferous tubules (Fig. 7 A and B), indicative of a post-meiotic pattern of expression of hMCA in late stages of spermiogenesis. No nuclear staining in spermatocytes or spermatids was noted. In the trachea, hMCA was restricted to the cilia (Fig. 7 E) at the apical surface of the tracheal epithelium lining the lumen (Fig. 7 C-F). Sections treated with the preimmune serum did not show any immunoreactivity, verifying specificity of the anti-hMCA antibody (Fig. 7C). Fig. 7 G-J shows the immunofluorescent localization of h-meichroacidin on paraffin sections of human spinal cord including a region of the central canal. The antibody localized h-meichroacidin to the apical surface of the epithelium (ependyma) of the central canal where staining was restricted to the ependymal cilia (Fig. 7 I). Sections of adult human liver treated with immune sera (Fig. 7 L) was comparable to the pre-immune control (Fig. 7 K) showing no immunoreactivity.
Fig. 7.
Indirect immmunofluorescent staining of sections of human testis (A and B), trachea (C-F) spinal cord (G-J) and liver (K and L). Sections were probed with either rat anti-meichroacidn sera (B, D and H) or pre-immune sera (A, C and G) and prepared for indirect immunofluorescence staining. The same sections were counterstained with DAPI II for nuclear staining. The antibody mainly stained the tails of the testicular spermatozoa (ts) in the lumen (L) of the seminiferous tubule. In the trachea, a region at the luminal surface of the epithelium corresponding to the location of ciliary structures was stained by the antibody (D and F). Similarly the cilia of the ependymal epithelium lining the central canal of the spinal cord were stained with the antibody (H and J). Figures E and I represent phase contrast images showing magnified views of a portion of the ciliated epithelium of the trachea and ependyma respectively, F and J depict the corresponding images stained with the RSP44 antibody showing staining of the cilia (CL). The liver section did not show any immunoreactivity when stained with the immune serum (L) and was comparable to the pre-immune control (K).
3.7. RT-PCR Analysis
In order to further demonstrate the possible expression of hMCA in different ciliated tissues and also to find out the level of expression, a semi-quantitative RT-PCR was performed. As shown in Fig. 8 A, an expected PCR product of 546 bp was formed in all the ciliated tissues tested. No product was generated from the cDNA derived from the liver RNA, confirming that message is not produced in liver. After normalizing against the level of G3PDH, expression level was highest in trachea and testis followed by fallopian tube, spinal cord, brain, lungs, and thyroid (Fig. 8 B).
Fig. 8.
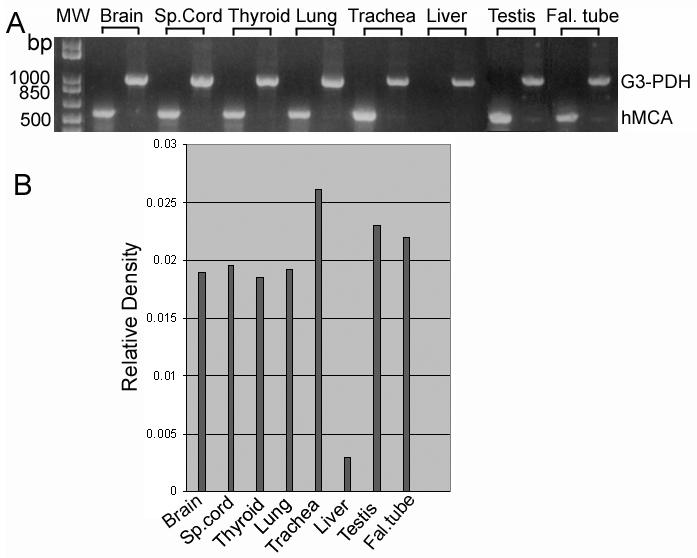
Semi-quantitative Reverse transcription polymerase chain reaction using total RNA from different ciliated tissues (A). Products generated by primer sets for hMCA were normalized against G3-PDH mRNA levels in the same set of samples. B) The relative densities of each band is plotted to show the level of message in different tissues.
