Abstract
Protein kinase casein kinase 1 (CK1) phosphorylates Ser-45 of β-catenin, “priming” the subsequent phosphorylation by glycogen synthase-3 of residues 41, 37, and 33. This concerted phosphorylation of β-catenin signals its degradation and prevents its function in triggering cell division. The sequence around Ser-45 does not conform to the canonical consensus for CK1 substrates, which prescribes either phosphoamino acids or acidic residues in position n-3 from the target serine. However, the β-catenin sequence downstream from Ser-45 is very similar to a sequence recognized by CK1 in nuclear factor for activated T cells 4. The common features include an SLS motif followed two to five residues downstream by a cluster of acidic residues. Synthetic peptides reproducing residues 38-65 of β-catenin were assayed with purified rat liver CK1 or recombinant CK1α and CK1αL from zebrafish. The results demonstrate that SLS and acidic cluster motifs are crucial for CK1 recognition. Pro-44 and Pro-52 are also important for efficient phosphorylation. Similar results were obtained with the different isoforms of CK1. Phosphorylation of mutants of full-length recombinant β-catenin from zebrafish confirmed the importance of the SLS and acidic cluster motifs. A search for proteins with similar motifs yielded, among other proteins, adenomatous polyposis coli, previously found to be phosphorylated by CK1. There is a strong correlation of β-catenin mutations found in thyroid tumors with the motifs recognized by CK1 in this protein.
Keywords: consensus sequence, nuclear factor for activated T cells 4, adenomatous polyposis coli, Wnt signaling, thyroid tumor mutations
Protein phosphorylation, catalyzed by hundreds of different protein kinases, plays a key role in signal transduction. Proteins that are phosphorylated in several residues by different protein kinases might, therefore, serve as signal integrators. Because the ultimate response to stimuli often involves changes in gene expression, particular significance may reside in the integrative role of transcription factors regulated by phosphorylation in multiple sites, as is the case with p53, a key tumor suppressor (1).
Signal Integration Through Multiple Phosphorylation: Hierarchical Phosphorylation
A particularly interesting example of signal integration through multiple phosphorylations is hierarchical phosphorylation (2, 3), also termed phosphoamino acid “primed” phosphorylation (4). Glycogen synthase-3 (GSK-3) and protein kinases casein kinases 1 (CK1) and 2 (CK2) are three kinases that recognize phosphorylated residues as specificity determinants in the proximity of their target hydroxylamino acid. Such substrates are primed by introducing the first phosphorylation followed by subsequent phosphorylation(s) by either the same or another kinase, thus establishing a “hierarchy” in the sequential phosphorylations.
GSK-3β plays a vital role in the regulation of metabolism, cell division, and embryogenesis and is known to phosphorylate primed substrates, as seen in the case of glycogen synthase that requires priming by CK2 for phosphorylation of Ser-652, Ser-648, and Ser-644 (site 3) (2).
The recent elucidation of the crystal structure of GSK-3β (5, 6) has provided evidence that a basic pocket composed of Arg-96, Arg-180, and Lys-205 binds the phosphate in the primed site and positions the substrate in the proper orientation to accept a new phosphate four amino acids upstream (7). Thus primed substrates have the structure S/T X X X S/T-PO4, where S/T corresponds to the target serine or threonine, X to any other amino acid, and S/T-PO4 to a previously phosphorylated serine or threonine. Although in vitro GSK-3 can phosphorylate proteins and peptides that are not primed, their efficiencies compared with primed substrates are several orders of magnitude lower (7). It is not clear whether in vivo GSK-3 can phosphorylate unprimed substrates.
CK2 is known to phosphorylate >300 protein substrates, be involved in the regulation of the cell cycle, and have antiapoptosis activity (8-12). Only 10 of 308 CK2 sites analyzed so far are strictly primed (i.e., “phosphate directed”) sites, that is having a phosphorylated residue acting as crucial determinant at position n+3 (10). The minimum substrate specificity for CK2 is S/T X X D/E for unprimed substrates or S/T XX S/T/Y-PO4 for primed substrates (D/E stands for aspartic or glutamic acid and Y-PO4 for phosphotyrosine), indicating that position n+3 from the target serine or threonine must be occupied by a side chain with either a carboxylic or a phosphoryl group (12).
CK1
CK1 is located in a separate group within the superfamily of eukaryotic protein kinases (13). Within the CK1 family, there are several genes that code for related kinases in all species. In mammals and vertebrates, there are seven CK1 genes coding for α, β, γ1, γ2, γ3, δ, and ε isoforms (14), and some of these genes generate several proteins through alternative splicing (15). The CK1 isoforms may have different functions; for instance, CK1ε is involved in regulating circadian rhythm through phosphoration of period (per) proteins (16), and it has also been linked to the activity of group I metabotropic glutamate receptors (17). CK1δ may be involved in neurodegenerative diseases (18). CK1α apparently participates in the phosphorylation of G protein-regulated receptors (19) and nuclear factor for activated T cells 4 (NF-AT4) phosphorylation (20). However, in vitro studies with model peptides using different isoforms of CK1 have not shown significant differences in the preferred target sequences (21). Differences in functions of isoforms most probably involve differential expression and cellular localization and specific docking sites that confer substrate selectivity. CK1ε and CK1δ have an interesting regulatory mechanism where autophosphorylation of their carboxyl-terminal extensions inhibits their catalytic activity (22, 23). Dephosphorylation of these carboxyl residues by calcineurin may be a mechanism for regulating the activity of these isoforms (17).
