Abstract
DNA topoisomerases solve topological problems during chromosome metabolism. We investigated where and when Top1 and Top2 are recruited on replicating chromosomes and how their inactivation affects fork integrity and DNA damage checkpoint activation. We show that, in the context of replicating chromatin, Top1 and Top2 act within a 600-base-pair (bp) region spanning the moving forks. Top2 exhibits additional S-phase clusters at specific intergenic loci, mostly containing promoters. TOP1 ablation does not affect fork progression and stability and does not cause activation of the Rad53 checkpoint kinase. top2 mutants accumulate sister chromatid junctions in S phase without affecting fork progression and activate Rad53 at the M–G1 transition. top1 top2 double mutants exhibit fork block and processing and phosphorylation of Rad53 and γH2A in S phase. The exonuclease Exo1 influences fork processing and DNA damage checkpoint activation in top1 top2 mutants. Our data are consistent with a coordinated action of Top1 and Top2 in counteracting the accumulation of torsional stress and sister chromatid entanglement at replication forks, thus preventing the diffusion of topological changes along large chromosomal regions. A failure in resolving fork-related topological constrains during S phase may therefore result in abnormal chromosome transitions, DNA damage checkpoint activation, and chromosome breakage during segregation.
Keywords: Replication forks, DNA damage checkpoint, chromosome replication, DNA topoisomerases
Chromosome replication is coordinated with transcription, recombination, chromatin remodeling, cohesion, and checkpoints. Several events can compromise the integrity of replicating chromosomes, such as misincorporation of dNTPs, DNA damage, interference between replication and transcription, and unscheduled recombination processes. Furthermore, the advancement of replication forks generates tremendous topological constraints that must be solved by DNA topoisomerases, specialized enzymes that act by nicking and closing the DNA strands. A failure to properly coordinate replication fork progression with the topoisomerase-mediated processes that relieve topological constrains may cause fork collapse and double-strand break (DSB) formation.
Several studies indicate that DNA topoisomerases catalyze strand passage reactions, thus changing the linkage of DNA molecules (Wang 1996; Champoux 2001), and can alleviate the topological problems generated during DNA replication (Fig. 1A). Unwinding of the DNA duplex by replicative helicases creates a compensatory increase in the intertwining of parental strands (Wang 2002; Postow et al. 2004). The mechanical strain thus generated can be converted into helical overwinding (positive supercoiling) of the unreplicated portions of the DNA ahead of the forks. It has been proposed that this mechanical strain can be also transmitted to replicated DNA by rotation at the replication fork branching point, thus generating intertwining of the daughter duplexes (known as precatenates). Accumulation of positive supercoiling thermodynamically counteracts the action of replicative DNA helicases. The precatenate nodes, if not resolved, would establish a physical link between the sister chromatids that can impede their accurate segregation during mitosis (Champoux and Been 1980; Wang 2002; Postow et al. 2004). Topological constraints generated by the tracking action of the replication machinery are thought to be confined to closed topological domains limited by physical barriers that restrict the diffusion of torsional changes to larger chromosomal regions (Postow et al. 2001, 2004). The mechanisms controlling the architecture of the topological domains of the chromosomes protect chromatin integrity and prevent unscheduled changes in the transcriptional program of the cell (Postow et al. 2004). DNA topoisomerases have been classified into two different categories: type I enzymes, which mediate the transient breakage of one DNA strand at a time; and type II enzymes, which generate nicks in both DNA strands. Among the first class, type IB DNA topoisomerases (like budding yeast Top1) are very efficient in relaxing positively supercoiled naked DNA molecules and could contribute to the removal of positive supercoils accumulating at unreplicated regions. Type II enzymes (like yeast Top2) can relax positively overwounded DNA molecules and could also reduce the torsional stress by resolving sister chromatid intertwining at precatenate nodes (Postow et al. 2001; Wang 2002). Type II enzymes are also involved in chromosome condensation and segregation during mitosis (Holm et al. 1985, 1989; Uemura et al. 1987). Type IA enzymes (such as budding yeast Top3) seem to be preferentially implicated in recombinational DNA repair (Wallis et al. 1989; Zhu et al. 2001), although it has been proposed that they could also act in replication termination (Postow et al. 2001; Wang 2002).
Figure 1.
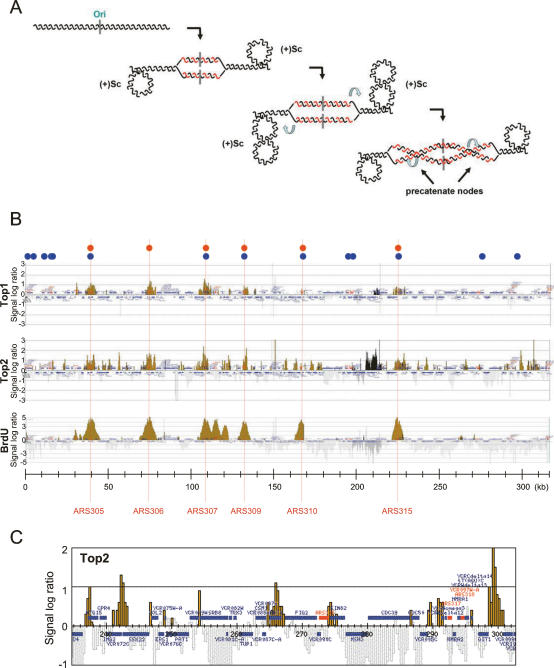
Top1 and Top2 localization at replicating chromosome III. (A) Schematic representation of the topological problems generated by advancing replication forks: Positive supercoiling [(+)Sc] at the unreplicated portions of a topological domain is generated by fork advance; precatenane nodes, at the replicated duplex, arise as a consequence of fork rotation. (B) Top1-3xFlag (CY6838), Top2-3xFlag (CY6839), and BrdU-incorporating (SY2201) cells were released from α-factor-induced G1 block, treated with 0.2 M HU for 1 h, and processed for ChIP with antibodies specific to the Flag epitopes or BrdU-substituted DNA. Black and yellow histogram bars in the Y-axis show the average signal ratio of loci significantly enriched in the immunoprecipitated fraction along chromosome III in log2 scale. Black bars correspond to a region represented at higher resolution in the oligonucleotide array, and the histograms are therefore more compacted. The X-axis shows kilobase units. Positions of ARS elements acting as efficient early-firing replication origins on the chromosome are indicated. Red dots correspond to ssDNA-accumulating regions in HU-treated cells (Feng et al. 2006). Blue dots mark the position of potential origins of replication as defined by ORC and MCM protein binding (Wyrick et al. 2001). (C) Magnification of a ∼70-kb region close to the right telomere of chromosome III showing non-origin-related Top2 binding. The horizontal bars indicate ORFs.
The current view assumes that positive supercoiling generated by helix unwinding is mainly relaxed by type IB enzymes, presumably anywhere within the unreplicated region of the topological domain (Postow et al. 2001; Schvartzman and Stasiak 2004). The contribution of the Type II enzymes to the resolution of topological stress during chromosome replication in vivo is less clear. Yeast cells can replicate DNA without Top1 or in top2 mutants, but not when both Top1 and Top2 functions are defective (Brill et al. 1987; Kim and Wang 1989), thus suggesting that, at least in the absence of Top1, Top2 performs an important role in assisting chromosome replication. Since Top2, in vitro, is not as efficient as Top1 in relaxing supercoils, it has been proposed that it could cooperate with the removal of torsional stress by resolving precatenates (Postow et al. 2001; Wang 2002). The latter possibility is supported by the finding that type II enzyme inhibition during plasmid replication in Xenopus egg extracts leads to the accumulation of highly linked replication intermediates (Lucas et al. 2001). These findings suggest that daughter chromatid decatenation may take place concomitantly with the progression of replication forks. Despite the tremendous effort in trying to elucidate how these important enzymes facilitate the process of DNA synthesis, their exact roles in DNA replication within the eukaryotic chromatin context is still elusive.
To investigate how DNA topoisomerases deal with the topological constrains of replicating chromosomes and their range of action within the topological domains generated by active replicons, we have analyzed the timing and the sites of recruitment of Top1 and Top2 on chromosomes. We have also investigated how Top1 and Top2 dysfunctions alter the dynamics of replication forks, and how the DNA damage checkpoint responds to the resulting topological abnormalities. We show that while both Top1 and Top2 localize in close proximity to replication forks, Top2 exhibits additional clusters at specific intergenic loci in unreplicated DNA regions, mostly at promoters. TOP1 ablation does not affect fork progression and stability and does not lead to activation of the Rad53-dependent DNA damage checkpoint. Conversely, top2 mutants accumulate during S-phase cruciform DNA structures tethering sister chromatids, without affecting fork progression, and activate Rad53 upon completion of mitosis. The simultaneous attenuation of Top1 and Top2 causes the block of replication forks, their aberrant mass reduction, and the generation of checkpoint signals leading to Rad53 activation in S phase. Ablation of the Exo1 exonuclease counteracts the mass reduction of replication intermediates in top1 top2 double mutants and delays the accumulation of active Rad53 in top1 top2 cells.
Altogether, our results indicate that Top1 and Top2 act coordinately within a 600-base-pair (bp) region spanning the moving forks to assist their progression and prevent their pathological processing. The fork-confined action of Top1 and Top2 would counteract the diffusion of topological changes along large chromosomal regions, thus protecting chromatin integrity, and would prevent the generation of aberrant replication intermediates causing DNA damage checkpoint activation and, possibly, chromosome breakage during segregation.