3.8. Recombinant hMCA Reacted with the ASA Positive Infertile Sera
To determine if epitopes recognized by ASA positive infertile serum were retained on recombinant hMCA, western blot analyses (Fig. 9) were performed with the sera that had reacted on 2-D gels with the native MCA spots at ∼44 kDa (Fig. 1B). The recombinant hMCA with a histidine tag shows a major molecular weight of ∼44 k Da accompanied by several lower forms as detected by Coomassie staining (Fig. 9 A) as well as by Western blotting with anti-histidine antibody (Fig. 9 B). The ASA positive infertile serum that was originally used to identify “native” hMCA for microsequencing bound the purified recombinant hMCA at ∼44kDa and also detected a few of the lower forms (Fig. 9 D), as did the rat anti hMCA antibody (Fig. 9 C). The absence of reactivity in the first lane in Fig. 9 D, which represents one of the blots probed with fertile sera without antisperm antibody, shows that reactivity of the infertile serum is specific for the recombinant hMCA. These results confirmed that native hMCA is an alloantigenic protein and that recombinant hMCA retained epitopes recognized by antibodies to the “native” molecule.
Fig. 9.
The antisperm antibody positive infertile serum that led to the identification of RSP44 also reacted with the recombinant protein. RSP44 with a C-terminal histidine tag was expressed in E.coli using the pET28b plasmid, induced by addition of 1.0 mM IPTG, and purified by nickel ion affinity chromatography followed by Prep cell. A) Bacterial extracts and purified rec RSP44 stained with Coomassie blue. B) Immunoblot of bacterial extracts and purified rec RSP44 with anti-His antibody. C) Immunoblot of purified rec RSP44 with rat antisera against rec RSP44. D) Immunoblot of purified rec RSP44 with the infertile male serum which was originally used to identify RSP44 in a 2-D immunoblot of human sperm proteins. The infertile serum recognized the recombinant protein showing a profile similar to that shown in panels A, B and C, with a major band at ∼44k Da and few lower forms. The fertile serum (D) did not result in any signal with the rec RSP44 confirming that reactivity of the infertile serum is specific.
4. Discussion
Our findings on human MCA show differences from those previously reported for the mouse that are of sufficient magnitude to indicate that a change in the designation of the human gene is warranted. At the outset of this study nucleotide and amino acid sequences for the protein under investigation were not present in the database. A mouse orthologue of this gene was subsequently deposited under the name TSGA2 to signify ‘testis specific gene A2’ after its discovery by using polyclonal antibodies raised against testicular antigens (Taketo et al., 1997). A report on the cloning and characterization of the mouse orthologue then appeared that applied the name meichroacidin, which signified male meiotic metaphase chromosome- associated acidic protein, due to its association with metaphase chromosomes and spindles during meiotic division (Tsuchida et al., 1998). Meichroacidin was reported to undergo changes in its subcellular distribution, cycling between the cytoplasm and the metaphase chromosomes during meiosis. Based upon the report on the mouse (Tsuchidaet al., 1998), the human orthologue of the mouse gene was then designated “TSGA2 homolog (mouse)” and the protein was named human meichroacidin. Mouse meichroacidin was reported to be germ cell specific, with expression exclusively in the testis and ovary, although there was much less in the ovary (Tsuchidaet al., 1998). In the testis, the protein was reported to be present in germ cells from pachytene spermatocytes through round spermatids. A study on the human orthologue of meichroacidin was later reported from the same group (Matsuka et. al., 2005) which provided evidence showing the localization of the protein in the sperm tail. It also reported that the gene was testis specific in expression and indicated that the protein is probably involved in the formation of the sperm flagellum.