CK1 has a substrate sequence consensus in which position n-3 is necessarily occupied by an acidic group or a phospho-amino acid. This consensus is D/E X X S/T for unprimed substrates or S/T-PO4 X X S/T for primed targets. It must be noted, however, that whereas CK2 carboxylic and phosphorylated side chains are equally potent as specificity determinants, CK1 by far prefers phosphorylated determinants, needing otherwise multiple acidic residues upstream from position n-2 to achieve catalytic efficiency comparable to that of primed sites (21, 24).
Regulation of NF-AT4 by Phosphorylation
NF-AT4 is a member of a family of transcription factors that activate T cells and participate in many cell processes (25). These proteins in unstimulated cells are phosphorylated at multiple serine and threonine residues in a region that masks a nuclear localization signal (NLS) and maintains NF-AT factors in a predominantly cytoplasmic localization. Specific external signals that increase intracellular Ca2+ activate calcineurin, a calmodulin-regulated protein phosphatase, which dephosphorylates NF-AT proteins, unmasking their NLS. NF-AT proteins then translocate to the nucleus altering gene expression.
Several kinases, including GSK-3β, c-Jun N-terminal kinase (JNK), p38, extracellularly regulated kinase (ERK), protein kinase A (PKA), and CK2 have been reported to participate in the phosphorylation of the NF-AT protein family (26-30). In 1998, however, Zhu et al. (20) showed that CK1α was bound to the regulatory region of NF-AT4, an interaction that enhanced its phosphorylation by this kinase. This observation prompted us to study this phosphorylation with the use of synthetic peptides derived from the NF-AT4 region reported to bind and be phosphorylated by CK1α. Although our results did not fully agree with those authors (20), we found that peptides corresponding to the A-2 domain of NF-AT4 could be phosphorylated by CK1 and that these peptides contained a motif that was different from the canonical consensus structure that had been determined for CK1 (31). Essentially, we found that an acidic cluster at n+4 to n+9 from the target serines was able to drive unprimed phosphorylation of these peptides. We postulated that this acidic cluster acted as a functional docking site that facilitated phosphorylation of residue Ser-177, which lacks the canonical consensus for CK1. Once the initial phosphorylation was attained on Ser-177, the primed peptides were then extensively phosphorylated by CK1α in a hierarchical and canonical fashion.
β-Catenin Phosphorylation by GSK-3 and CK1
β-Catenin also participates in transcription and is regulated through multiple phosphorylations that control its access to the cell nucleus. β-Catenin is degraded by the ubiquitin proteasome pathway in unstimulated cells. When β-catenin is phosphorylated in Ser-33 and Ser-37 it is recognized by β-Trcp, a component of the ubiquitination apparatus, and degraded. Much evidence supports the idea that GSK-3β is the kinase responsible for the phosphorylation in these β-catenin residues (32). This phosphorylation is favored by the formation of a multiprotein complex bound to the scaffold protein axin, which includes GSK-3, β-catenin, and adenomatous polyposis coli (APC) among other proteins. Phosphorylation of β-catenin at these N-terminal residues was held as a typical example of nonprimed phosphorylation by GSK-3 (7, 32).
The regulation of the degradation of β-catenin plays a key role in embryogenesis and cell proliferation. Extracellular Wnt signal proteins are bound to a membrane receptor called “frizzled,” which causes an activation of the protein dishevelled (Dvl). It is interesting that Dvl can be phosphorylated by CK1ε and this phosphorylation apparently enhances its interaction with other regulatory factors (33). Activated Dvl inhibits, by a mechanism not yet understood, the phosphorylation of β-catenin in the multiprotein complex. As a result, β-catenin is not phosphorylated and therefore is not recognized and ubiquitinated by the Skip1-Cullin-F box complex β-Tcrp E3 ubiquitin ligase o degraded by the proteasome with the consequence that its intracellular concentration increases, especially in the nucleus. Nuclear β-catenin binds and activates the T cell factor transcription factors, eliciting the transcription of Wnt-responsive genes. Extensive work has established that in addition to Ser-33 and Ser-37 the phosphorylation of Thr-41 and Ser-45 is also critical for β-catenin regulation. The importance of these four amino acid sites is highlighted by the frequent finding in tumors of mutations that convert these phosphorylatable residues into nonphosphorylatable ones (34-36), a finding that correlates well with abundant nuclear β-catenin in transformed cells. In 1999, reports appeared that indicated that CK1 was an important participant in Wnt signaling. Initial reports disagreed about the specificity of the isoforms of CK1 in potentiating Wnt signals in the embryogenesis of Xenopus laevis and Caenorhabditis elegans (37, 38). However, recent work demonstrated that overexpression of any of the α, β, δ, or ε isoforms of CK1 could potentiate Wnt signals and cause double axis formation in Xenopus embryos (39). On the other hand, a year ago, several groups reported that CK1 can oppose Wnt, being directly involved in the phosphorylation of β-catenin, in a dual kinase concerted action together with GSK-3 (40-43). Liu et al. (40) reported that the anti-Wnt effect was caused by the CK1α isoform that phosphorylated Ser-45. Confirming these results, they observed that depletion of CK1α but not CK1ε through double-stranded RNA-mediated interference caused a significant decrease in Ser-45 phosphorylated β-catenin in human embryonic kidney 293 cells. The results of Sakanaka (41), however, indicate that CK1ε is able to phosphorylate Ser-45 in β-catenin. Yanagawa et al. (42), using a Drosophila system to study the phosphorylation of Armadillo (the β-catenin homologue of Drosophila), also confirmed the participation of CK1 in this phosphorylation. RNA interference experiments showed that both CK1α and CK1ε are involved in Armadillo phosphorylation and regulation. Those authors recognized the importance of the acidic cluster downstream of the target serine for the phosphorylation of this protein by CK1α. Finally, Amit et al. (43) reported that CK1 isoforms α, δ, and ε bound to axin were responsible for the priming phosphorylation at Ser-45 of β-catenin, which is subsequently followed by GSK-3 phosphorylation of Thr-41, Ser-37, and Ser-33. Interestingly, those authors found that Wnt3A and Dvl overexpression in cells completely abrogates Ser-45 phosphorylation of β-catenin. This finding implies that the Wnt cascade would preferentially act by inhibiting the CK1 activity that phosphorylates β-catenin.