Results
Top1 and a subpopulation of Top2 localize at chromosomal regions undergoing replication
We have analyzed the chromosomal localization of DNA topoisomerases by chromatin immunoprecipitation (ChIP) followed by hybridization to a high-density oligonucleotide array (Katou et al. 2003, 2006). This method allows the detection of protein-binding sites on yeast chromosomes at a resolution of at least 600 bp (Katou et al. 2006).
Cells were presynchronized in G1 and then released from the G1 block in the presence of hydroxyurea (HU) for 1 h to slow down replication fork progression. We then analyzed the binding of Top1 and Top2 to chromosome III (Fig. 1B). Top1 binding was enriched specifically at those regions containing replication origins that fire efficiently and early during DNA synthesis (ARS305, ARS306, ARS307, ARS309, ARS310, and ARS315) (Poloumienko et al. 2001; Raghuraman et al. 2001; Feng et al. 2006). We did not detect any specific association of Top1 at those chromosomal regions that associate with initiation factors such as ORC and MCM proteins (Wyrick et al. 2001) but either fire late during S phase or remain dormant (Poloumienko et al. 2001). We found that the chromosomal binding of Top2 overlapped the Top1 signal around the same origin regions, although additional clusters were also detected along both chromosome arms, preferentially at intergenic regions (Fig. 1C). To correlate the chromosomal binding of Top1 and Top2 to regions exhibiting ongoing DNA synthesis, we first labeled newly synthesized DNA with BrdU and then performed ChIP analysis (Katou et al. 2003, 2006). We found that all the regions showing Top1 and Top2 binding and containing early replication origins also incorporated BrdU (Fig. 1B), thus indicating that the concomitant chromosomal recruitment of Top1 and Top2 correlates with the sites where DNA synthesis is ongoing.
Inactivating mutations in the ARS consensus sequences (ACSes) do not affect the Top2 intergenic clusters but prevent the recruitment of Top1 and Top2 at the origin regions
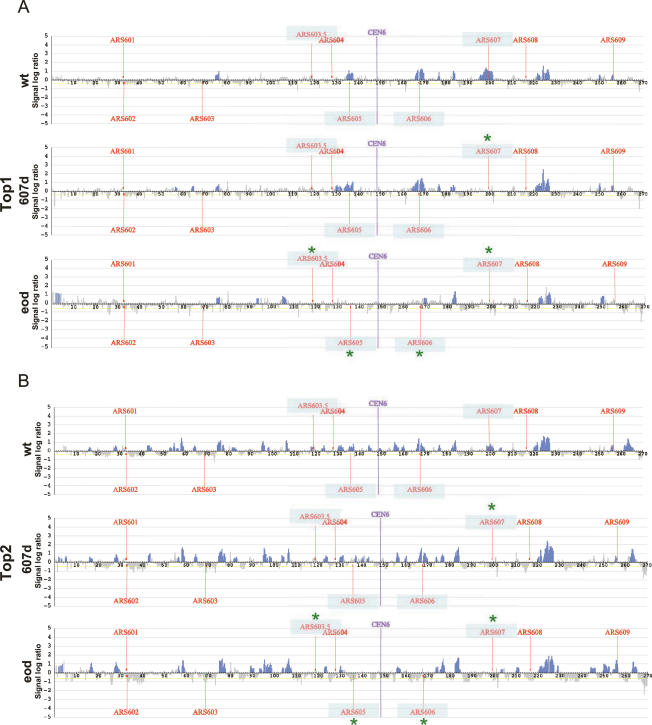
To firmly ascertain the specificity of Top1 and Top2 binding to replicating regions, and to address whether the additional Top2 clusters were depending on replicon dynamics, we have analyzed Top1 and Top2 localization in wild-type cells and in cells bearing either the disruption of ARS607 only (607d) or of all of the described early origins (eod, early origin disrupted) (ARS603.5, ARS605, ARS606, and ARS607) on chromosome VI (Fig. 2; Friedman et al. 1997). Inactivating mutations in the ACS (Shirahige et al. 1993; Yamashita et al. 1997) of the indicated origins prevent the recruitment of the ORC complex and origin firing (K. Shirahige, unpubl.). We found that, as in the case of chromosome III, Top1 and Top2 concomitantly localized around regions containing early replication origins (ARS605, ARS606, and ARS607) in HU-treated wild-type cells (Fig. 2A,B, wt panels). Additional Top2 clusters were detected along the chromosome, again preferentially at intergenic locations that did not contain obvious replication origins. Disruption of the ARS607 sequence abolished Top1 and Top2 binding within the ARS607 site and the origin-surrounding regions (Fig. 2A,B). We observed the same effect in the eod strain with disruptions in the ARS607, ARS606, and ARS605 origins (Fig. 2A,B). We also found that the ablation of early origins did not lead to the recruitment of Top1 or Top2 to additional chromosomal sites, thus indicating that a reduction in the number of replicons does not generate novel chromosomal spots requiring Top1 or Top2 activities.
Figure 2.
Top1 and Top2 binding at replicating regions is dependent on efficient origin firing. Top1-10xFlag (A) and Top2-10xFlag (B) cells bearing a wild-type (wt) chromosome VI (CY7179 and CY7315, respectively), disrupted ARS607 (CY7363 and CY6379), or mutations in all early origins (shaded in blue) (CY7366 and CY7372) were released from α-factor-induced G1 block, treated with 0.2 M HU for 1 h, and processed for ChIP with antibodies specific to the Flag epitopes. Blue histogram bars in the Y-axis show the average signal ratio of loci significantly enriched in the immunoprecipitated fraction along chromosome VI in log2 scale. The X-axis shows kilobase units. Positions of all ARS elements acting as replication origins on the chromosome and the CEN sequence are indicated. Blue asterisks indicate the mutated ARSs.
Altogether, these data indicate that Top1 and Top2 recruitment at early replicating chromosomal sites depends on initiation events occurring at functional origins of replication. We can also conclude that the additional clusters observed in the Top2-binding profile are not influenced by the size and number of the replicons, as they remain unaltered in the strains lacking the early origins.
Top1 and Top2 associate with the origin regions after START, and their localization is influenced by the movement of replication forks
We then addressed whether the recruitment of Top1 and Top2 at origin-containing regions already occurred in G1. We analyzed chromosome VI, and we did not observe any significant enrichment of Top1 and Top2 at origin-containing regions and of Top2 at the origin-unrelated clusters in cells arrested at START by α-factor treatment (Supplementary Fig. 1). These findings do not rule out the possibility that in G1 cells, at START, either Top1 or Top2 stochastically and/or occasionally associate with chromosome VI or other chromosomes to deal with potential topological problems. However, based on these results, and on the previous findings that neither Top1 or Top2 associated with late-firing origins in early S phase, our data suggest that these enzymes are not recruited at replication origins at START or prior to origin unwinding and are unlikely to be part of the prereplicative complex.
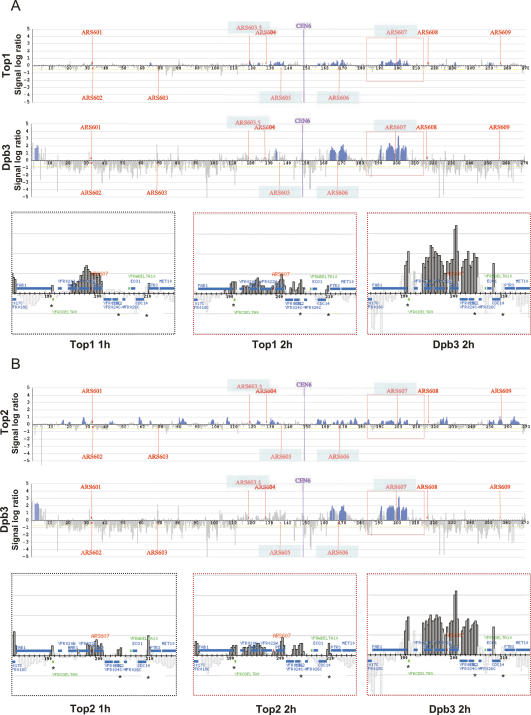
We next analyzed the Top1 and Top2 recruitment with respect to the dynamics of the replication forks. In the presence of HU, replication forks arising from early origins proceed slowly for several kilobases before stalling (Lopes et al. 2001). ChIP analysis of replisome components showed that the movement of the two forks outward of the origin split the ChIP signal of fork-associated proteins into two separate peaks (Katou et al. 2003). We thus carried out parallel ChIP analysis of Dpb3, a subunit of the DNA polymerase ε (polε) complex, and either Top1 or Top2 in cells experiencing a prolonged treatment with HU following release from a G1 block (Fig. 3A,B). As expected, the signal corresponding to Dpb3 was detected in the ARS606 and ARS607 regions and was resolved into two well-defined peaks, marking the position of the fraction of Dpb3 associated with the two forks that proceeded slowly outwards from the origin sites (cf. Supplementary Fig. 2 and Fig. 3). When Top1 and Top2 were analyzed by ChIP in the same cells, we observed a similar bimodal distribution of the corresponding signals, which was different from the one-peak profile observed at the 1-h time point (Fig. 3A,B; cf. also Supplementary Fig. 2). These findings indicate that both topoisomerases bind in close proximity of replication forks and suggest that their movement along the replicating chromosome is influenced by fork progression. We did not observe a significant change in the distribution of the Top2 origin-unrelated clusters (Fig. 3B).
Figure 3.
