Abstract
Chromosome abnormalities resulting in gene fusions are commonly associated with acute myeloid leukemia (AML), however, the molecular mechanism(s) responsible for these defects are not well understood. The partial tandem duplication of the ALL1 (MLL) gene is found in patients with AML and trisomy 11 as a sole cytogenetic abnormality and in 11% of patients with AML and normal cytogenetics. This defect results from the genomic fusion of ALL1 intron 6 or intron 8 to ALL1 intron 1. Here, we examined the DNA sequence at the genomic fusion in nine cases of AML with a tandem duplication of ALL1 spanning exons 2–6. Each breakpoint occurred within intron 6 of the ALL1 breakpoint cluster region and within a discrete 3.8-kb region near the 3′ end of intron 1. In seven cases, a distinct point of fusion of intron 6 with intron 1 could not be identified. Instead, the sequence gradually diverged from an Alu element in intron 6 to an Alu element in intron 1 through a heteroduplex fusion. Thus, these rearrangements appear to be the result of a recombination event between homologous Alu sequences in introns 6 and 1. In two cases, the genomic junction was distinct and involved the fusion of a portion of an Alu element in intron 6 with non-Alu sequence in intron 1. These data support the hypothesis that a recombination event between homologous Alu sequences is responsible for the partial tandem duplication of ALL1 in the majority of AML cases with this genetic defect. Although Alu element-mediated homologous recombination events in germline cells are thought to be responsible for partial gene duplications or deletions in many inherited diseases, this appears to be the first demonstration identifying Alu element-mediated recombination as a consistent mechanism for gene rearrangement in somatic tissue.
In patients with acute leukemia, cytogenetic translocations involving chromosome region 11q23 usually result in the fusion of a gene known as ALL1 (1, 2), also referred to as MLL, HRX, and HTRX1 (3–5), with a variety of different genetic partners (6). In patients with acute myeloid leukemia (AML) but without cytogenetic abnormalities involving 11q23, we discovered a molecular defect involving ALL1 in which the gene undergoes a partial tandem duplication (PTD) of itself (7, 8). The PTD of ALL1 is present in the majority of patients with AML and trisomy 11 (+11) as a sole cytogenetic abnormality (9, 10) and in ≈11% of patients with AML and normal cytogenetics (11, 12).
Chromosome abnormalities resulting in gene fusions are commonly associated with AML, however, the molecular mechanism(s) responsible for these defects are not well understood (13). In many of the lymphoid malignancies, the presence of heptamer-nonamer recombination signals adjacent to the genomic breakpoints on both chromosomes has suggested that V(D)J recombinase activity is involved in illegitimate recombination events leading to these translocations (14). A similar mechanism has not been identified in the majority of molecular defects found in AML (13). Chromosome translocations involving ALL1 often are present in patients who develop secondary leukemia following treatment of a primary malignancy with drugs that inhibit topoisomerase II (15, 16). Although several topoisomerase II DNA binding sites have been identified within the 11q23 breakpoint cluster region (bcr) (17), and in vitro treatment of cell lines with topoisomerase II inhibitors induces double-stranded breaks within this region (18), the mechanism of translocation associated with these drugs has not been elucidated.
In two cases of ALL1 PTD, we and others have provided evidence to suggest that the genomic rearrangement involved a recombination event between Alu sequences (19, 20). In this study, we examined the DNA sequence at the genomic fusion in nine additional cases of primary AML, each with an ALL1 PTD spanning exons 2–6. The results of this study support the hypothesis that a recombination event between homologous Alu sequences is responsible for the ALL1 PTD in the majority of cases with this defect. Although Alu-mediated homologous recombination events in germline cells are thought to be responsible for genetic defects in many inherited diseases (21–26), this appears to be the first demonstration identifying Alu-mediated homologous recombination as a consistent mechanism for gene rearrangement in somatic tissue.
MATERIALS AND METHODS
Patients.
Nine patients with primary AML were studied. The presence of ALL1 PTD has been described for patients 23 (8), 24 (8), 101 (11), 166 (11), 211 (11), and 300 (11). For patients 316, 340, and 350, the presence of ALL1 PTD was determined during this study. DNA was extracted from the leukemic specimens, digested with EcoRI or HindIII and probed by Southern blot analysis using the B859 and SAS1 probes (8). By this method, each sample was found to have a rearranged ALL1 gene (data not shown). By using described amplification conditions (9), further analysis of total cellular RNA from each case by reverse transcription-PCR revealed the presence of an ALL1 PTD (data not shown). All cases in this study had a tandem duplication of ALL1 spanning exons 2–6.