In summary, although CK1 may have other effects, there is substantial agreement that this protein kinase is involved in the initial phosphorylation of β-catenin at Ser-45, and this phosphorylation results in the priming of GSK-3 for targeting residues Thr-41, Ser-37, and Ser-33, which are required for the degradation of this protooncogenic protein.
Corroborating that CK1 opposes Wnt signaling, Rubinfield et al. (44) demonstrated that the CK1ε isoform phosphorylates APC and that this phosphorylation strengthens its binding to axin and β-catenin degradation.
The Present Work
The evidence that CK1 was responsible for Ser-45 phosphorylation in the protein β-catenin attested to a second instance in which CK1 could phosphorylate a transcription factor at a noncanonical site and in which CK1 and GSK-3 could cooperate in multisite phosphorylations. Comparison of the sequence surrounding Ser-45 in β-catenin and the noncanonical site of phosphorylation described in NF-AT4 peptides (31) (Fig. 1) disclosed intriguing coincidences. Notably, both sequences contain an acidic cluster downstream from the target serines, and the motif SLS involving Ser-45 of β-catenin is present in both sequences. This similarity prompted us to carry out in vitro experiments to test the relevance of these common features for the recognition by CK1 of the sequences neighboring Ser-45 of β-catenin.
Fig. 1.
Alignment of sequences from NF-AT4 (human), β-catenin (human), and Armadillo (the β-catenin homologue of Drosophila) to highlight the similarities that may be recognized by CK1.
The results presented in this article demonstrate that both the acidic cluster in positions n+7 and the SLS motif are very important for this type of unprimed noncanonical recognition by CK1 both in synthetic peptides and the full-length β-catenin protein. Another protein that contains these motifs is APC, which had been previously shown to be phosphorylated by CK1 (44). The previous finding of mutations of the amino acids present in the motifs important for CK1 recognition in the β-catenin genes of thyroid tumors (45, 46) lends strong physiological significance to these observations.
Materials and Methods
Expression and Purification of Proteins. Recombinant CK1α and CK1αL from zebrafish (Danio rerio) with amino-terminal (His)6 tags were expressed in Escherichia coli BL21 (DE3) and purified through Ni2+-NTA-agarose columns as described (15).
Rat liver CK1 was purified from fresh tissue as described (47) (specific activity 11,000 units/mg, one unit being the amount of enzyme that can transfer 1 pmol of phosphate per min into the specific peptide R R K H A A I G D D D D D A Y S I T A). This preparation is free of other kinases such as GSK-3, CK2, and proline-directed kinases.
β-Catenin was cloned by PCR from a cDNA library of zebrafish using as primers 5′-TAT ATA CAT ATG GCT ACC CAG TC-3′ and 5′-TAT ATA GAA TTC TTA CAG ATC GGT GT-3′. The amplified product was introduced in a pT7H6 vector for expression in E. coli. The β-catenin coding insert was sequenced with an Applied Biosystems Prism 377 automatic sequencer, confirming the sequence published in GenBank (accession no. BC047815).
Mutations of β-catenin to generate S45A, L46A, and the mutant with all acidic groups changed to alanine (E53, D54, E55, D56, D58 to A) were prepared by an overlapping primer PCR method (48) using the following primers: for S45A, 5′-CTG CCC CGG CCC TGA GTG G-3′ and 5′-CCA CTC AGG GCC GGG GCA-3′; for L46A, 5′-CTG CCC CGT CCG CGA GTG GC-3′ and 5′-GCC ACT CGC GGA CGG GGC AG-3′, and for the acid cluster primers, 5′-CAA CCC GGC GGC TGC CGC TGT GGC TAA TCA G-3′ and 5′-CTG ATT AGC CAC AGC GGC AGC CGC CGG GTT G-3′.
Peptide Synthesis. The synthesis of peptides derived from X. laevis β-catenin, NF-AT4, and APC was carried out as described (49).
Peptide Phosphorylation Assay. The assay has been described in detail (31). Briefly, synthetic peptide substrates (0.2 mM) were phosphorylated by incubation in a medium (25 μl final volume) containing 50 mM Tris·HCl buffer (pH 7.5), 10 mM MgCl2, 100 mM NaCl, and 40 μM [γ-32P]ATP (specific radioactivity 500-1,000 cpm/pmol). The reaction was started with addition of the protein kinase and stopped by cooling in ice and absorption onto phosphocellulose paper for scintillation counting. Kinetic constants were determined by regression analysis of double-reciprocal plots constructed from initial-rate measurements.
β-Catenin Phosphorylation. For β-catenin phosphorylation, 5 pmol of recombinant full-length zebrafish β-catenin was phosphorylated for 15 min at 37°C in kinase buffer containing 50 mM Tris·HCl buffer (pH 7.5), 10 mM MgCl2, 100 mM NaCl, and 10 μM [γ-32P]ATP (specific radioactivity 8,000-10,000 cpm/pmol). The reaction was started with the addition of CK1α and stopped by addition of 5× Laemmli buffer. Samples were separated by SDS/PAGE, transferred to poly(vinylidene difluoride) membranes for Western blots, and developed with anti-His mAb. The membranes were also subjected to autoradiography using x-ray film (Kodak).