Top1 and Top2 colocalize with Dbp3 at replication forks. Top1-10xFlag/Dpb3-3xHA (A) and Top2-10xFlag/Dpb3-3xHA (B) cells (CY7340 and CY7343 strains, respectively) were released from α-factor-induced G1 block and treated with 0.2 M HU for 2 h; cultures were then split and processed for ChIP with antibodies specific to either Flag or HA epitopes. Blue histogram bars in the Y-axis show the average signal ratio of loci significantly enriched in the immunoprecipitated fraction along chromosome VI in log2 scale. The X-axis shows kilobase units. Positions of all ARS sequences acting as replication origins on the chromosome and CEN sequence are indicated. The bottom panels show Top1/Top2 (after 1- and 2-h HU treatment) and Dpb3 (after 2-h HU treatment) clusters surrounding ARS607 element in detail. Blue horizontal bars represent ORFs. Asterisks indicate the position of tRNA genes.
Top1 and Top2 influence cell cycle progression and Rad53-dependent checkpoint response
To investigate how Top1 and Top2 influence chromosome replication, we have analyzed S-phase progression, origin firing, fork movement, and fork integrity in top1, top2, and top1 top2 mutants.
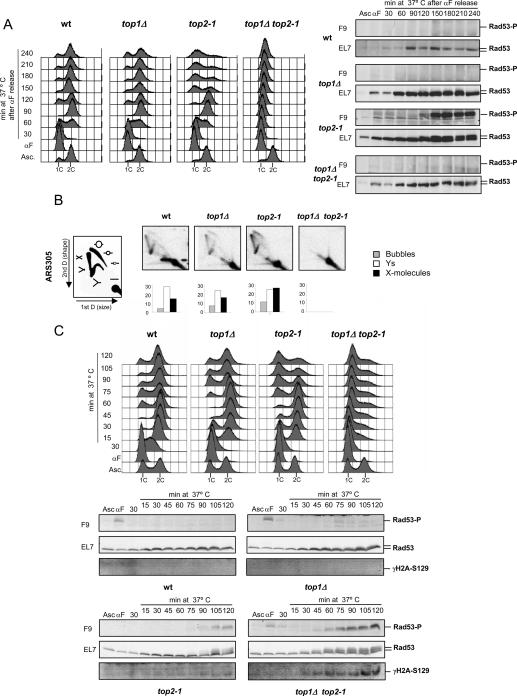
Cells carrying a TOP1 (top1Δ) deletion, a temperature-sensitive allele of TOP2 (top2-1), or both the top1Δ and top2-1 mutations were presynchronized in G1 at the permissive temperature for the top2-1 mutation and then released into S phase at the restrictive temperature. We analyzed the transition from G1 into S phase by FACS (fluorescence-activated cell sorting) analysis, which measures the bulk of DNA synthesis, and by examining the bud-emergence profile, which is a late G1 event. We found that the FACS and budding profiles of top1Δ cells were undistinguishable from the ones of wild-type cells (Fig. 4A; data not shown). top2-1 mutants passed through the G1–S boundary and completed S phase with kinetics similar to wild-type cells but, following the completion of the first cell cycle, progressively accumulated unbudded cells with a 1C DNA content (Fig. 4A; data not shown). In agreement with previous observations (Goto and Wang 1985; Brill et al. 1987), top1Δ top2-1 mutants failed to proceed through S phase and accumulated in G1 with unbudded cells and a 1C DNA content (Fig. 4A; data not shown). A gradual shift toward 2C DNA content was observed in top1Δ top2-1 arrested cells. This effect is most likely due to the persistence of mitochondrial DNA replication, as it was not observed in ρ0 derivatives of top1Δ top2-1 mutants (data not shown). Mutants defective in DNA replication often delay or arrest cell cycle progression due to the accumulation of DNA lesions that cause the activation of the Mec1/Rad53 DNA damage checkpoint response (Hartwell and Weinert 1989). Checkpoint activation ultimately delays cell cycle progression. We therefore addressed whether the cell cycle abnormalities of top2-1 and of top1Δ top2-1 mutants were caused by checkpoint activation by measuring the phosphorylation state of the Rad53 protein kinase.
Figure 4.
Effect of Top1 and/or Top2 activity attenuation on chromosomal replication and checkpoint activation. (A) Wild-type (SY359), top1Δ (CY2279), top2-1 (SY2183), and top1 Δtop2-1 (CY7039) cells were arrested in G1 by α-factor treatment at permissive temperature and then released into fresh medium at 37°C. Samples were collected at the indicated time points for FACS analysis and TCA protein precipitation and further immunodetection of Rad53 protein using antibodies recognizing the protein backbone (EL7) or the phosphorylated epitopes only (F9). (B) Wild-type, top1Δ, top2-1, and top1Δ top2-1 cells were arrested in G1 and released into fresh medium containing 0.2 M HU at 37°C. Samples were collected after 2 h, genomic DNA was extracted, and replication intermediates (RI) were analyzed by 2D gel electrophoresis using a probe specific to ARS305 region. (C) Wild-type, top1Δ, top2-1, and top1Δ top2-1 cells were arrested in G1, released into fresh medium for 30 min at 25°C, and then transferred to 37°C prewarmed medium. Samples were collected at the indicated time points for FACS analysis, TCA protein precipitation, and immunodetection of Rad53 and S129-phosphorylated H2A proteins.
Cycling wild-type and top1Δ cells did not exhibit any checkpoint activation (Fig. 4A). Conversely, top2-1 mutants progressively showed a high level of phosphorylated Rad53 when cells with a 1C DNA content started to accumulate in the second cell cycle. We did not observe phosphorylated Rad53 in top1Δ top2-1 cells released from G1 at the restrictive temperature; further, the same mutant cells, when shifted back at the permissive temperature, were able to complete S phase without activating the checkpoint (data not shown).
We conclude that top2-1 cells, following the incubation at the restrictive temperature, activate the DNA damage checkpoint response at the M–G1 transition.
Top1 and Top2 are required for replication fork progression and stability
We then addressed whether the relative amount and the quality of the replication intermediates arising at a specific origin of replication during the initial steps of DNA synthesis were affected by mutations in the TOP1 and/or TOP2 genes. Origin firing generates intermediates that on two-dimensional (2D) gels migrate as a bubble arc, containing forks proceeding bidirectionally, and large Y molecules, resulting from forks migrating asymmetrically outside of the origin fragment (Friedman and Brewer 1995). Cruciform DNA structures can be also visualized by 2D gels (Brewer and Fangman 1988). Different types of X-shaped molecules can migrate on 2D gels as cruciform structures (Schwacha and Kleckner 1994) and have been implicated in various processes such as DNA recombination, sister chromatid cohesion, and replication-coupled repair processes (Zou and Rothstein 1997; Benard et al. 2001; Segurado et al. 2002; Lopes et al. 2003; Liberi et al. 2005; Robinson et al. 2007).
Wild-type, top1Δ, top2-1, and top1Δ top2-1 cells were presynchronized in G1 and released at the restrictive temperature for the top2-1 mutation in the presence of HU to enrich for replication intermediates. Under these conditions, wild-type cells fire only a subset of early origins, and forks proceed for short tracts (Lopes et al. 2001; Katou et al. 2003; Feng et al. 2006). We analyzed the replication intermediates arising at ARS305, which is an efficient origin of replication that fires early during S phase (Lopes et al. 2001; Poloumienko et al. 2001; Raghuraman et al. 2001). We found that wild-type, top1Δ, and top2-1 cells accumulated bubbles, Y structures, and X-shaped molecules (Fig. 4B), thus indicating that ARS305 was fired in these genetic backgrounds. However, we noticed that while the relative distribution of the different species of replication intermediates was comparable in wild-type and top1Δ cells, in top2-1 mutants, we found an accumulation of X-shaped intermediates (Fig. 4B; Supplementary Fig. 3). The accumulation of these cruciform structures in top2-1 cells did not depend on the presence of HU, as it could be observed also when cells were released at the restrictive temperature without HU (Supplementary Fig. 3). top1Δ top2-1 mutants did not accumulate replication intermediates under these conditions, thus implicating that ARS305 was not fired in this background. The inability of top1Δ top2-1 cells to fire ARS305 is likely a consequence of the failure to complete the G1 phase of the cell cycle. We then analyzed the requirement of these topoisomerases specifically for replication progression. Wild-type, top1Δ, top2-1, and top1Δ top2-1 mutants were presynchronized in G1, released at the permissive temperature for 30 min to allow entry into S phase and origin firing, and then shifted at the restrictive temperature for the top2-1 mutation. FACS analysis showed that wild-type, top1Δ, and top2-1 cells were able to complete the first cell cycle under these conditions, although top1Δ and top2-1 mutants exhibited a slight delay in traversing the G1–S boundary (Fig. 4C). Further, top2-1 mutants accumulated cells with a 1C DNA content at the second cell cycle (Fig. 4C, 75-min time point). top1Δ top2-1 mutants under these conditions were able to bud and enter S phase (data not shown) but failed to complete replication and arrested with a near 1C DNA content (Fig. 4C). We have also analyzed, in the same experiment, the Rad53 and γH2A phosphorylation state, which correlates with the presence of DNA breaks (Lydall and Whitehall 2005; Vidanes et al. 2005). top2-1 mutants accumulated phosphorylated Rad53 and γH2A ∼15 min after G1 cells started to increase (Fig. 4C, 90-min time point; data not shown). top1Δ top2-1 mutants already showed phosphorylated Rad53 and γH2A during S phase 45 min after the shift at the restrictive temperature. This result, together with the previous finding that top1Δ top2-1 mutants experiencing a G1 block at the restrictive temperature do not activate Rad53, suggests that Top1 and Top2 likely cooperate not only to facilitate the completion of S phase but also to prevent the formation of chromosome breaks and intra-S checkpoint activation.