Compared to the earlier findings on the mouse and human MCA our studies of human MCA show several important differences and also some additional information on the characterization of the protein. First, hMCA was not germ cell specific at either the mRNA or protein level, being present in humans in brain, spinal cord, thyroid, oviduct, trachea, lung in addition to testes. Second, hMCA was not present in the nucleus of male germ cells to any appreciable extent as reported for its mouse counterpart. It was localized to the developing flagella of spermatids and testicular sperm, indicating that the protein is expressed during the steps of spermiogenesis. Immunocytochemistry of hMCA both at the light and electron microscopic levels precisely localized the protein to the region of the radial spokes in the axonemal compartment of the tail of ejaculated sperm. Third, the antibody to the recombinant hMCA stained tracheal and ependymal cilia, confirming that this protein is common to both cilia and flagella. RT-PCR experiments also confirmed these findings showing the mRNA message in all the ciliated tissues tested. We believe that in humans meichroacidin's primary role is in the axoneme of cilia and flagella and suggest that the protein will be expressed in those tissues is which cilia and flagella are prominent. We find no evidence in humans for a role in the meiotic nucleus as was proposed for m-meichroacidin in the mouse testis. Our experimental observations are supported by bioinformatic analyses which show that the protein is conserved among members of different plant and animal phyla, including unicellular ciliates and flagellates. Taken together, our findings call for adoption of a different name for h-meichroacidin. We propose radial spoke protein 44 (RSP44) as a more accurate designation for the human meichroacidin (MCA) protein and suggest that RSP44 or a similar term be assigned by the Human Genome Nomenclature Committee to the gene since the current name TSGA2 suggests incorrectly that it is a testis specific gene. Our observations differed from the previous observations of of h-MCA (Matsuka et. Al., 2005) who reported the testis specific expression of the protein at both transcriptional and translational level. However our observation is in agreement with theirs showing that the protein is localized to the tail in the human sperm and extend their observations to reveal that the protein is associated with the radial spokes.
The occurrence of MORN repeat structures may provide clues as to the role of RSP44 in cilia and flagella. This repeat sequence of Y-x-G-x-x-x-x-x-x-x-(x)H-G-x-G-x-x-x-x-x-x-x-x-x (Fig. 2A) with regular appearance of glycine residues may constitute a functional domain as proposed earlier (Tsuchidaet al., 1998). The presence of a highly acidic region towards the C-terminus of RSP44 with multiple glutamic acid residues at positions 181-300 (Fig. 2A) may facilitate interaction with basic proteins. The potential phosphorylation sites for casein kinase II (positions 88 – 91, 189 – 192, 229 – 232, 243 – 246, and 297 – 300) and tyrosine tyrosine kinase at position 37-44 suggest that the function of RSP 44 might be regulated by these protein kinases and that RSP44 may be involved in one or more paths of signal transduction.
RSP44 also showed sequence similarities to phoshatidyl inositol 4 phosphate 5-kinase (PIP5K) and junctophilins, mainly because of the presence of the MORN repeat structures. However, RSP44 lacked the kinase homology domain of PIP5K (Anderson et al., 1999) and the c-terminal transmembrane domain found in all the known junctophilins (Takeshima et al., 2000). In junctophilins, a family of proteins that are components of junctional membrane complexes, the MORN motifs are known to contribute to the plasma membrane binding capacity of the protein and are believed to mediate cross-talk between cell surface and intracellular ion channels in the endoplasmic/sarcoplasmic reticulum (Takeshima et al., 2000). Interestingly a data base search using the InterProScan tool (European Bioinformatic Institute) to find proteins with MORN domains revealed that the majority of proteins with this domain are uncharacterized molecules belonging to species from both plant and animal kingdoms including unicellular organisms. Analysis of those proteins that show significant identity to the RSP44 in their MORN domains indicates the existence of orthologues of RSP44 in primates, ruminants, carnivores, amphibians, teleosts (Ju et al., 2004) and ascidians (Satouh et al., 2005) among chordates, and echinoderms, arthropods, and flat worms among non-chordates. In addition, a wide range of unicellular organisms also show significant identity to RSP44, including Leishmania, Tetrahymena, Trypanosoma, Plasmodium, Chlamydomonas (Yang et al., 2006), and Paramecium, all of which have motile cilia or flagella in at least part of their life cycle. These observations suggest that RSP44 has a functional role in ciliary and flagellar motility that has been conserved through evolution.
The fine structural localization of RSP44 to the axonemal complexes in the flagella of ejaculated sperm and fluorescent staining of testicular sperm flagella (Figs.5, 6, 7A&B) coupled with its localization by immunofluorescence to cilia of the trachea (Fig. 7 C-F) and ependyma (Fig. 7 G-J) confirm that RSP44 is a previously unrecognized protein component of cilia in several tissues. This is supported by Western blot data demonstrating RSP44 in protein extracts of trachea, thyroid and spinal cord which have ciliated epithelia. Although the text book descriptions of the thyroid gland generally do not report this epithelium to be ciliated, one or more cilia are indeed present at the center of almost every epithelial cell in the normal human thyroid gland as well as in most goiters (Nesland et al., 1987) where they are thought to function to keep the colloid from stratifying.