Homology Search for a New CK1 Consensus Motif. canprosite searches (available at http://ca.expasy.org/tools/scanprosite) were performed by using the pattern SLS-[CRQWFMTVSGKNPLAHIY](2,5)-[DE](2,6)-[ACDEFGHILMNPQSTVWY](0,1)— [DE](0,6) as query, in the SwissProt database. Searches were performed in the Homo sapiens taxon and all other parameters at default values. See Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
Specialized Reagents. Oligonucleotides were prepared by the Oligopeptido Core Facility, University of Chile. Ni2+-NTA Sepharose was from Novagen. [γ-32P]ATP was from New England Nuclear, and anti-His mAb was from CLONTECH.
Results
Phosphorylation of β-Catenin-Derived Peptides by CK1. Peptides reproducing with suitable modifications the sequence of X. laevis β-catenin between amino acids 38 and 65 and containing three additional arginine residues in the N terminus to facilitate the phosphocellulose paper assay were synthesized and purified as described (49).
These peptides contained permutations in the sequence motifs common to NF-AT4 and β-catenin and in positions neighboring Ser-45, the known target of CK1 (Table 1). Several modifications introduced into these peptides were inspired by naturally occurring mutations of β-catenin found in thyroid tumors (45, 46). Peptides were phosphorylated by using CK1 purified from rat liver (47), which is essentially free of other kinase activities and with recombinant CK1α and CK1αL (15) from zebrafish. Table 1 provides the values for approximate Km, Vmax, and phosphorylation efficiency as reflected in the ratio of Vmax/Km. Values obtained with zebrafish splice variant CK1αL (15) were essentially similar to those found with zebrafish CK1α (data not shown). It is clear that the SLS motif involving amino acids 45-47 is important for CK1 recognition. Replacement of Ser-45 with alanine almost completely eliminated phosphorylation in peptide (S45A) consistent with the previous evidence that Ser-45 and not Ser-47 is the target of CK1 in β-catenin (40-43). This finding also agrees with the analysis of phosphoamino acids (data not shown) after incubation of WT peptide with CK1 that showed no detectable phosphothreonine. Interestingly, replacement of Ser-45 with threonine (S45T) also lowered the phosphorylation efficiency, reflecting the known preference of CK1 for serine over threonine. Replacement of Ser-45 with phosphoserine in the synthetic peptide (pS45) completely eliminates the capacity of this peptide to function as a substrate, demonstrating that the enzyme cannot use this primed substrate for subsequent phosphorylation and demonstrating that Ser-47 is not a target for CK1.
Table 1. Phosphorylation by protein kinase CK1 of peptides derived from β-catenin, NF-AT4, and APC and canonical peptides from β-casein and inhibitor-2 of protein phosphatase 1.
|
Rat liver CK1
|
CK1 α zebrafish
|
||||||
|---|---|---|---|---|---|---|---|
| Short name | Sequence | Km, μM | Vmax, cpm | Vmax/km | Km, μM | Vmax, cpm | Vmax/km |
| BC-WT or parent | RRRGATTTAPSLSGKGNPEDEDVDTNQVLYE | 714 | 43,478 | 61 | 1,260 | 44,604 | 35 |
| BC S45A | RRRGATTTAPALSGKGNPEDEDVDTNQVLYE | 715 | 3,644 | 5 | nd | nd | 0 |
| BC S45T | RRRGATTTAPTLSGKGNPEDEDVDTNQVLYE | 357 | 4,347 | 12 | 821 | 5,352 | 6 |
| BC pS45 | RRRGATTTAPpSLSGKGNPEDEDVDTNQVLYE | nd | nd | 0 | nd | nd | 0 |
| BC L46A | RRRGATTTAPSASGKGNPEDEDVDTNQVLYE | 988 | 4,166 | 4 | 2,159 | 1,784 | 1 |
| BC L46V | RRRGATTTAPSVSGKGNPEDEDVDTNQVLYE | 1,060 | 31,279 | 29 | 821 | 25,870 | 31 |
| BC L46I | RRRGATTTAPSISGKGNPEDEDVDTNQVLYE | 1,330 | 25,575 | 19 | 780 | 28,546 | 36 |
| BCL46(d-Leu) | RRRGATTTAPS(D-L)SGKGNPEDEDVDTNQVLYE | 1,050 | 2,857 | 3 | nd | nd | 0 |
| BCL46(±t-Bu-Gly) | RRRGATTTAPS(tBu-Gly)SGKGNPEDEDVDTNQVLYE | 800 | 93,940 | 117 | — | — | — |
| BC S47A | RRRGATTTAPSLAGKGNPEDEDVDTNQVLYE | 714 | 16,666 | 23 | 1,413 | 22,748 | 16 |
| BC S47N | RRRGATTTAPSLNGKGNPEDEDVDTNQVLYE | 1,330 | 10,869 | 8 | 585 | 8,028 | 14 |
| BCS47T | RRRGATTTAPSLTGKGNPEDEDVDTNQVLYE | 1,031 | 17,391 | 17 | 505 | 10,285 | 20 |