To further characterize the roles of Top1 and Top2 during fork elongation, we analyzed the replication intermediates by 2D gel using the experimental conditions described in Figure 4C. In particular, we followed the fate of those forks that arise at ARS305 and progressively invade adjacent chromosomal regions. Thirty minutes after release from the G1 block, wild-type cells accumulated bubbles, large Y molecules, and X intermediates at the ARS305 region, thus indicating that the origin had fired (Fig. 5A; Supplementary Fig. 3). At the same time point, a fraction of the forks arising from the ARS305 origin have already invaded the chromosomal regions A, B, and C and, few of them, also region D (Supplementary Fig. 4). Following the shift at the restrictive temperature, at 60 min from the G1 release, most of the cells have reached G2 as shown by FACS analysis (Supplementary Fig. 4). Accordingly, very few cells, perhaps representing either daughter cells that entered S phase late or cells experiencing already the second cell cycle, showed replication intermediates at the ARS305 region but not at the flanking fragments. At 120 min, a fraction of cells that went through cell division experienced a new round of replication as judged by FACS. This is consistent with the 2D gel analysis showing that these cells accumulated intermediates at the ARS305, A, and C regions. At 180 min, the culture was mostly asynchronous, and a fraction of cells exhibited replication intermediates at the ARS305 and A regions. We therefore conclude that these experimental conditions allowed wild-type cells to experience at least two rounds of origin firing and normal fork progression.
Figure 5.
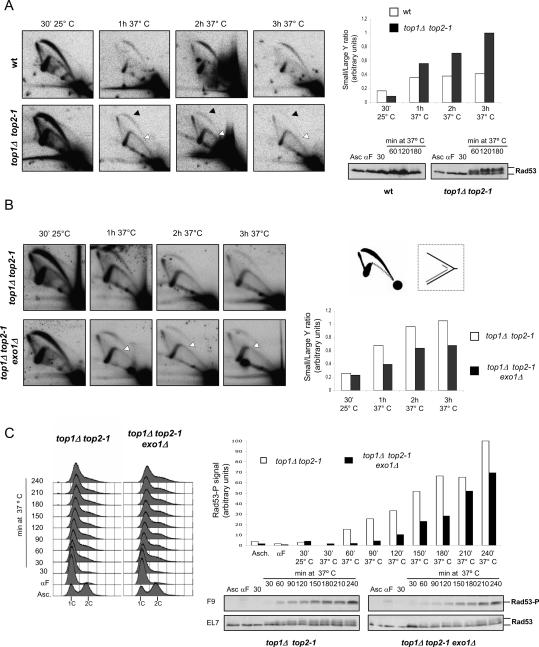
Aberrant replication intermediates accumulate in top1 top2 mutants. (A) Wild-type (SY359) and top1Δtop2-1 (CY7039) cells were arrested in G1, released into fresh medium for 30 min at 25°C, and then transferred to 37°C prewarmed medium. Samples were collected at the indicated time points, and genomic DNA was extracted. Replication intermediates were analyzed by 2D gel electrophoresis with ARS305 probe. Black and white arrowheads indicate bubble and small-Y arcs, respectively, in arrested Δtop1 top2-1 cells. TCA protein precipitation of the same samples and immunodetection of Rad53 protein with EL7 antibodies are shown. (B) top1Δ top2-1 (CY7039) and top1Δ top2-1 exo1Δ (CY8002) cells where arrested in G1, released into fresh medium for 30 min at 25°C, and then transferred to 37°C prewarmed medium. Samples were collected at the indicated time points, and genomic DNA was extracted. Replication intermediates were analyzed by 2D gel electrophoresis with ARS305 probe. A schematic representation of replication intermediates (RIs) and small Ys arising at ARS305 region is shown. (C) top1Δ top2-1 and top1Δ top2-1 exo1Δ cells were arrested in G1, released into fresh medium for 30 min at 25°C, and then transferred to 37°C prewarmed medium. Samples were collected at the indicated time points for FACS analysis, TCA protein precipitation, and immunodetection of Rad53 with EL7 and F9 antibodies. Quantization of the abundance of Rad53-phosphorylated species detected by F9 antibody immunoblot is shown.
We then analyzed top1Δ top2-1 mutants. At 30 min after release from G1 at the permissive temperature, the 2D gel profile of top1Δ top2-1 mutants was similar to the one of wild-type cells, thus indicating that forks proceeded normally (Fig. 5A; Supplementary Fig. 4). FACS analysis showed that, following the shift at the restrictive temperature, top1Δ top2-1 cells were unable to complete the bulk of DNA synthesis. 2D gels showed that the relative amount of the replication intermediates visualized at regions ARS305, A, B, C, and D remained comparable throughout the shift at the restrictive temperature. Moreover, when we measured the relative amount of the different replication intermediates at the ARS305 region, we found that, with time, the relative percentage of bubbles decreased, while the small Ys increased (Fig. 5A). The progressive accumulation of small Ys correlates with checkpoint activation (Fig. 5A). The accumulation of small Ys and checkpoint signals in top1Δ top2-1 mutants is diagnostic of progressive fork degeneration that might cause the erosion and the reduction in mass of the Y intermediates (Lopes et al. 2001; Cotta-Ramusino et al. 2005). We therefore conclude that, in top1Δ top2-1 mutants, replication fork progression and stability were impaired.
Genetic pathways influencing fork stability and checkpoint activation in top1Δ top2-1 mutants
The previous set of data indicates that top1Δ top2-1 mutants at the restrictive temperature exhibit a block in fork progression, associated with the mass reduction of replication intermediates, and activation of the DNA damage checkpoint response.
We then tested whether the mass reduction of the Y intermediates was mediated by the Exo1 exonuclease, as in the case of checkpoint-defective cells (Cotta-Ramusino et al. 2005). We found that the ablation of EXO1 in top1Δ top2-1 mutant cells counteracts the accumulation of small Y intermediates (Fig. 5B). A possible interpretation of this result is that Exo1 mediates the erosion of nascent chains at the forks, thus leading to the formation of single-stranded DNA (ssDNA) regions, as in the case of rad53 mutants (Lopes et al. 2001; Cotta-Ramusino et al. 2005). If that is the case, a logical expectation is that the ablation of EXO1 in top1Δ top2-1 mutants should also affect Rad53 activation through the reduction of ssDNA-mediated checkpoint signals (Zou and Elledge 2003; Lucca et al. 2004). We found that in top1Δ top2-1 exo1Δ mutants, compared with top1Δ top2-1 cells, Rad53 phosphorylation was significantly delayed although not abolished (Fig. 5C). We therefore conclude that fork stability and checkpoint activation in top1Δ top2-1 cells are influenced by the presence of Exo1.
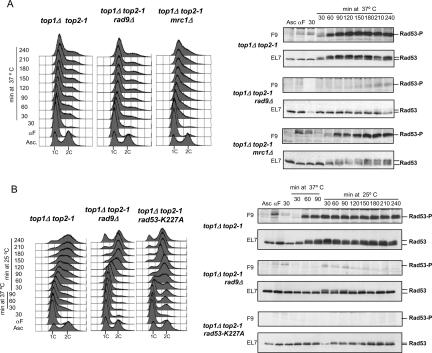
Rad53-dependent checkpoint activation in response to replication stress causing fork stalling is mediated by Mrc1, while Rad9 seems to be dispensable under these conditions (Paulovich et al. 1997; Pellicioli et al. 1999; Alcasabas et al. 2001; Pan et al. 2006). Rad9, however, is needed for checkpoint activation in response to intra-S DNA damage (Paulovich et al. 1997; Pellicioli et al. 1999) and DNA breaks and for the formation of Rad53 foci (Lisby et al. 2004). Stalled forks are thought to activate the checkpoint by generating ssDNA at the fork, as in the case of HU-treated cells (Branzei and Foiani 2005). Checkpoint activation in response to intra-S DNA damage is likely mediated by ssDNA generated at collapsed forks, by gaps arising when forks replicate across DNA lesions, or by DNA breaks (Branzei and Foiani 2005). We therefore tested whether the checkpoint response induced in top1Δ top2 mutants was dependent on the Mrc1 or on the Rad9 subpathways. top1Δ top2-1, top1Δ top2-1 rad9Δ, and top1Δ top2-1 mrc1Δ mutants were shifted at the restrictive temperature after a 30-min preincubation at the permissive temperature in S phase. FACS analysis showed that the top1Δ top2-1, top1Δ top2-1 rad9Δ, and top1Δ top2-1 mrc1Δ mutants exhibited cells with a DNA content close to 1C throughout the incubation at the restrictive temperature (Fig. 6A). We obtained the same result in top1Δ top2-1 rad53-K227A mutants (data not shown). Thus, the inability of top1Δ top2-1 cells to complete the bulk of replication does not depend on Rad9, Mrc1, or Rad53. We then analyzed the Rad53 phosphorylation state in the same experiments (Fig. 6A). In both top1Δ top2-1 and top1Δ top2-1 mrc1Δ mutants, phosphorylated Rad53 accumulated following the shift at the restrictive temperature with similar kinetics. Conversely, a dramatic decrease of Rad53 phosphorylation was observed in top1Δ top2-1 rad9Δ cells. A possible interpretation for this result is that in top1Δ top2-1 mutants, the progressive degeneration of the forks causes accumulation of intra-S DNA damage that is channeled into a Rad9-mediated signaling. Hence, the signaling pathway leading to checkpoint activation in top1Δ top2-1 mutants differs from the one of HU-treated cells, which relies on Mrc1 but not on Rad9. The finding that top1Δ top2-1 mutants exhibit phosphorylated γH2A suggests that these cells accumulate DNA breaks that trigger the Rad9–Rad53-dependent checkpoint. We then tested whether the genome damage accumulated in top1Δ top2-1 mutants at the restrictive temperature was preventing S-phase completion following a shift back at the permissive temperature. We found that top1Δ top2-1 cells were still able to complete the bulk of replication under these conditions although very slowly, but then arrested in G2 (Fig. 6B). Furthermore, the slow S-phase progression and the G2 arrest correlated with the presence of phosphorylated Rad53 (Fig. 6B). We also found that, under the same conditions, top1Δ top2-1 rad9Δ and top1Δ top2-1 rad53-K227A mutants completed replication faster than top1Δ top2-1 cells and did not arrest in G2. We therefore conclude that the lesions accumulated by top1Δ top2-1 mutants at the restrictive temperature persist after the shift back at the permissive temperature and sustain a Rad9–Rad53-mediated checkpoint response that causes a cell cycle block in G2. The slow completion of S phase in top1Δ top2-1 mutants under these conditions may result from the extensive and irreversible fork degeneration experienced at the restrictive temperature and from the consequent Rad53-mediated inhibition of late replicons following the shift back at the permissive temperature. Rad53 attenuation in this background would cause a faster completion of DNA synthesis through the unscheduled firing of additional replicons (Tercero and Diffley 2001).