Ultrastructural examination of RSP44 precisely localized the protein to the region of the radial spokes of the human sperm axomene. This observation correlates with the identification of a possible orthologue of RSP44 in the radial spokes of the axonemes of sperm from the ascidian Ciona intestinalis (Satouh et al., 2005). The ascidian protein, named CiMORN40, was suggested to participate in the Hsp40-mediated molecular chaperone system in the assembly and motility of axonemes as well as in signal transduction for the modulation of dynein activity in the radial spokes. A possible orthologue of RSP44 has also been identified in the carp Cyprinus carpio as a sperm axonemal component (Ju et al., 2004). Identification of at least two radial spoke proteins from Chlamydomonas reinhardtii (radial spoke protein 1 and radial spoke protein 10 (Yang et al., 2006) showing similarity to RSP44, particularly in the regions containing multiple MORN repeats, underscores the functional significance of such proteins in the radial spokes of axonemes across species. RSP1 is reported to be localized to the radial spoke head in Chlamydomonas (Yang et al., 2006).
The deduced molecular weight of RSP44 (35.1 k Da) is lower than the estimated molecular weight (44) observed for the native protein. One reason may be the acidic character of the protein, a feature known to lead to overestimation of molecular weight due to the interference of negatively charged molecules with SDS binding (Tsuchida et al., 1998). Differences between western blot profiles of testicular and sperm proteins (Fig. 4) suggest that the RSP44 undergoes post-translation modification, with the addition of a lower molecular weight band in the case of sperm. The hypothesis that this change might be related to capacitation, however, was not confirmed by Western blot analysis which revealed no difference between capacitated and non-capacitated specimens.
Reports of human phenotypes showing immotile sperm in conjunction with defective mucociliary clearance (Sturgess et al., 1979) and the lack of radial spokes in both the sperm tail and tracheal cilia help to establish the common function of radial spoke proteins. The importance of the radial spokes in flagellar motility is further highlighted by several reports of paralyzed or abnormal motility in certain mutants of Chlamydomonas reinhardtii, in which abnormal motility is related to the lack of the entire radial spoke or a part of radial spoke head (Witman et al., 1978; Huang et al., 1981).
In our study RSP44 (hMCA) was discovered in ejaculated spermatozoa by microsequencing following identification of the alloantigenic properties of the protein in 2 of 14 ASA positive serum samples. The reactivity of anti-recombinant RSP44 to the same immunoreactive spot as immunoinfertile serum confirmed that the authentic alloantigenic protein had been cloned and characterized. Patient sera recognized both native RSP44 as well as recombinant RSP44 indicating that immunogenic epitopes in humans were retained in the recombinant protein. This suggests that the recombinant protein might be a suitable biomarker for monitoring the anti-sperm responses of some patients. However, irrespective of the potential of the recombinant protein as a diagnostic tool for ASA, it is difficult to envisage how antibodies to RSP44 cause infertility in patients due to the likely inaccessibility of the antigen deep within the axonemal complex. Although several human sperm auto- and iso-antigens are recognized by infertile patient sera containing ASA (e.g., SAGA-1, SPAG2, SP17, FA1, AKAP3) (Diekman et al., 1997, 1998; Lea et al., 1997;Naz et al., 1984; Mandal et al., 1999) only a very few have been found on the spermatozoal surface (e.g., SAGA-1) (Diekman et al., 1997) a critical criterion for considering its potential involvement in fertility. RSP44 belongs to the group of internal alloantigens which is probably not involved in antibody-mediated infertility based on the current understanding of the physiology of fertilization.
In summary, RSP44 is a member of the growing group of axonemal component proteins now estimated to comprise > 250 members (Ostrowski et al., 2002; Azour et al., 2005; Nicastro et al., 2006). Bioinformatic comparisons support the conclusion that RSP44 is an “ancestral gene” conserved during evolution and present in mammalian sperm. The central role of the axoneme in flagellar movement and the localization of RSP44 to the radial spokes suggest sperm motility and ciliary movement are important new dimensions in considering the functions of this recently annotated protein.