| BC E53A,E55A,D58A | RRRGATTTAPSLSGKGNPADADVATNQVLYE | 2,000 | 9,803 | 5 | 2,802 | 29,884 | 11 |
| BC acidic all A | RRRGATTTAPSLSGKGNPAAAAVATNQVLYE | 1,612 | 2,337 | 1.5 | — | — | — |
| BC E55K | RRRGATTTAPSLSGKGNPEDKDVDTNQVLYE | 2,850 | 21,739 | 8 | 2,956 | 25,478 | 9 |
| BC N-terminal (Δ52-65) | RRRGATTTAPSLSGKGN————- | 2,127 | 5,955 | 3 | 4,581 | 5,352 | 1 |
| BC C-terminal (Δ38-48) | RRR———-KGNPEDEDVDTNQVLYE | 1,282 | 988 | 1 | nd | nd | 0 |
| BC P44S | RRRGATTTASSLSGKGNPEDEDVDTNQVLYE | 1,200 | 15,202 | 13 | — | — | — |
| BC P52A | RRRGATTTAPSLSGKGNAEDEDVDTNQVLYE | 1,372 | 20,869 | 15 | 2,992 | 31,222 | 10 |
| BCΔ48-51 | RRRGATTTAPSLS—PEDEDVDTNQVLYE | 2,000 | 26,191 | 13 | 3,205 | 21,588 | 7 |
| BC 48-51 all A | RRRGATTTAPSLSAAAAPEDEDVDTNQVLYE | 833 | 6,793 | 8 | 1,544 | 4,461 | 3 |
| BC K49R | RRRGATTTAPSLSGRGNPEDEDVDTNQVLYE | 500 | 38,476 | 77 | — | — | — |
| β-casein (Ser-22) | ...pSpSpSEES22ITRI... | 44 | 72,608 | 1,650 | — | — | — |
| Inhibitor-2 peptide | RRKHAAIGDDDDAYSITA | 363 | 373,910 | 1,030 | — | — | — |
| APC (1275-1290) | RRRASSLSSLSSAEDEIGCN | 140 | 24,844 | 177 | — | — | — |
| APC L (1278, 1281)A | RRRASSASSASSAEDEIGCN | nd | nd | 0 | |||
| NF-AT4 S194A (183-202) | RRRAESLSHIYDDVDAELNEAAAR | 909 | 14,187 | 16 | — | — | — |
Peptides preparation is described in Materials and Methods. The parent or WT peptide of β-catenin (BC) contained the sequence of X. laevis β-catenin corresponding to residues 38-65. The NF-AT4 peptide contained the sequence of the human NF-AT4 protein from residues 183-202 with a sequence RRRA attached to its amino end. The APC peptides contain the sequence of the human APC protein from residues 1275 to 1290 or mutants with a sequence RRRA attached to its amino end. Peptides from β-casein and protein phosphatase 1 inhibitor-2 had been studied (21, 24). All other peptides contain three arginines in their amino terminus to facilitate the phosphocellulose paper assay. nd, nondetectable phosphorylation; —, assay not performed.
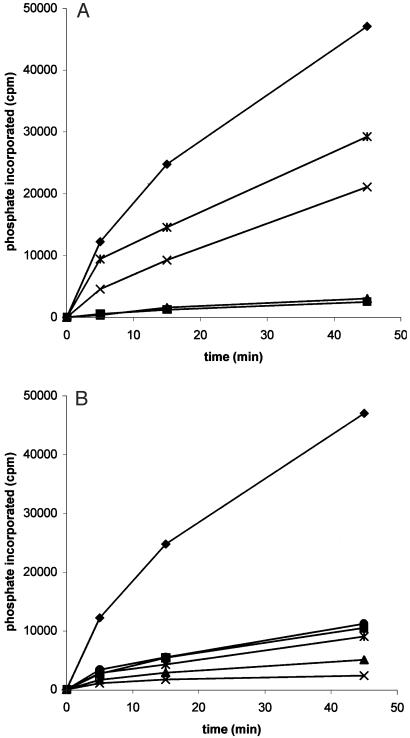
Replacement of Leu-46 with alanine (L46A), valine (L46V), or isoleucine (L46I) gave different results. Alanine replacement nearly completely suppressed phosphorylation with both the rat liver and zebrafish CK1. With the rat liver enzyme the peptides with the valine or isoleucine were partially active as compared with the parent peptide, whereas with the recombinant CK1, isoleucine substitution was as efficient as the parent peptide, whereas valine was slightly less active. This result suggested that the hydrophobicity of the side chain in position 46 may play an important role in determining the efficiency of CK1 phosphorylation. To explore this further, peptides with d-leucine or t-butyl-Gly in this position were synthesized and tested. The peptide containing d-leucine was completely inactive, whereas the compound that carries the t-butyl-Gly substitution was twice as good a substrate as the parent peptide. Replacement of Ser-47 with alanine (S47A) caused a significant reduction of activity even though Ser-47 is not phosphorylated. Substitution of Ser-47 with asparagine (S47N) severely reduced the phosphorylation, while its replacement for threonine (S47T) yielded a partially active peptide. Fig. 2A shows the time course of phosphorylation of several peptides with changes in the SLS motif, and the results corroborate the effects deduced from Table 1.
Fig. 2.
Time course of phosphorylations of β-catenin-derived peptides by CK1. The phosphorylations were carried out as described in Materials and Methods using peptides at 200μM concentration and rat liver CK1. (A) The incidence of the SLS motif is analyzed, with peptide ♦, parent or WT; *, L46V; X, S47A; ▴, L46A, and ▪, S45A. (B) The relevance of the acidic cluster is analyzed, with peptides ♦, parent or WT; *, E53A, E55A, and D58A; X, C terminal (Δ38-48); ▴, N terminal (Δ 52-65); •, E55K; and ▪, acidic all A, a peptide in which E53, D54, E55, D56, and D58 all have been changed to A.