Figure 6.
Lack of Top1 and Top2 activity after replication onset induces persistent Rad9-dependent DNA damage checkpoint activation. (A) top1Δ top2-1 (CY7039), top1Δ top2-1rad9Δ (CY7642), and top1Δ top2-1mrc1Δ (CY7701) cells were arrested in G1, released into fresh medium for 30 min at 25°C, and then transferred to 37°C prewarmed medium. Samples were collected at the indicated time points for FACS analysis, TCA protein precipitation, and immunodetection of Rad53 protein. (B) top1Δ top2-1, top1Δ top2-1 rad9Δ, and Δtop1 top2-1 rad53-K227A (CY7295) cells were arrested in G1, released into fresh medium for 30 min at 25°C, then transferred for 90 min to 37°C prewarmed medium and subsequently back into fresh medium at permissive temperature. Samples were collected at the indicated time points for FACS analysis, TCA protein precipitation, and immunodetection of Rad53 protein.
Discussion
Top1 and Top2 associate with replicating chromosomal regions
Our results, besides emphasizing a fundamental role for Top1 in the process of chromosome replication, demonstrate that Top2 acts directly at replication forks and provide a high-resolution analysis of Top1 and Top2 distribution on eukaryotic replicating chromosomes. Furthermore, we show that unresolved topological constraints during S phase cause pathological rearrangements at the forks, chromosome breaks, DNA damage checkpoint activation, and, likely, chromosome segregation defects. Based on our data, we conclude that Top1 and Top2 bind active replication origins sometime after START, and their association correlates with one of the replisomes that is assembled at the initial stage of DNA synthesis after origin unwinding. This is in agreement with recent data showing that Top1 associates with the ARS305 region and copurifies with the GINS–MCM complex (Gambus et al. 2006). However, the situation is somehow different in human cells, as recent observations obtained by treating cells with Top1 and Top2 inhibitors suggest that DNA topoisomerase I interacts with replication origins in M, G1, and G1/S, while DNA topoisomerase II does so in M and G1 (Abdurashidova et al. 2007). Following origin firing, the localization of Top1 and Top2 is influenced by fork movement. A possible interpretation of this result is that Top1 and Top2 migrate with the forks in proximity to the replisome–fork complexes to resolve the topological problems generated during replication elongation (Wang 2002). Furthermore, our data suggest that the topological constraints requiring Top1 and Top2 activity arise within an ∼600-bp region spanning the moving fork. It has been suggested that the eukaryotic topological domain may extend for 15 kb (Poirier and Marko 2002) and that DNA topoisomerases may act anywhere within the topological domains to change their linking state (Champoux 2001; Postow et al. 2001; Wang 2002). Our findings rather seem to imply that the action of Top1 and Top2 is coordinated with replisome progression within small chromosomal regions spanning the moving forks. By solving the topological constraints directly at the fork, the cell might counteract the diffusion along large chromosomal domains of topological changes that otherwise would cause chromatin distortions and nucleosome disruption, possibly followed by the deregulation of certain transcription units.
Origin-unrelated Top2 chromosome localization sites
Unlike Top1, Top2 also localizes at certain intergenic regions. These origin-unrelated Top2 clusters are specifically visualized in S-phase-blocked cells, as they are not detected in α-factor-arrested cells and differ from the clusters observed in nocodazole-blocked cells (data not shown). However, these Top2 clusters remain unaltered when forks move and when the number of replicons is reduced, thus suggesting that their formation is not influenced by replicon dynamics. This is also in agreement with the finding that specifically the intergenic Top2 clusters (but not the origin-related ones) are detectable in cdc7 mutants that arrest at the initial stage of S phase due to limiting DDK activity that prevents origin firing (Supplementary Fig. 5). Top2 has been implicated in chromosome condensation (Uemura et al. 1987; Wang 2002) and, in Drosophila, colocalizes with certain condensing proteins such as the ortholog of the yeast Brn1 protein (Lupo et al. 2001). However, we found that the origin-unrelated Top2 clusters that we observed in HU-treated cells do not overlap with the Brn1-binding sites (our unpublished observations), and neither colocalizes with condensins, cohesions, nor the Smc5/6 complex (Lengronne et al. 2004; Lindroos et al. 2006), thus suggesting that the Top2 intergenic clusters do not reflect a concerted action of Top2 and the SMC proteins.
An alternative possibility is that, during S phase, Top2 is specifically recruited at promoters. This is consistent with the findings that out of the 18 Top2 origin-unrelated clusters visualized on chromosome VI, 17 are located upstream of transcription units, and only one is located between two genes whose transcription is convergent. Furthermore, in 50% of these clusters there is at least one gene whose expression is regulated in response to S-phase stress (Jelinsky et al. 2000). This hypothesis is also in accordance with the observations that Top2 inhibitors cause notable changes at or near promoters (Collins et al. 2001) and that a critical role for Top2 in enhancing transcription has been demonstrated at a specific type of promoters (Ju et al. 2006). These clusters remain excluded from the transcription unit, and, therefore, it is unlikely that they reflect a need for Top2 activity in the progression of the transcription bubble. The simplest explanation is that during transcription initiation, Top2 has to deal with the unwinding stresses generated at promoters or in their proximity (Collins et al. 2001).
Eukaryotic DNA topoisomerase II is a main component of the chromosome scaffold (Gasser et al. 1986), and Top2-like activities have been implicated in remodeling chromatin organization both locally and in a global context (Varga-Weisz et al. 1997; Ju et al. 2006) and in DNA looping (Li et al. 1999). In this view, it is possible that the intergenic Top2 clusters have a role in coordinating chromatin organization with transcription through the formation of chromosome loops containing transcription units. It has been proposed that chromosome loops create the conditions for high local concentrations of binding sites for transcription factors and chromatin remodeling proteins that favor the productive interaction between these proteins and promoters (Chambeyron and Bickmore 2004 and references therein). However, the intergenic Top2 localization specifically observed in cells arrested in S phase by HU treatment raises the question as to whether these Top2 clusters play a role only in transcription or whether they also influence chromosome replication. Transcription factors may regulate DNA replication by facilitating the temporal control of replication and transcription during the cell cycle and by integrating the origin selection with the transcriptional program (Goldman et al. 1984). Furthermore, cells have evolved specialized replication fork barriers to avoid collision between replication forks and transcription at the rDNA locus (Brewer and Fangman 1988), and, in general, a lack of coordination between replication and transcription may lead to dangerous and unscheduled recombination events (Aguilera 2002). A tantalizing possibility is that the Top2 clusters might be needed to coordinate replication fork progression with the enhancement of transcription of certain genes perhaps by insulating, through DNA looping, the transcription units from the incoming replication forks or to ensure that, following the passage of the forks, DNA looping may occur at those regions.
Another possible scenario is that the Top2 intergenic clusters represent Top2-mediated chromosome organization centers that somehow influence chromosome replication. Intriguingly, in Xenopus embryos, the S-phase nuclei are organized in loops and replicons, and the Top2-dependent resetting of the loop size affects the replicon dynamics (Lemaitre et al. 2005).
Replication fork dynamics and checkpoint activation in top2 mutants
At the restrictive temperature, top2-1 and top2-4 (data not shown) mutants accumulate cruciform molecules at the forks, complete chromosome replication, and, after cell division, progressively accumulate in G1 with an active checkpoint.
Top2 has been implicated in the resolution of precatenates (Wang 2002; Postow et al. 2004). Conversely, type I topoisomerases cannot pass an intact double helix through another, unless one of the duplexes contains nicks/gaps or unpaired bubbles (Wang 2002; Postow et al. 2004). Since restriction digestion will resolve the cruciform precatenate structure characterized by the helix–helix juxtaposition into two linear fragments, it is unlikely that the cruciform structures accumulating in top2 cells represent precatenates. However, we cannot rule out the possibility that unresolved precatenates might be converted into interlocked cruciform derivatives, perhaps through topological transitions even mediated by type I topoisomerases; these interlocked structures would migrate on a 2D gel like X-shaped molecules.