Acknowledgements
We thank Dr. M. Beatriz Lopes, Department of Pathology, University of Virginia, for providing paraffin sections of adult human spinal cord for the immunolocalization of RSP44.
This work was supported by grants from D43 TW/HD 00654 from the Fogarty International Center, National Institutes of Health Grant HD U54 29099, the Andrew W. Mellon Foundation, Schering A.G., and Office of Justice Programs, National Institute of Justice, United States Department of Justice Grant 2000-IJ-CX-K013.
Abbreviations
- RSP44
radial spoke protein 44
- TSGA2
Testis specific gene A2
- hMCA
human meichroacidin
- MORN
membrane occupation recognition nexus
- EM
electron microscopy
- PCR
polymerase chain reaction
- BWW
Biggers, Whitten and Whittingham
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson RA, Boronenkov IV, Doughman SD, Kunz J, Loijens JC. Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J Biol Chem. 1999;274:9907–9910. doi: 10.1074/jbc.274.15.9907. [DOI] [PubMed] [Google Scholar]
- Bouchard P, Cosson MP. Molecular basis of sperm movement. Ann. Endocrinol. (Paris) 1981;42:398–406. [PubMed] [Google Scholar]
- Bronson RA, Cooper GW, Rosenfeld DL. Correlation between regional specificity of antisperm antibodies to the spermatozoan surface and complement-mediated sperm immobilization. Am. J. Reprod. Immunol. 1982;2:222–224. doi: 10.1111/j.1600-0897.1982.tb00170.x. [DOI] [PubMed] [Google Scholar]
- Bronson RA, Tung KSK. Human sperm antibodies and their detection. In: Rose NR, de Macario ER, editors. Am. Soc. for Microbiol. 4th edition Manual of Clinical Laboratory Immunology; Washington, DC: 1992. pp. 1013–1017. [Google Scholar]
- Caron CP, Saling PM. Sperm antigens and immunological interference of fertilization. In: Wassarman PM, editor. Elements of Mammalian Fertilization. CRC Press; Boca Raton, FL: 1991. pp. 147–176. [Google Scholar]
- Celis JE, et al. The human keratinocyte two-dimensional gel protein database (update 1992): towards an integrated approach to the study of cell proliferation, differentiation and skin diseases. Electrophoresis. 1992;13:893–959. doi: 10.1002/elps.11501301198. [DOI] [PubMed] [Google Scholar]
- D'Cruz OJ, Hass GG, Wang B, DeBault LE. Activation of human complement by IgG antisperm antibody and the demonstration of C3 and C5b–9-mediated immune injury to human sperm. J. Immunol. 1991;146:611–620. [PubMed] [Google Scholar]
- Diekman AB, Westbrook-Case VA, Naaby-Hansen S, Klotz KL, Flickinger CJ, Herr JC. Biochemical characterization of sperm agglutination antigen-1, a human sperm surface antigen implicated in gamete interactions. Biol. Reprod. 1997;57:1136–1144. doi: 10.1095/biolreprod57.5.1136. [DOI] [PubMed] [Google Scholar]
- Diekman AB, Olson G, Goldberg E. Expression of the human antigen SPAG2 in the testis and localization to the outer dense fibers in spermatozoa. Mol. Reprod. Dev. 1998;50:284–293. doi: 10.1002/(SICI)1098-2795(199807)50:3<284::AID-MRD4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Egydio de Carvalho C, Tanaka H, Iguchi N, Ventela S, Nojima H, Nishimune Y. Molecular cloning and characterization of a complementary DNA encoding sperm tail protein SHIPPO 1. Biol Reprod. 2002;66:785–795. doi: 10.1095/biolreprod66.3.785. [DOI] [PubMed] [Google Scholar]
- Fawcett DW. The mammalian spermatozoon. Dev Biol. 1975;44:394–436. doi: 10.1016/0012-1606(75)90411-x. [DOI] [PubMed] [Google Scholar]
- Hao Z, Wolkowicz MJ, Shetty J, Klotz K, Bolling L, Sen B, Westbrook VA, Coonrod S, Flickinger CJ, Herr JC. SAMP32, a testis-specific, isoantigenic sperm acrosomal membrane-associated protein. Biol Reprod. 2002;66:735–744. doi: 10.1095/biolreprod66.3.735. [DOI] [PubMed] [Google Scholar]