The other major common feature found in both NF-AT4 and β-catenin is the cluster of acidic residues downstream from the SLS motif. In this regard, we found that substitution of three acidic residues by alanines (E53A, E55A, and D58A) greatly reduced the efficiency of phosphorylation, whereas substitution of all five acidic residues in the cluster (E53A, D54A, E55A, D56A, and D58A), designated in Table 1 as acidic all A, completely eliminated the phosphorylation of this peptide. Substitution of the single glutamic acid 55 with lysine (E55K) was sufficient to cause a drastic drop in the phosphorylation of this peptide by CK1. Total deletion of the acidic cluster (Δ52-65) also eliminated the substrate capacity of this peptide, as did deletion of residues 38-48 in the C-terminal peptide. Fig. 2B shows the time course of the phosphorylation of peptides containing changes in the acidic cluster and demonstrates the very stringent requirements for this motif.
Assessments of other features that are present in the β-catenin sequence in the vicinity of Ser-45 but not in the NF-AT4 protein, are also presented in Table 1. Replacement of proline residues immediately upstream of the SLS (P44S) and the acidic cluster (P52A) also resulted in a reduction of the phosphorylation by 70-80%.
Elimination of four amino acids in the linker region separating the SLS and acidic cluster (Δ48-51) or their replacement by alanines (48-51 all A) also decreased the substrate efficiency significantly. However, replacement of Lys-49 with arginine (K49R) caused an important increase in the substrate efficiency through a reduction of the Km.
It is noteworthy that alterations or deletions of the acidic cluster affect both the apparent Km and Vmax, whereas the changes in the SLS motif decrease the Vmax of the reaction without greatly affecting the apparent Km.
Qualitatively, the changes in the peptides caused similar effects on the recognition of the CK1 obtained either from rat liver or recombinant D. rerio CK1α. In general, the Km for the peptides with the recombinant CK1α is higher than those found with the mammalian enzyme. Most of these peptides also were checked with the splice variant D.r. CK1αL (15), with no significant difference from the results obtained with CK1α being observed. In Table 1, for comparison, data obtained with peptides that contain a canonical consensus for CK1 are presented. It can be seen that a β-casein-derived peptide with phosphoamino acids (Ser-22) and a protein phosphatase inhibitor-2 peptide, which is not primed but contains several acidic residues upstream, have phosphorylation efficencies that are 15- to 25-fold greater than the parent β-catenin peptide.
Phosphorylation of Full-Length β-Catenin in Vitro. The CK1 recognition motifs determined with synthetic peptides were also analyzed by using full-length recombinant β-catenin from D. rerio. This protein interacted very well with an antibody specific for mammalian β-catenin and showed a mobility in SDS/PAGE corresponding to the expected molecular mass of 80,000 Da. Three mutants were also generated in which Ser-45, Leu-46, and the five acidic residues in positions 53, 54, 55, 56, and 58 were replaced by alanine. This latter mutant showed a slightly faster mobility on SDS/PAGE.
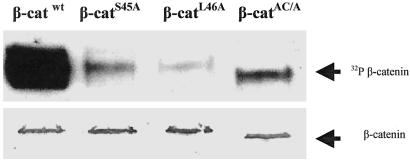
β-catenin and mutants were phosphorylated with CK1α, CK1αL, and CK1δ from zebrafish, and the products were analyzed by Western blot and autoradiography. Fig. 3 shows that the amounts of β-catenin and mutant proteins detected with antibodies against the His6 tag are apparently equivalent. On the other hand, the phosphorylation of the β-catenin with CK1αL was >10- to 20-fold higher than that observed with the S45A, L46A, and the acidic cluster mutant. These results are in very good agreement with the data obtained with the peptides. They corroborate the concept that Ser-45 is the major phosphoacceptor site for CK1 in the full-length protein and that both the SLS and acidic cluster motifs play an important role in the recognition of the protein substrate by this kinase. The results obtained with CK1α and CK1δ were qualitatively similar to those found with CK1αL; however, CK1δ seems to be much less efficient in phosphorylating β-catenin than casein (results not shown). The apparent Km for β-catenin with CK1α was estimated to be in the 100-nM range, which is several orders of magnitude lower than the peptides.
Fig. 3.
The phosphorylation of full-length β-catenin WT and mutants by CK1α. The recombinant, bacterially expressed, (His)6-tagged β-catenin WT and mutants from zebrafish were phosphorylated in vitro with the homologous CK1αL enzyme as described in Materials and Methods. The mutants included Ser-45 replaced by alanine (S45A), Leu-46 replaced by alanine (L46A), and all components of the acidic cluster (E53, D54, E55, D56, and D58) changed to alanine (AC/A). (Upper) Bands are autoradiography of the 32P incorporated into β-catenin after SDS/PAGE fractionation. (Lower) Band represents the Western blot of the (His)6 β-catenin present that was developed with monoclonal anti-(His)6 antibody.
Search for Proteins That Contain the SLS Motif Upstream from an Acidic Cluster. The importance of the SLS and acidic cluster motifs in CK1 phosphorylation prompted us to search for other proteins that contain these recognition features and to test the predictive capacity of our findings in regard to other proteins that may also be phosphorylated by CK1 in this noncanonical mode. The Swiss-Prot human protein sequences were searched for proteins that contain the SLS motif followed by an intervening sequence fluctuating in length between 2 and 5 aa and a downstream acidic cluster with six residues, of which at least three are aspartic or glutamic acid and without the presence of basic residues.