PCR.
Genomic DNA was extracted by using a standard isolation procedure (27). PCR was performed by using Taq Extender PCR Additive (Stratagene) according to the manufacturer’s instructions. The upstream primer was 6.1 from exon 6 and the downstream primer was 2.0R from exon 2 (9). Reactions were carried out for 35 cycles (95°C for 1 min, 60°C for 1 min, and 72°C for 3.5 min), followed by a 10 min extension at 72°C. All PCR products were subsequently analyzed on 0.8% agarose gels stained with ethidium bromide.
Breakpoint Subcloning.
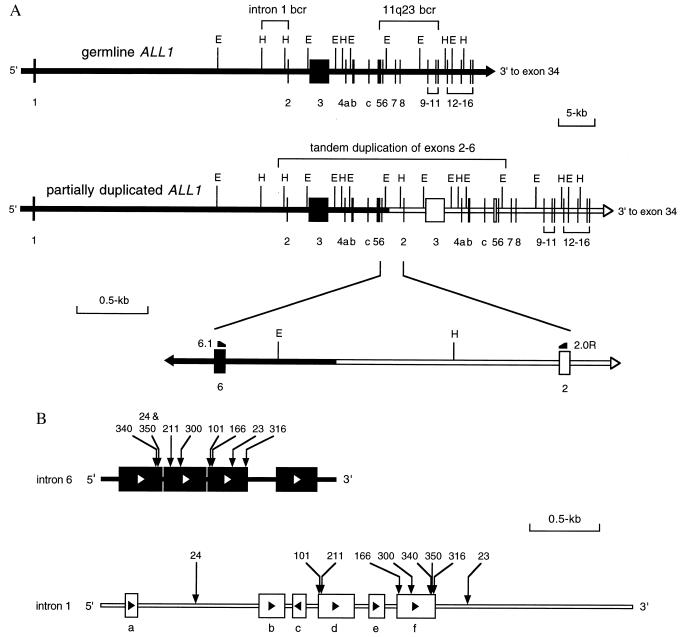
PCR amplification products were purified (Qiagen, Chatsworth, CA) and then digested by using restriction enzymes EcoRI and HindIII. In all cases, the presence of an EcoRI restriction site in intron 6, 363-bp downstream of exon 6, and a HindIII restriction site in intron 1, 724-bp upstream of exon 2, allowed for the subcloning of a fragment containing the intronic fusion into pBluescript (Stratagene) (Fig. 1A).
Figure 1.
Cloning the genomic breakpoints involved in the tandem duplication of ALL1 exons 2–6. (A) The wild-type ALL1 gene is normally composed of 36 exons. For the purposes of this figure, only exons 1–16 are shown. In this study, all breakpoints occurred within intron 6 of the 11q23 bcr and within a discrete region near the 3′ end of intron 1, indicated as intron 1 bcr. Tandem duplication of ALL1 exons 2–6 results from a fusion of intron 6 with intron 1 in a 5′ to 3′ direction. This is indicated by the fusion of the solid bar with the open bar. The fusion region is enlarged to show the location of PCR primers 6.1 and 2.0R and restriction sites used for cloning the breakpoints. E, EcoRI; H, HindIII. (B) Alu elements and breakpoints involved in the tandem duplication of ALL1 exons 2–6. Intron 6 is 1.7 kb long and contains four full-length Alu elements, as represented by the solid boxes. Open arrows in the boxes indicate the orientation of the Alu elements. The location of the breakpoint for each case is indicated by a vertical arrow and corresponding patient number. The breakpoints of patients 24 and 350 are only one nucleotide apart and are indicated by the same arrow. The intron 1 bcr is 3.8 kb and contains 2 Alu elements (boxes d and f), and four shorter portions of Alu elements (boxes a, b, c, and e). Solid arrows in the boxes indicate their orientation. The location of the breakpoint for each case is indicated by the vertical arrow and corresponding patient number.
DNA Sequencing and Analysis.