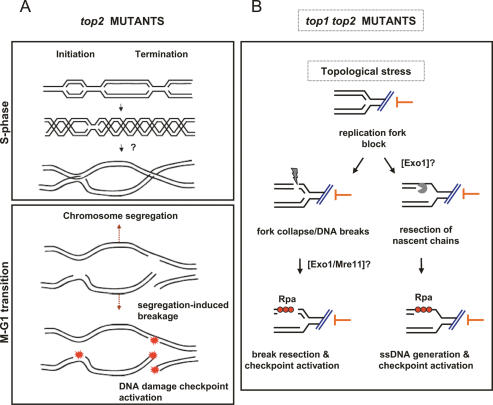
The accumulation of cruciform junctions behind the forks in top2 mutants does not seem to affect fork progression but might have dramatic consequences for sister chromatid separation, particularly if the X-shaped structures are interlocked. At least part of the problems exhibited by top2 mutants during chromosome segregation might be inherited from the previous S phase (Fig. 7A): The abnormal accumulation of interlocked cruciform junctions at the forks during the initial stage of S phase would cause sister chromatid entangling. The mechanical stress generated in the attempt to pull apart the sister chromatids that are still topologically linked would inevitably lead to chromosome nondisjunction and/or the generation of chromosome breaks followed by DNA damage checkpoint activation, as our data also suggest. This model implies that the entangled sister chromatids in top2-1 mutants do not trigger any specific DNA decatenation checkpoint such as the one acting in mammalian cells exposed to the action of Topoisomerase II catalytic inhibitors (Downes et al. 1994). The identification of certain top2 mutants that delay the anaphase onset suggests that a Top2-dependent decatenation checkpoint may also act in yeast (Andrews et al. 2006). However, it is formally possible that those top2 mutants somehow exhibit an allele-specific loss of function of Top2 affecting the anaphase onset. Previous observations support the view that in top2 mutants the attempt to segregate entangled chromatids would cause chromosome breaks: First, top2 mutants experiencing S phase at the restrictive temperature accumulate plasmids that appear as multiply intertwined catenated dimers (DiNardo et al. 1984); second, cell division in the absence of Top2 activity leads to nondisjunction and chromosome breakage (Holm et al. 1985, 1989). Our data suggest that sister chromatid entangling might indeed occur in early S phase, although we cannot rule out that interlocked structures might also form in top2 mutants at replication termination, especially considering that precatenates might also arise at regions where two forks converge during termination of DNA synthesis (Fig. 7A). The following findings support the previous hypothesis: (1) Delaying anaphase onset in top2 mutants released from a G1 block at the restrictive temperature by nocodazole treatment prevents Rad53 activation (data not shown), and (2) Rad53 activation at the M–G1 transition in top2 mutants at the restrictive temperature occurs only if cells have previously experienced S phase at the nonpermissive conditions (data not shown). Hence, it is reasonable to think that at least part of the problems exhibited by top2 mutants during chromosome segregation can be ascribed to a faulty replication process that causes sister chromatid entangling.
Figure 7.
Hypothetical models to explain the chromosomal abnormalities that arise in top2 and top1 top2 mutants. (A) Schematic representation of the phenotypes observed in top2-1 mutants. Precatenate formation during initiation and/or termination of DNA replication would require Top2 activity for resolution. Although other possibilities can be envisaged, it is possible that, in the absence of a fully functional Top2 activity, unscheduled strand passage reactions, perhaps mediated by type I DNA topoisomerases, may lead to the generation of sister chromatid interlocking during S phase. Upon anaphase onset, the mechanical tension generated by the mitotic spindle (dashed arrows) might force the separation of the entangled chromatids, leading to the formation of DNA breaks and DNA damage checkpoint signals. (B) Schematic representation of the phenotypes observed in top1Δ top2-1 mutants. The topological constrains arising in the double mutants cause the block of fork progression. Stalled forks could either suffer DNA breaks, perhaps as a consequence of fork collapse at nicks, or resection of nascent chains. Exo1 is likely implicated in the resection of nascent chains (Cotta-Ramusino et al. 2005) and perhaps also in DSB resection together with Mre11 (Nakada et al. 2004). In both cases, RPA–ssDNA filaments could form, leading to checkpoint activation.
Fork abnormalities and checkpoint activation in top1 top2 double mutants
We found that the concomitant attenuation of Top1 and Top2 during DNA synthesis causes an immediate block of fork progression. This block is most likely due to the accumulation of topological constraints (positive supercoiling) to such high levels that further DNA unwinding by the replicative helicase would be impeded. Unlike HU-induced fork stalling, the top1 top2 replication block does not activate the Mrc1-dependent checkpoint response. At least in theory, a failure in resolving the topological constraints arising during fork elongation should counteract the movement of the replicative helicase, thus preventing the generation of ssDNA regions at the forks as a result of uncoupling between the helicase and the DNA polymerases. Hence, the checkpoint signals in top1 top2 mutants might arise through different mechanisms. At least two, not necessarily mutually exclusive, possibilities can be envisaged (Fig. 7B): fork collapse followed by DSB formation (Kuzminov 1995), and resection that would generate ssDNA filaments (Pellicioli et al. 2001) and/or ssDNA regions resulting from resection of the newly synthesized strands (Cotta-Ramusino et al. 2005). Both hypotheses are consistent with our data, as the Rad9 dependency and the phosphorylation of γH2A might account for DNA break formation, while the Exo1-mediated progressive reduction in mass of Y intermediates supports the hypothesis that a fraction of nascent chains may undergo extensive resection with the consequent accumulation of ssDNA and checkpoint signals. In any case, we can rule out that in top1 top2 mutants the checkpoint signals arise due to premature attempts to segregate partially replicated chromosomes, as nocodazole treatment does not prevent checkpoint activation (data not shown). Future work will be required to further investigate the exact mechanisms leading to activation of the DNA damage checkpoint in top1 top2 mutants.
Materials and methods
Saccharomyces cerevisiae strains and growing conditions
The strains used in this study are isogenic derivatives of W303-1A and are listed in Supplementary Table 1. Cells were grown in YPD at the indicated temperatures. Synchronization in G1 was carried out by adding 2 μg/mL α−factor. Cells were then released into fresh YPD medium after removal of α−factor.
ChIP-on-chip analysis
S. cerevisiae chromosomes III–V and chromosome VI high-density oligonucleotide microarrays were provided by Affymetrix Custom Express Service (SC3456a520015F, P/N 520015; rikDACF, P/N 510,636, respectively). Sequence and position of oligonucleotides on the microarrays are available from Affymetrix. ChIP was carried out as previously described (Katou et al. 2003, 2006): We disrupted 1.5 × 108 cells by Multi-beads shocker (MB400U, Yasui Kikai) using glass beads. Anti-HA monoclonal antibody HA.11 (16B12, CRP Inc.) and anti-Flag monoclonal antibody M2 (Sigma-Aldrich) were used for ChIP. Chromatin-immunprecipitated DNA was purified and amplified by random priming as described (Katou et al. 2003): A total of 10 μg of amplified DNA was digested with DNaseI to a mean size of 100 bp and purified, and the fragments were end-labeled with biotin-N6-ddATP. Hybridization, washing, staining, and scanning were performed according to the manufacturer’s instructions (Affymetrix). Primary data analyses were carried out using the Affymetrix microarray Suite version 5.0 software to obtain hybridization intensity, fold change value, change P-value, and detection P-value for each locus. For the discrimination of positive and negative signals for the binding, we compared the chromatin-immunprecipitated fraction with the supernatant fraction by using three criteria. First, the reliability of strength of signal was judged by detection P-value of each locus (P ≥ 0.025). Second, the reliability of binding ratio was judged by change P-value (P ≥ 0.025). Third, clusters consisting of at least three contiguous loci that filled the above two criteria were selected, because it was known that a single site of protein–DNA interaction will result in immunoprecipitation of DNA fragments that hybridized not only to the locus of the actual binding site but also to its neighbors. For the analyses of BrdU incorporation, cells were fixed by ice-cold buffer containing 0.1% azide, and then total DNA from 3 × 108 cells was purified. DNA was sheared to 300 bp by sonication, denatured, and mixed with 2 μg of anti-BrdU monoclonal antibody (2B1D5F5H4E2; MBL). Antibody-bound and unbound fractions were subsequently purified, amplified, labeled, and hybridized to the DNA chip. Microarray data presented in this study can be obtained from Gene Expression Omnibus with accession number GSE8305.
2D gel analysis of replication intermediates
Total genomic DNA was isolated and digested with EcoRV and HindIII restriction enzymes (New England Biolabs), and 2D gel electrophoresis was carried out as previously described (Lopes et al. 2003).
FACS analysis
FACS analysis was performed using a Beckton Dickinson fluorescence-activated cell analyzer. Cells were fixed and processed as previously described (Pellicioli et al. 1999).
Western blot analysis of Rad53
Preparation of yeast protein extracts from TCA-treated cells, SDS-PAGE electrophoresis, and Western blotting were performed as previously described (Pellicioli et al. 1999). For immunodetection of Rad53 we used two antibodies: EL7, which recognizes the Rad53 protein backbone, and F9, which recognizes phosphorylated Rad53. The antibodies were produced and characterized by A. Pellicioli and the IFOM antibody facility. Phospho-S129 Histone H2A polyclonal antibodies (Abcam) were used to score γH2A phosphorylation.