Table 2 gives a partial list of the proteins and the sequences identified in that search. In addition to NF-AT4 and β-catenin, there are many very interesting signaling proteins that contain these features. The most striking result, however, was to find APC protein on the list. This protein is related in function with β-catenin because it is an important component of the multiprotein complex assembled around axin, the scaffold protein that facilitates the phosphorylation of β-catenin by CK1 and GSK-3 (32). The CK1 recognition motif occurs in APC within three 20-aa repeats that are known to regulate β-catenin. Our prediction that these sequences of APC might be phosphorylated by CK1 had been validated by Rubinfield et al. (44), who observed that CK1ε can phosphorylate these same sequences and that these phosphorylations are required for the regulatory effect of APC on β-catenin function.
A peptide corresponding to APC residues 1275-1290 was synthesized and tested for phosphorylation by CK1. This peptide is an excellent substrate of CK1, 3-fold more efficient than the WT β-catenin peptide (Table 1), possibly because it has two SLS motifs in tandem and because noncanonical phosphorylation of S1276 could prime subsequent canonical phosphorylation of S1279. Substitution of the two leucines in this peptide completely eliminates its phosphorylation by CK1. In Table 1 we also include the assays of phosphorylation of residues 183-202 of an NF-AT peptide. This peptide is also phosphorylated by CK1 on the SLS motif, albeit with lower efficiency than the peptides from the other proteins.
Discussion
There is agreement in this area of study that CK1 is responsible for the in vivo phosphorylation of Ser-45 of β-catenin (40-43). This phosphorylation is crucial to prime the subsequent phosphorylation of this protein at residues Thr-41, Ser-37, and Ser-33 by GSK-3. Analysis of the sequence surrounding Ser-45 in β-catenin does not reveal the well-known features of the consensus sequence that are generally recognized by CK1 in a variety of proteins and peptides that serve as its substrates. This canonical consensus depends on the presence of a side chain containing a negative charge derived from either a phosphoamino acid or one or more acidic residues at position n-3 relative to the target serine or threonine (2, 21, 24). These features are absent in the sequence surrounding Ser-45 of β-catenin. A cluster of acidic residues is indeed found in the vicinity but it is located on the “wrong” side, beginning at n+7 downstream from Ser-45.
Early work on the specificity of CK1 suggested that another positive determinant could be a hydrophobic residue in position n+1 relative to the hydroxylamino acid to be phosphorylated (50). Interestingly, an isoleucine at position n+1 was also selected along with an acidic residue in position n-3 in a peptide library used to investigate the specificity of CK1 (51). The relevance seen in this work of Leu-46 in β-catenin does validate the importance of this feature.
The sequence surrounding Ser-45 of β-catenin did show considerable similarity to a noncanonical motif present in the case of NF-AT4 peptides phosphorylated by CK1 (31). These studies with synthetic peptides, which reproduce NF-AT4 sequences, and the studies described here using peptides with β-catenin sequences, have allowed us to determine the common features of these two important proteins related to their recognition by CK1. The acidic cluster downstream from the target serine is definitely important, yet, there is still some question as to how many acidic residues are necessary. Replacement of three of the five acidic residues with alanine causes a very important reduction in phosphorylation, whereas a similar replacement of all five residues or their deletion results in totally unphosphorylatable peptides. Replacement of Glu-55 with lysine also greatly diminishes the substrate capacity of the peptides.
There is some flexibility regarding the number of residues that can separate the cluster from the SLS motif. In naturally occurring proteins there are examples such as Armadillo and APC in which there are two intervening residues, whereas β-catenin has a 5-aa separation between the SLS and the acidic cluster. In our studies, the reduction of the intervening sequence to one proline or its replacement by alanines significantly affected their substrate capacity.
Comparison of the NF-AT4 and β-catenin sequences points to the SLS motif that is present in both proteins and involves the first serine (Ser-45) as a target hydroxylamino acid for CK1 in β-catenin. The importance of this feature was not apparent to us in our studies with the NF-AT4 peptides, because focus was on another serine (Ser-177) upstream from the SLS motif. However, the results obtained with the peptides of β-catenin in which Leu-46 or Ser-47 are mutated to alanine unambiguously demonstrate that both amino acids play an important role in CK1 recognition, the most critical being that of the leucine residue adjacent to the C terminus of the target serine. The results obtained with substitution of Leu-46 by alanine, valine, isoleucine, and t-butyl-glycine demonstrate that in general increasing the hydrophobicity of the side chain in this position results in an increase in the efficiency of recognition by CK1. However, results with the isoleucine substitution are somewhat different with the two enzymes used, whereas with rat liver enzyme L46I is less efficient than L46V, and with zebrafish enzyme L46I is as efficient as the parent peptide. The serine in position 47 also plays an important role because its replacement by threonine, asparagine, or alanine has a strong negative effect on its phosphorylation. A peptide corresponding to the NF-AT4 sequence, which includes the SLS motif as the only phosphorylatable target, together with the acidic cluster does act as a substrate of CK1, albeit with lower efficiency than the β-catenin peptides.
There are other features of the β-catenin peptides that are not conserved in NF-AT4 or APC and that influence their substrate efficiency. In particular, the Pro-52 replacement by alanine caused a significant decrease in the phosphorylation of the peptide as did the replacement of Pro-44 for serine. These proline residues may be specific features that influence the recognition of that particular sequence. Supporting this notion is the fact Armadillo, the β-catenin homologue of Drosophila melanogaster, conserves the residue corresponding to Pro-44 but does not contain a proline corresponding to Pro-52 of β-catenin. Comparison of the efficiency of the phosphorylation of the peptides with the noncanonical motif with those that have a cononical motif and that are derived from β-casein and protein phosphatase inhibitor-2 demonstrate that the noncanonical peptides are 15- to 25-fold less efficient than the canonical peptides of either the primed or unprimed varieties.