DNA sequencing of plasmids was performed by using an Applied Biosystems model 373 Stretch DNA Sequencing System. To avoid inclusion of PCR generated point mutations in the sequences, three different subclones from each case were sequenced. All fusion sequences were compared with germline sequences by using the geneworks computer program (IntelliGenetics, Campbell, CA). The ALL1 intron 6 germline sequence was obtained from GenBank. A genomic DNA fragment containing germline ALL1 intron 1 was provided by Rolf Marschalek and Iris Nilson (University of Erlangen-Nürnberg, Germany). The intron 1 bcr extends 3.8 kb upstream from exon 2 to a HindIII site. This region was sequenced and compared against the GenBank database. Alu mapping was performed by using the basic local alignment search tool (blast).
RESULTS AND DISCUSSION
The ALL1 PTD spanning exons 2–6 has two breakpoints within ALL1; one within intron 6 and one within intron 1 (8). The result is a fusion of intron 6 with intron 1 in a 5′ to 3′ direction (Fig. 1A). To determine the precise location of the intronic breakpoints, fusion sequence was compared with the germline sequences of introns 6 and 1. The sequence of intron 6 has been previously reported and has been shown to contain four Alu elements (17). Although intron 1 is ≈35 kb long (28), the intron 1 breakpoints all clustered within a discrete 3.8-kb region near the 3′ end of the intron, which we have designated as the intron 1 bcr (Fig. 1A). Sequence analysis of this region identified two Alu elements and four shorter portions of Alu elements (Fig. 1B). Characteristically, Alu elements are composed of a tandem repeat of two highly homologous sequences separated by a short A-rich region and are 300 bp long (29). Four distinct segments in the intron 1 bcr do not have these standard Alu features, but instead consist of portions of Alu sequence ranging from 97 to 186 bp (Fig. 1B).
We analyzed the fusion sequence in genomic DNA extracted from the blasts of nine patients with AML and ALL1 PTD of exons 2–6. In all nine cases, the genomic breakpoints were cloned by using PCR (Fig. 2). All nine cases had the intron 6 breakpoint directly within an Alu element. In seven cases (patients 101, 166, 211, 300, 316, 340, and 350) the intron 1 breakpoints were located within one of the two full-length Alu elements (Fig. 1B). Six of these cases had normal karyotypes, and one case had a karyotype of 46,XY, der(5)t(5;8)(q35;q11.2). The two remaining cases (patients 23 and 24) had intron 1 breakpoints located within non-Alu sequence (Fig. 1B). Both cases had +11 as a sole cytogenetic abnormality on karyotype analysis (data not shown).
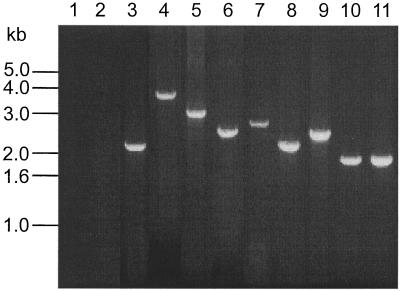
Figure 2.
PCR products amplified from genomic DNA of nine patients with tandem duplication of ALL1 exons 2–6 by using primers 6.1 and 2.0R. Lanes 1 and 2 are reagent and normal controls, respectively. Lanes 3–11 show the amplified products from patients 23, 24, 101, 166, 211, 300, 316, 340, and 350, respectively. Bands ranged in size from 1.9 to 3.6 kb.