- Haas GG., Jr. How should sperm antibody tests be used clinically? Am.J. Reprod. Immunol. Microbiol. 1987;15:106–111. doi: 10.1111/j.1600-0897.1987.tb00166.x. [DOI] [PubMed] [Google Scholar]
- Huang B, Piperno G, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Bio. 1981;88:80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju TK, Huang FL. MSAP, the meichroacidin homolog of carp (Cyprinus carpio), differs from the rodent counterpart in germline expression and involves flagellar differentiation. Biol Reprod. 2004;71:1419–1429. doi: 10.1095/biolreprod.104.030346. [DOI] [PubMed] [Google Scholar]
- Lea IA, Adoyo P, O′Rand MG. Autoimmunogenicity of the human sperm protein SP17 in vasectomized men and identification of linear B cell epitopes. Fertil. Steril. 1997;67:355–361. doi: 10.1016/S0015-0282(97)81923-1. [DOI] [PubMed] [Google Scholar]
- Lee C-YG, Liu M-S, Su M-W, Zhu J-B. Studies of sperm antigens reactive to HS-11 and HS-63 monoclonal antibodies. In: Alexander NJ, Griffin D, Spieler JM, Waites GMH, editors. Gamete Interaction: Prospects for Immunocontraception. Wiley-Liss, Inc.; New York: 1990. pp. 37–52. [Google Scholar]
- Luconi M, Krausz C, Forti G, Baldi E. Extracellular calcium negatively modulates tyrosine phosphorylation and tyrosine kinase activity during capacitation of human spermatozoa. Biol Reprod. 1996;55:207–216. doi: 10.1095/biolreprod55.1.207. [DOI] [PubMed] [Google Scholar]
- Mandal A, Naaby-Hansen S, Wolkowicz MJ, Klotz K, Shetty J, Retief JD, Coonrod SA, Kinter M, Sherman N, Cesar F, Flickinger CJ, Herr JC. FSP95, a testis-specific 95-kilodalton fibrous sheath antigen that undergoes tyrosine phosphorylation in capacitated human spermatozoa. Biol Reprod. 1999;61:1184–1197. doi: 10.1095/biolreprod61.5.1184. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Nishimura H, Numazawa K, Tsuchida J, Miyagawa A, Tsujimura A, Matsumiya K, Okuyama A, Nishimune Y, Tanaka H. Sperm glagella protein components: Human meichroacidin constructed by the membrane occupation and recognition nexus motif. Reprod. Med. and Biol. 2005;4:213–219. doi: 10.1111/j.1447-0578.2005.00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Tanaka H, Koga M, Miyagawa Y, Iguchi N, Egydio de Carvalho C, Yomogida K, Nozaki M, Nojima H, Matsumiya K, Okuyama A, Nishimune Y. Molecular cloning and characterization of oppo 1: a haploid germ cell-specific complementary DNA encoding sperm tail protein. Biol Reprod. 2002;67:1–7. doi: 10.1095/biolreprod67.1.1. [DOI] [PubMed] [Google Scholar]
- Nakano I, Kobayashi T, Yoshimura M, Shingyoji C. Central-pair-linked regulation of microtubule sliding by calcium in flagellar axonemes. J. Cell Sci. 2003;116:1627–1636. doi: 10.1242/jcs.00336. [DOI] [PubMed] [Google Scholar]
- Naz RK, Alexander NJ, Isahakia M. Hamilton MS. Monoclonal antibody to a human germ cell membrane glycoprotein that inhibits fertilization. Science. 1984;225:342–344. doi: 10.1126/science.6539947. [DOI] [PubMed] [Google Scholar]
- Nesland JM, Sobrinho-Simoes M, Johannessen JV. Scanning electron microscopy of the human thyroid gland and its disorders. Scanning Microsc. 1987;1:1797–1810. [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;18:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- O'Farrel PZ, Goodman HM, O'Farrel PH. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1142. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol. Cell Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennarun G, Bridoux AM, Escudier E, Dastot-Le Moal F, Cacheux V, Amselem S, Duriez B. Isolation and expression of the human hPF20 gene orthologous to Chlamydomonas PF20: evaluation as a candidate for axonemal defects of respiratory cilia and sperm flagella. Am. J. Respir. Cell Mol. Biol. 26:362–370. doi: 10.1165/ajrcmb.26.3.4738. [DOI] [PubMed] [Google Scholar]