Acknowledgments
We thank J. Diffley for discussions and for sharing unpublished observations; all members of our laboratories for helpful discussions; D. Branzei, M. Lopes, and G. Liberi for critical reading of the manuscript; R. Rothstein and E. Schwob for providing yeast strains; A. Pellicioli for EL7 and F9 antibodies; and R. Jossen for assistance in producing yeast strains. This work was supported by grants form the Associazione Italiana per la Ricerca sul Cancro, Association for International Cancer Research and, partially, from European Union, Telethon-Italy, and the Italian Ministry of Education to M.F., and Grant-in-Aid for Scientific Research to K.S.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.432107
References
- Abdurashidova G., Radulescu S., Sandoval O., Zahariev S., Danailov M.B., Demidovich A., Santamaria L., Biamonti G., Riva S., Falaschi A., Radulescu S., Sandoval O., Zahariev S., Danailov M.B., Demidovich A., Santamaria L., Biamonti G., Riva S., Falaschi A., Sandoval O., Zahariev S., Danailov M.B., Demidovich A., Santamaria L., Biamonti G., Riva S., Falaschi A., Zahariev S., Danailov M.B., Demidovich A., Santamaria L., Biamonti G., Riva S., Falaschi A., Danailov M.B., Demidovich A., Santamaria L., Biamonti G., Riva S., Falaschi A., Demidovich A., Santamaria L., Biamonti G., Riva S., Falaschi A., Santamaria L., Biamonti G., Riva S., Falaschi A., Biamonti G., Riva S., Falaschi A., Riva S., Falaschi A., Falaschi A. Functional interactions of DNA topoisomerases with a human replication origin. EMBO J. 2007;26:998–1009. doi: 10.1038/sj.emboj.7601578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcasabas A.A., Osborn A.J., Bachant J., Hu F., Werler P.J., Bousset K., Furuya K., Diffley J.F., Carr A.M., Elledge S.J., Osborn A.J., Bachant J., Hu F., Werler P.J., Bousset K., Furuya K., Diffley J.F., Carr A.M., Elledge S.J., Bachant J., Hu F., Werler P.J., Bousset K., Furuya K., Diffley J.F., Carr A.M., Elledge S.J., Hu F., Werler P.J., Bousset K., Furuya K., Diffley J.F., Carr A.M., Elledge S.J., Werler P.J., Bousset K., Furuya K., Diffley J.F., Carr A.M., Elledge S.J., Bousset K., Furuya K., Diffley J.F., Carr A.M., Elledge S.J., Furuya K., Diffley J.F., Carr A.M., Elledge S.J., Diffley J.F., Carr A.M., Elledge S.J., Carr A.M., Elledge S.J., Elledge S.J. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Andrews C.A., Vas A.C., Meier B., Gimenez-Abian J.F., Diaz-Martinez L.A., Green J., Erickson S.L., Vanderwaal K.E., Hsu W.S., Clarke D.J., Vas A.C., Meier B., Gimenez-Abian J.F., Diaz-Martinez L.A., Green J., Erickson S.L., Vanderwaal K.E., Hsu W.S., Clarke D.J., Meier B., Gimenez-Abian J.F., Diaz-Martinez L.A., Green J., Erickson S.L., Vanderwaal K.E., Hsu W.S., Clarke D.J., Gimenez-Abian J.F., Diaz-Martinez L.A., Green J., Erickson S.L., Vanderwaal K.E., Hsu W.S., Clarke D.J., Diaz-Martinez L.A., Green J., Erickson S.L., Vanderwaal K.E., Hsu W.S., Clarke D.J., Green J., Erickson S.L., Vanderwaal K.E., Hsu W.S., Clarke D.J., Erickson S.L., Vanderwaal K.E., Hsu W.S., Clarke D.J., Vanderwaal K.E., Hsu W.S., Clarke D.J., Hsu W.S., Clarke D.J., Clarke D.J. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/securin. Genes & Dev. 2006;20:1162–1174. doi: 10.1101/gad.1367206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard M., Maric C., Pierron G., Maric C., Pierron G., Pierron G. DNA replication-dependent formation of joint DNA molecules in Physarum polycephalum. Mol. Cell. 2001;7:971–980. doi: 10.1016/s1097-2765(01)00237-4. [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M., Foiani M. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 2005;17:568–575. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Brewer B.J., Fangman W.L., Fangman W.L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Brill S.J., DiNardo S., Voelkel-Meinman K., Sternglanz R., DiNardo S., Voelkel-Meinman K., Sternglanz R., Voelkel-Meinman K., Sternglanz R., Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Chambeyron S., Bickmore W.A., Bickmore W.A. Does looping and clustering in the nucleus regulate gene expression? Curr. Opin. Cell Biol. 2004;16:256–262. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Champoux J.J. DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Champoux J., Been M., Been M. Topoisomerases and the swivel problem. In: Alberts B., editor. Mechanistic studies of DNA replication and genetic recombination: ICN-UCLA symposia on molecular and cellular biology. Academic Press; New York: 1980. pp. 809–815. [Google Scholar]
- Collins I., Weber A., Levens D., Weber A., Levens D., Levens D. Transcriptional consequences of topoisomerase inhibition. Mol. Cell. Biol. 2001;21:8437–8451. doi: 10.1128/MCB.21.24.8437-8451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C., Fachinetti D., Lucca C., Doksani Y., Lopes M., Sogo J., Foiani M., Fachinetti D., Lucca C., Doksani Y., Lopes M., Sogo J., Foiani M., Lucca C., Doksani Y., Lopes M., Sogo J., Foiani M., Doksani Y., Lopes M., Sogo J., Foiani M., Lopes M., Sogo J., Foiani M., Sogo J., Foiani M., Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K., Sternglanz R., Voelkel K., Sternglanz R., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: Topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl. Acad. Sci. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes C.S., Clarke D.J., Mullinger A.M., Gimenez-Abian J.F., Creighton A.M., Johnson R.T., Clarke D.J., Mullinger A.M., Gimenez-Abian J.F., Creighton A.M., Johnson R.T., Mullinger A.M., Gimenez-Abian J.F., Creighton A.M., Johnson R.T., Gimenez-Abian J.F., Creighton A.M., Johnson R.T., Creighton A.M., Johnson R.T., Johnson R.T. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- Feng W., Collingwood D., Boeck M.E., Fox L.A., Alvino G.M., Fangman W.L., Raghuraman M.K., Brewer B.J., Collingwood D., Boeck M.E., Fox L.A., Alvino G.M., Fangman W.L., Raghuraman M.K., Brewer B.J., Boeck M.E., Fox L.A., Alvino G.M., Fangman W.L., Raghuraman M.K., Brewer B.J., Fox L.A., Alvino G.M., Fangman W.L., Raghuraman M.K., Brewer B.J., Alvino G.M., Fangman W.L., Raghuraman M.K., Brewer B.J., Fangman W.L., Raghuraman M.K., Brewer B.J., Raghuraman M.K., Brewer B.J., Brewer B.J. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 2006;8:148–155. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K.L., Brewer B.J., Brewer B.J. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- Friedman K.L., Brewer B.J., Fangman W.L., Brewer B.J., Fangman W.L., Fangman W.L. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:667–678. doi: 10.1046/j.1365-2443.1997.1520350.x. [DOI] [PubMed] [Google Scholar]
- Gambus A., Jones R.C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K., Jones R.C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K., Kanemaki M., van Deursen F., Edmondson R.D., Labib K., van Deursen F., Edmondson R.D., Labib K., Edmondson R.D., Labib K., Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Gasser S.M., Laroche T., Falquet J., de Boy la Tour E., Laemmli U.K., Laroche T., Falquet J., de Boy la Tour E., Laemmli U.K., Falquet J., de Boy la Tour E., Laemmli U.K., de Boy la Tour E., Laemmli U.K., Laemmli U.K. Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 1986;188:613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Goldman M.A., Holmquist G.P., Gray M.C., Caston L.A., Nag A., Holmquist G.P., Gray M.C., Caston L.A., Nag A., Gray M.C., Caston L.A., Nag A., Caston L.A., Nag A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Goto T., Wang J.C., Wang J.C. Cloning of yeast TOP1, the gene encoding DNA topoisomerase I, and construction of mutants defective in both DNA topoisomerase I and DNA topoisomerase II. Proc. Natl. Acad. Sci. 1985;82:7178–7182. doi: 10.1073/pnas.82.21.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Weinert T.A., Weinert T.A. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J.C., Botstein D., Goto T., Wang J.C., Botstein D., Wang J.C., Botstein D., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Holm C., Stearns T., Botstein D., Stearns T., Botstein D., Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol. Cell. Biol. 1989;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky S.A., Estep P., Church G.M., Samson L.D., Estep P., Church G.M., Samson L.D., Church G.M., Samson L.D., Samson L.D. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 2000;20:8157–8167. [Google Scholar]
- Ju B.G., Lunyak V.V., Perissi V., Garcia-Bassets I., Rose D.W., Glass C.K., Rosenfeld M.G., Lunyak V.V., Perissi V., Garcia-Bassets I., Rose D.W., Glass C.K., Rosenfeld M.G., Perissi V., Garcia-Bassets I., Rose D.W., Glass C.K., Rosenfeld M.G., Garcia-Bassets I., Rose D.W., Glass C.K., Rosenfeld M.G., Rose D.W., Glass C.K., Rosenfeld M.G., Glass C.K., Rosenfeld M.G., Rosenfeld M.G. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K., Kanoh Y., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K., Tanaka H., Ashikari T., Sugimoto K., Shirahige K., Ashikari T., Sugimoto K., Shirahige K., Sugimoto K., Shirahige K., Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kaneshiro K., Aburatani H., Shirahige K., Kaneshiro K., Aburatani H., Shirahige K., Aburatani H., Shirahige K., Shirahige K. Genomic approach for the understanding of dynamic aspect of chromosome behavior. Methods Enzymol. 2006;409:389–410. doi: 10.1016/S0076-6879(05)09023-3. [DOI] [PubMed] [Google Scholar]