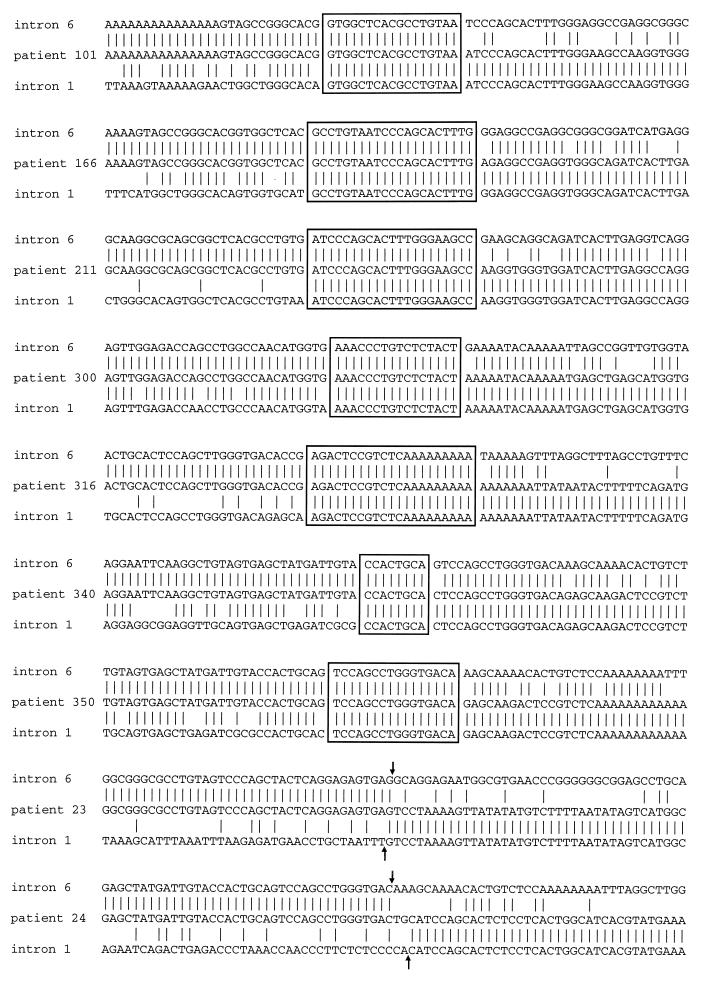
In seven cases (patients 101, 166, 211, 300, 316, 340, and 350), a distinct point of fusion of intron 6 with intron 1 could not be identified. Instead, the sequence gradually diverged through an Alu element from intron 6 to intron 1 via an overlapping or heteroduplex fusion region having an average length of 18 bp (range, 8 to 21 bp) (Fig. 3). In each instance, nucleotide sequences were not altered at the site of exchange, and the fusion maintained the natural length and structure of the Alu element (Fig. 4). Thus, these rearrangements appear to result from recombination between homologous Alu sequences in introns 6 and 1. In two cases (patients 23 and 24), the genomic fusion involved a portion of an Alu element in intron 6 with a non-Alu sequence in intron 1 (Fig. 3). In both instances, the Alu element within intron 6 was disrupted near its 3′ end and resulted in the formation of a truncated Alu element (Fig. 4).
Figure 3.
DNA sequence of genomic fusion from nine cases with tandem duplication of ALL1 exons 2–6. Fusion sequences are compared with germline intron 6 and intron 1 sequence. Vertical lines indicate sites of nucleotide homology. Patients 101, 166, 211, 316, 340, and 350 showed evidence of recombination between Alu elements from introns 6 and 1. Boxed sequences indicate overlapping regions of homology, consistent with heteroduplex formation during homologous recombination. Patients 23 and 24 had fusion of Alu sequence from intron 6 to non-Alu sequence from intron 1. For these two cases, the arrows indicate the breakpoints within introns 6 and 1. Sequences are 5′ to 3′.
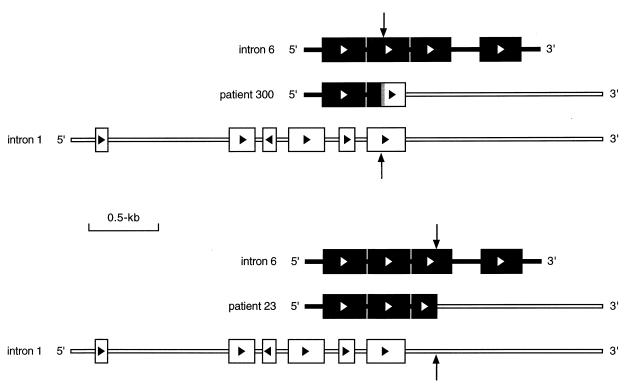
Figure 4.
Genomic structures involved in ALL PTD. Patient 300 had breakpoints within Alu elements in introns 6 and 1. This fusion resulted in the formation of a full-length Alu element. The vertical arrows indicate all breakpoints. Solid and open boxes represent Alu elements from introns 6 and 1, respectively. The gray box at the fusion represents the region of overlapping homology. Patient 23 had the intron 6 breakpoint near the 3′ end of an Alu element and the intron 1 breakpoint within non-Alu sequence. This fusion resulted in the formation of a truncated Alu element.