- Satouh Y, Padma P, Toda T, Satoh N, Ide H, Inaba K. Molecular characterization of radial spoke subcomplex containing radial spoke protein 3 and heat shock protein 40 in sperm flagella of the ascidian Ciona intestinalis. Mol. Biol. Cell. 2005;16:626–636. doi: 10.1091/mbc.E04-09-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Tarnasky HA, Lee JP, Oko R, van der Hoorn FA. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev. Biol. 1999;211:109–123. doi: 10.1006/dbio.1999.9297. [DOI] [PubMed] [Google Scholar]
- Shetty J, Diekman AB, Jayes FC, Sherman NE, Naaby-Hansen S, Flickinger CJ, Herr J,C. Differential extraction and enrichment of human sperm surface proteins in a proteome: identification of immunocontraceptive candidates. Electrophoresis. 2001;22:3053–3066. doi: 10.1002/1522-2683(200108)22:14<3053::AID-ELPS3053>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Shetty J, Naaby-Hansen S, Shibahara H, Bronson R, Flickinger CJ, Herr JC. Human sperm proteome: immunodominant sperm surface antigens identified with sera from infertile men and women. Biol. Reprod. 1999;61:61–69. doi: 10.1095/biolreprod61.1.61. [DOI] [PubMed] [Google Scholar]
- Shetty J, Wolkowicz MJ, Digilio LC, Klotz KL, Jayes FL, Diekman AB, Westbrook VA, Farris EM, Hao Z, Coonrod SA, Flickinger CJ, Herr JC. SAMP14, a novel, acrosomal membrane-associated, glycosylphosphatidylinositol-anchored member of the Ly-6/urokinase-type plasminogen activator receptor superfamily with a role in sperm-egg interaction. J Biol. Chem. 2003;278:30506–30515. doi: 10.1074/jbc.M301713200. [DOI] [PubMed] [Google Scholar]
- Sturgess JM, Chao J, Wong J, Aspin N, Turner JA. Cilia with defective radial spokes: a cause of human respiratory disease. N. Engl. J. Med. 1979;300:53–56. doi: 10.1056/NEJM197901113000201. [DOI] [PubMed] [Google Scholar]
- Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- Taketo, et al. Mapping of eight testis-specific genes to mouse chromosomes. Genomics. 1997;46:138–142. doi: 10.1006/geno.1997.5014. [DOI] [PubMed] [Google Scholar]
- Tsuchida J, Nishina Y, Wakabayashi N, Nozaki M, Sakai Y, Nishimune Y. Molecular cloning and characterization of meichroacidin (male meiotic metaphase chromosome-associated acidic protein) Dev. Biol. 1998;197:67–76. doi: 10.1006/dbio.1998.8885. [DOI] [PubMed] [Google Scholar]
- Witkin SS, Kligman I, Bongiovanni AM. Relationship between an asymptomatic male genital tract exposure to Chlamydia trachomatis and an autoimmune response to spermatozoa. Hum. Reprod. 1995;10:2952–2955. doi: 10.1093/oxfordjournals.humrep.a135827. [DOI] [PubMed] [Google Scholar]
- Witman GB, Plummer J, Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. J. Cell Biol. 1978;76:729–747. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowicz MJ, Shetty J, Westbrook A, Klotz K, Jayes F, Mandal A, Flickinger CJ, Herr JC. Equatorial segment protein defines a discrete acrosomal subcompartment persisting throughout acrosomal biogenesis. Biol. Reprod. 2003;69:735–745. doi: 10.1095/biolreprod.103.016675. [DOI] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, Rosenbaum JL, Witman GB. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]