- Kim R.A., Wang J.C., Wang J.C. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J. Mol. Biol. 1989;208:257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- Lemaitre J.M., Danis E., Pasero P., Vassetzky Y., Mechali M., Danis E., Pasero P., Vassetzky Y., Mechali M., Pasero P., Vassetzky Y., Mechali M., Vassetzky Y., Mechali M., Mechali M. Mitotic remodeling of the replicon and chromosome structure. Cell. 2005;123:787–801. doi: 10.1016/j.cell.2005.08.045. [DOI] [PubMed] [Google Scholar]
- Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Watanabe Y., Shirahige K., Uhlmann F., Shirahige K., Uhlmann F., Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.K., Chen A.Y., Yu C., Mao Y., Wang H., Liu L.F., Chen A.Y., Yu C., Mao Y., Wang H., Liu L.F., Yu C., Mao Y., Wang H., Liu L.F., Mao Y., Wang H., Liu L.F., Wang H., Liu L.F., Liu L.F. Activation of topoisomerase II-mediated excision of chromosomal DNA loops during oxidative stress. Genes & Dev. 1999;13:1553–1560. doi: 10.1101/gad.13.12.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Lopes M., Pellicioli A., Haber J.E., Foiani M., Pellicioli A., Haber J.E., Foiani M., Haber J.E., Foiani M., Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes & Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos H.B., Strom L., Itoh T., Katou Y., Shirahige K., Sjogren C., Strom L., Itoh T., Katou Y., Shirahige K., Sjogren C., Itoh T., Katou Y., Shirahige K., Sjogren C., Katou Y., Shirahige K., Sjogren C., Shirahige K., Sjogren C., Sjogren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Lisby M., Barlow J.H., Burgess R.C., Rothstein R., Barlow J.H., Burgess R.C., Rothstein R., Burgess R.C., Rothstein R., Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M., Muzi-Falconi M., Newlon C.S., Foiani M., Newlon C.S., Foiani M., Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Liberi G., Foiani M., Cotta-Ramusino C., Liberi G., Foiani M., Liberi G., Foiani M., Foiani M. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell. 2003;12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- Lucas I., Germe T., Chevrier-Miller M., Hyrien O., Germe T., Chevrier-Miller M., Hyrien O., Chevrier-Miller M., Hyrien O., Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J. 2001;20:6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucca C., Vanoli F., Cotta-Ramusino C., Pellicioli A., Liberi G., Haber J., Foiani M., Vanoli F., Cotta-Ramusino C., Pellicioli A., Liberi G., Haber J., Foiani M., Cotta-Ramusino C., Pellicioli A., Liberi G., Haber J., Foiani M., Pellicioli A., Liberi G., Haber J., Foiani M., Liberi G., Haber J., Foiani M., Haber J., Foiani M., Foiani M. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene. 2004;23:1206–1213. doi: 10.1038/sj.onc.1207199. [DOI] [PubMed] [Google Scholar]
- Lupo R., Breiling A., Bianchi M.E., Orlando V., Breiling A., Bianchi M.E., Orlando V., Bianchi M.E., Orlando V., Orlando V. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol. Cell. 2001;7:127–136. doi: 10.1016/s1097-2765(01)00161-7. [DOI] [PubMed] [Google Scholar]
- Lydall D., Whitehall S., Whitehall S. Chromatin and the DNA damage response. DNA Repair (Amst.) 2005;4:1195–1207. doi: 10.1016/j.dnarep.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Nakada D., Hirano Y., Sugimoto K., Hirano Y., Sugimoto K., Sugimoto K. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 2004;24:10016–10025. doi: 10.1128/MCB.24.22.10016-10025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D., Yuan D.S., Wang X., Bader J.S., Boeke J.D., Wang X., Bader J.S., Boeke J.D., Bader J.S., Boeke J.D., Boeke J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Paulovich A.G., Margulies R.U., Garvik B.M., Hartwell L.H., Margulies R.U., Garvik B.M., Hartwell L.H., Garvik B.M., Hartwell L.H., Hartwell L.H. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P.P., Foiani M., Lucca C., Liberi G., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P.P., Foiani M., Liberi G., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P.P., Foiani M., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P.P., Foiani M., Lopes M., Plevani P., Romano A., Di Fiore P.P., Foiani M., Plevani P., Romano A., Di Fiore P.P., Foiani M., Romano A., Di Fiore P.P., Foiani M., Di Fiore P.P., Foiani M., Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lee S.E., Lucca C., Foiani M., Haber J.E., Lee S.E., Lucca C., Foiani M., Haber J.E., Lucca C., Foiani M., Haber J.E., Foiani M., Haber J.E., Haber J.E. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Poirier M.G., Marko J.F., Marko J.F. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc. Natl. Acad. Sci. 2002;99:15393–15397. doi: 10.1073/pnas.232442599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloumienko A., Dershowitz A., De J., Newlon C.S., Dershowitz A., De J., Newlon C.S., De J., Newlon C.S., Newlon C.S. Completion of replication map of Saccharomyces cerevisiae chromosome III. Mol. Biol. Cell. 2001;12:3317–3327. doi: 10.1091/mbc.12.11.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L., Crisona N.J., Peter B.J., Hardy C.D., Cozzarelli N.R., Crisona N.J., Peter B.J., Hardy C.D., Cozzarelli N.R., Peter B.J., Hardy C.D., Cozzarelli N.R., Hardy C.D., Cozzarelli N.R., Cozzarelli N.R. Topological challenges to DNA replication: Conformations at the fork. Proc. Natl. Acad. Sci. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L., Hardy C.D., Arsuaga J., Cozzarelli N.R., Hardy C.D., Arsuaga J., Cozzarelli N.R., Arsuaga J., Cozzarelli N.R., Cozzarelli N.R. Topological domain structure of the Escherichia coli chromosome. Genes & Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Davis R.W., Brewer B.J., Fangman W.L., Brewer B.J., Fangman W.L., Fangman W.L. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Robinson N.P., Blood K.A., McCallum S.A., Edwards P.A., Bell S.D., Blood K.A., McCallum S.A., Edwards P.A., Bell S.D., McCallum S.A., Edwards P.A., Bell S.D., Edwards P.A., Bell S.D., Bell S.D. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 2007;26:816–824. doi: 10.1038/sj.emboj.7601529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman J.B., Stasiak A., Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Segurado M., Gomez M., Antequera F., Gomez M., Antequera F., Antequera F. Increased recombination intermediates and homologous integration hot spots at DNA replication origins. Mol. Cell. 2002;10:907–916. doi: 10.1016/s1097-2765(02)00684-6. [DOI] [PubMed] [Google Scholar]
- Shirahige K., Iwasaki T., Rashid M.B., Ogasawara N., Yoshikawa H., Iwasaki T., Rashid M.B., Ogasawara N., Yoshikawa H., Rashid M.B., Ogasawara N., Yoshikawa H., Ogasawara N., Yoshikawa H., Yoshikawa H. Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:5043–5056. doi: 10.1128/mcb.13.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J.A., Diffley J.F., Diffley J.F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ohkura H., Adachi Y., Morino K., Shiozaki K., Yanagida M., Ohkura H., Adachi Y., Morino K., Shiozaki K., Yanagida M., Adachi Y., Morino K., Shiozaki K., Yanagida M., Morino K., Shiozaki K., Yanagida M., Shiozaki K., Yanagida M., Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Wilm M., Bonte E., Dumas K., Mann M., Becker P.B., Wilm M., Bonte E., Dumas K., Mann M., Becker P.B., Bonte E., Dumas K., Mann M., Becker P.B., Dumas K., Mann M., Becker P.B., Mann M., Becker P.B., Becker P.B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Vidanes G.M., Bonilla C.Y., Toczyski D.P., Bonilla C.Y., Toczyski D.P., Toczyski D.P. Complicated tails: Histone modifications and the DNA damage response. Cell. 2005;121:973–976. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Wallis J.W., Chrebet G., Brodsky G., Rolfe M., Rothstein R., Chrebet G., Brodsky G., Rolfe M., Rothstein R., Brodsky G., Rolfe M., Rothstein R., Rolfe M., Rothstein R., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Wang J.C. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wyrick J.J., Aparicio J.G., Chen T., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Aparicio J.G., Chen T., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Chen T., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M., Young R.A., Bell S.P., Aparicio O.M., Bell S.P., Aparicio O.M., Aparicio O.M. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- Yamashita M., Hori Y., Shinomiya T., Obuse C., Tsurimoto T., Yoshikawa H., Shirahige K., Hori Y., Shinomiya T., Obuse C., Tsurimoto T., Yoshikawa H., Shirahige K., Shinomiya T., Obuse C., Tsurimoto T., Yoshikawa H., Shirahige K., Obuse C., Tsurimoto T., Yoshikawa H., Shirahige K., Tsurimoto T., Yoshikawa H., Shirahige K., Yoshikawa H., Shirahige K., Shirahige K. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:655–665. doi: 10.1046/j.1365-2443.1997.1530351.x. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Pongpech P., DiGate R.J., Pongpech P., DiGate R.J., DiGate R.J. Type I topoisomerase activity is required for proper chromosomal segregation in Escherichia coli. Proc. Natl. Acad. Sci. 2001;98:9766–9771. doi: 10.1073/pnas.171579898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S.J., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou H., Rothstein R., Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]