Homologous recombination is generally considered to be a natural mechanism for homologous chromosomes in meiotic cells to exchange large segments of DNA (30). Occasionally, however, these events are imperfect and unequal crossing-over between homologous Alu elements in meiotic cells has been shown to be the cause of several inherited diseases including predisposition to colon cancer (21), predisposition to breast cancer (22), adenosine deaminase deficiency (23), hypobetalipoproteinemia (24), familial hypercholesterolemia (25), and lipoprotein lipase deficiency (26). In these disorders, Alu-mediated homologous recombination events cause either partial deletions or PTDs of a specific gene. In the disorders that result from PTDs, the genetic defects appear to have the same general features as the ALL1 PTD in that an internal portion of the gene is duplicated in a tandem fashion with an overlap of Alu sequence at the genomic fusion. However, while these are inherited germline mutations that occur in meiotic cells, the ALL1 PTD occurs in somatic cells that only undergo mitosis. Thus, it is reasonable to believe that if the ALL1 PTD was caused by an unequal cross-over event between homologous chromosomes, then two chromosome 11s would be rearranged; one with a PTD and one with a partial deletion. This does not appear to be the case, however, as Southern blot analysis of ALL1 PTD consistently identifies a single germline band and a single rearranged band (8–12, 19, 31). Further, in AML cases with +11 or normal cytogenetics that have been studied, only one of the chromosome 11s carry the ALL1 PTD (32).
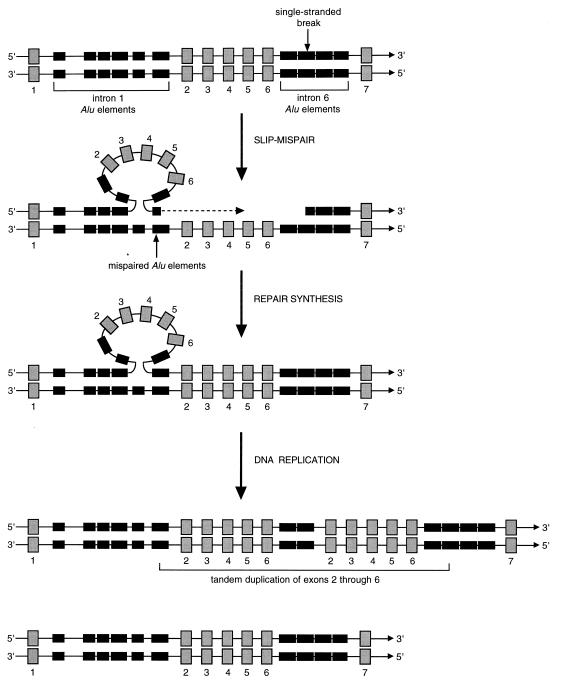
A possible mechanism of ALL1 PTD may therefore involve an intrastrand recombination event. This process, called intrastrand slipped-mispairing, was described by Roth et al. (33) to explain tandem duplications found in minisatellite regions of chromosomes and within the simian virus 40 genome. The intrastrand slipped-mispairing model suggests that upon breakage of a single strand of double-stranded DNA, mispairing can occur between Alu elements in a slipped region and homologous Alu elements in another region. Following repair synthesis, an additional round of DNA replication will generate a tandem duplication with a short overlap at the point of fusion, and a wild-type allele. Theoretically, a PTD of ALL1 could be generated by slipped-mispairing if the initial break was within intron 6 of ALL1. The slipped region would span exons 2–6 and mispairing would occur between homologous Alu sequences from introns 6 and 1. Such a structure could provide a primer for the synthesis of a tandem duplication 3′ of the slipped region (Fig. 5). Alternatively, if the initial break was within intron 1 and mispairing occurred between homologous Alu sequences from introns 1 and 6, exons 2 though 6 could be synthesized immediately 5′ and tandem to the trapped structure (data not shown). The cause of the initial single strand DNA breaks in this model is unknown.
Figure 5.
The intrastrand slipped-mispairing model as applied to the ALL1 PTD. Shown at the top of the figure is a segment of the wild-type ALL1 gene as double-stranded DNA. Gray numbered boxes represent exons. Solid boxes represent Alu elements from introns 1 and 6. In the model, slipped mispairing of a single strand of double-stranded DNA generates a structure that could prime the synthesis of the slipped region, resulting in a tandem duplication. In this example, the break in a single strand occurs in the middle of the second Alu element in intron 6 (vertical arrow). The resultant slipped region mispairs with an Alu element in intron 1. Repair synthesis produces a single strand with a tandem duplication of exons 2–6. A subsequent round of DNA replication produces and one chromosome with the partial tandem duplication of ALL1 and one chromosome with a wild-type ALL1. Adapted from Roth et al. (33).
Two cases (patients 23 and 24) presented here did not have a heteroduplex at the intronic fusion, and did not appear to result from intrastrand recombination between Alu elements. These were the only two cases in this series with +11 as a sole cytogenetic abnormality. Interestingly, one other case of AML with +11 and the PTD of ALL1 spanning exons 2–6 has been shown to fuse a portion of an Alu element in intron 6 with a non-Alu sequence in intron 1 (10). Further analysis of the upstream and downstream sequences flanking the breakpoints in both cases reported here revealed the presence of Alu elements in introns 6 and 1 that were aligned. An example of this can be seen for patient 23 in the lower portion of Fig. 4 where, although the breakpoint occurred within the third Alu element in intron 6, the second Alu element aligns with one of the full-length Alu elements from intron 1. Although neither of the PTDs in these cases appear to directly result from homologous recombination, it is plausible that the alignment of Alu elements may help to stabilize a putative recombination complex. The significance of these findings only in cases of AML with the PTD of ALL1 and +11 is currently unclear.
In summary, the results of this study support the hypothesis that the ALL1 PTD spanning exons 2–6 in AML is caused by a homologous recombination event between Alu sequences within introns 6 and 1. This appears to be the first demonstration identifying Alu-mediated homologous recombination as a consistent mechanism for gene rearrangement in somatic tissue.
Acknowledgments
We thank Drs. Rolf Marschalek and Iris Nilson for kindly providing a genomic clone containing the germline ALL1 intron 1. This work was supported, in part, by a Translational Research Grant from the Leukemia Society of America, the Lady Tata Memorial Trust, National Cancer Institute Grant P30CA16058, National Cancer Institute National Research Service Award CA-09338, and the Coleman Leukemia Research Fund.
ABBREVIATIONS
- AML
acute myeloid leukemia
- PTD
partial tandem duplication
- +11
trisomy 11
- bcr
breakpoint cluster region
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF033349).
References
- 1.Cimino G, Moir D T, Canaani O, Williams K, Crist W M, Katzav S, Cannizzaro L, Lange B, Nowell P C, Croce C M, et al. Cancer Res. 1991;51:6712–6714. [PubMed] [Google Scholar]
- 2.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 3.Ziemin-van der Poel S, McCabe N R, Gill H J, Espinosa R, III, Patel Y, Harden A, Rubinelli P, Smith S D, LeBeau M M, Rowley J D, et al. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 5.Djabali M, Selleri L, Parry P, Bower M, Young B D, Evans G A. Nat Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 6.Bernard O A, Berger R. Genes Chromosomes Cancer. 1995;13:75–85. doi: 10.1002/gcc.2870130202. [DOI] [PubMed] [Google Scholar]
- 7.Caligiuri M A, Schichman S A, Strout M P, Mrozek K, Baer M R, Frankel S R, Barcos M, Herzig G P, Croce C M, Bloomfield C D. Cancer Res. 1994;54:370–373. [PubMed] [Google Scholar]
- 8.Schichman S A, Caligiuri M A, Gu Y, Strout M P, Canaani E, Bloomfield C D, Croce C M. Proc Natl Acad Sci USA. 1994;91:6236–6239. doi: 10.1073/pnas.91.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caligiuri M A, Strout M P, Schichman S A, Mrozek K, Arthur D C, Herzig G P, Baer M R, Schiffer C A, Heinonen K, Knuutila S, et al. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]
- 10.Bernard O A, Romana S P, Schichman S A, Mauchauffe M, Jonveaux P, Berger R. Leukemia. 1995;9:1487–1490. [PubMed] [Google Scholar]
- 11.Caligiuri M A, Strout M P, Lawrence D, Arthur D C, Baer M R, Yu F, Knuutila S, Mrozek K, Oberkircher A R, Marcucci G, et al. Cancer Res. 1998;58:55–59. [PubMed] [Google Scholar]
- 12.Yu M, Honoki K, Andersen J, Paietta E, Nam D K, Yunis J J. Leukemia. 1996;10:774–780. [PubMed] [Google Scholar]
- 13.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 14.Thandla S, Aplan P D. Semin Oncol. 1997;24:45–56. [PubMed] [Google Scholar]
- 15.Gill-Super H J, McCabe N R, Thirman M J, Larson R A, Le Beau M M, Pederson-Bjergaard J, Preben P, Diaz M O, Rowley J D. Blood. 1993;82:3705–3711. [PubMed] [Google Scholar]
- 16.Felix C A, Hosler M R, Winick N J, Masterson M, Wilson A E, Lange B J. Blood. 1995;85:3250–3256. [PubMed] [Google Scholar]
- 17.Gu Y, Alder H, Nakamura T, Schichman S A, Prasad R, Canaani O, Saito H, Croce C M, Canaani E. Cancer Res. 1994;54:2326–2330. [PubMed] [Google Scholar]
- 18.Aplan P D, Chervinsky D S, Stanulla M, Burhans W C. Blood. 1996;87:2649–2658. [PubMed] [Google Scholar]
- 19.Schichman S A, Caligiuri M A, Strout M P, Carter S L, Gu Y, Canaani E, Bloomfield C D, Croce C M. Cancer Res. 1994;54:4277–4280. [PubMed] [Google Scholar]
- 20.So C W, Ma Z G, Price C M, Dong S, Chen S J, Gu L J, So C K C, Wiedemann L M, Chan L C. Cancer Res. 1997;57:117–122. [PubMed] [Google Scholar]
- 21.Nystrom-Lahti M, Kristo P, Nicolaides N C, Chang S, Aaltonen L A, Moisio A, Jarvinen H J, Mecklin J, Kinzler K W, Vogelstein B, et al. Nat Med. 1995;1:1203–1206. doi: 10.1038/nm1195-1203. [DOI] [PubMed] [Google Scholar]
- 22.Puget N, Torchard D, Serova-Sinilnikova O M, Lynch H T, Feunteun J, Lenoir G M, Mazoyer S. Cancer Res. 1997;57:828–831. [PubMed] [Google Scholar]
- 23.Markert M L, Hutton J J, Wiginton D A, States J C, Kaufman R E. J Clin Invest. 1988;81:1323–1327. doi: 10.1172/JCI113458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Ripps M E, Korman S H, Deckelbaum R J, Breslow J L. J Biol Chem. 1989;264:11394–11400. [PubMed] [Google Scholar]
- 25.Lehrman M A, Goldstein J L, Russell D W, Brown M S. Cell. 1987;48:827–835. doi: 10.1016/0092-8674(87)90079-1. [DOI] [PubMed] [Google Scholar]
- 26.Devlin R H, Deeb S, Brunzell J, Hayden M R. Am J Hum Genet. 1990;46:112–119. [PMC free article] [PubMed] [Google Scholar]
- 27.Gustincich S, Manfioletti G, Del Sal G, Schneider C, Carninci P. BioTechniques. 1991;11:298–300. [PubMed] [Google Scholar]
- 28.Rasio D, Schichman S A, Negrini M, Canaani E, Croce C M. Cancer Res. 1996;56:1766–1769. [PubMed] [Google Scholar]
- 29.Schmid C W. Prog Nucleic Acid Res. 1996;53:283–318. doi: 10.1016/s0079-6603(08)60148-8. [DOI] [PubMed] [Google Scholar]
- 30.Darnell J, Lodish H, Baltimore D. Molecular Cell Biology. New York: Scientific American Books; 1990. [Google Scholar]
- 31.Poirel H, Rack K, Delabesse E, Radford-Weiss I, Troussard X, Debert C, Leboeuf D, Bastard C, Picard F, Veil-Buzyn A, et al. Blood. 1996;87:2496–2505. [PubMed] [Google Scholar]
- 32.Caligiuri M A, Strout M P, Oberkircher A R, Yu F, de la Chapelle A, Bloomfield C D. Proc Natl Acad Sci USA. 1997;94:3899–3902. doi: 10.1073/pnas.94.8.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth D B, Porter T N, Wilson J H. Mol Cell Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]