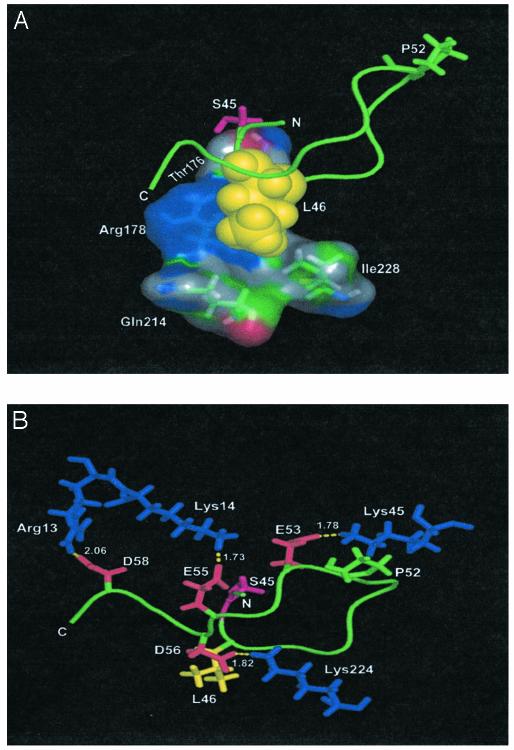
To gain information about the structural features possibly accounting for the relevance of the recurrent motifs found in the new class of CK1 substrates, a modeling study was performed based on the crystal structure of rat CK1δ (52) in which a peptide encompassing the 44-58 sequence of β-catenin has been inserted with the side chain of the phosphoacceptor serine (Ser-45) facing the putative position of the ATP γ phosphate. After energy minimization, the peptide adopts a bent conformation (Fig. 4), which is stabilized by a number of features also known to be required for its efficient phosphorylation by CK1. Especially noteworthy is the embedding of the bulky side chain at position n+1 (Leu-46) into a cavity formed by two residues of the P+1 loop (Thr-176 and Arg-178) and by Gln-214 and Ile-228. This model also illustrates the role of Pro-52 in favoring the folding back of the peptide backbone (Fig. 4A). Such a backward orientation of the C-terminal moiety of the peptide makes possible several electrostatic interactions between the acidic cluster and a number of basic residues of the kinase, namely Arg-13, Lys-14, Lys-45, and Lys-224, as illustrated in Fig. 4B. Interestingly, all of the residues of the kinase implicated in the main interactions with the peptide are conserved or conservatively replaced in the different isoforms of CK1, and Lys-224, which interacts with a tungstate ion in the crystal structure of rat CK1δ, has been already suggested to be implicated in the binding of primed CK1 substrates in position n-3 (52). Mutational analysis and/or the solution of the crystal structure of complexes between CK1 and peptide substrates will be required to validate the mode of binding emerging from our modeling study. Note, however, that the modeling accounts very well for all of the biochemical data available, especially the relevance of Pro-52, the acidic cluster downstream from it, and the bulky hydrophobic residue adjacent to the C-terminal side of the phosphoacceptor serine Leu-46. This latter is oriented toward the lower lobe of the kinase, entering a cavity where it makes van der Waals interactions with four residues surrounding it at distances varying between 2.64 and 3.79 Å. Consistent with this scenario the replacement of Leu-46 with an artificial bulkier side chain (t-butyl-Gly) can be expected to make even closer interactions, improving the phosphorylation efficiency, whereas its replacement with smaller residues (notably Ala) or, more so, with d-Leu, whose side chain is oriented in the opposite direction, abrogates phosphorylation (see Table 1).
Fig. 4.
Modelization of the mode of binding of a peptide encompassing the 44-58 sequence of β-catenin to the active site of CK1δ. The backbone of the peptide and the side chain of Pro-52 are shown in green. Phosphorylatable Ser-45 is indicated in magenta. (A) The spheres representing Leu-46 (yellow) are embedded in a cavity formed by four residues of CK1 shown as molecular surface. The minimum distances between peptide Leu-46 and CK1 Thr-176, Arg-178, Gln-214, and Ile-228 are 2.55, 2.64, 3.79, and 3.78 Å, respectively. (B) The interactions among four acidic residues of the peptide (red) clustered in its C-terminal part and four basic residues of CK1 (blue) are highlighted. N and C denote the amino- and carboxyl-terminal ends of the peptide, respectively. The residues of the β-catenin peptide and CK1 are indicated by the one- and three-letter codes, respectively. The β-catenin peptide structure was first built from the amino acid sequence (PS45LSGKGNPEDEDVD) using the protein program of the tinker molecular modeling software (53) with Amber98 force field. The structure was minimized with the program optimize until the energy reached a minimum and manually superimposed to the CK1δ structure (Protein Data Bank ID code 1CK-J.pdb). An energy minimization of the complex CK1-peptide was performed with the program minimize and Amber98 force field until the energy reached a minimum. The figure was drawn by using the pymol program (K. L. DeLano, www.pymol.org).
Further work is required to explain how peptides from APC and NF-AT4, which lack a proline residue upstream of the acidic cluster, can be modeled to bind and be phosphorylated by CK1.
Experiments with full-length β-catenin mutated in some of the key features of the SLS and acidic clusters detected in the peptides corroborate that those structural motifs maintain their influence when the whole protein is used as a substrate. These results also tell us that Ser-45 is the major phosphorylation target of CK1 in β-catenin. The much lower Km of the full-length protein as compared with the peptides suggests that CK1 may also recognize a secondary binding site in β-catenin that is not in the peptides.
We cannot presume that these in vitro results reflect in vivo behavior. This is specially true in the case of β-catenin phosphorylation, which is regulated within the context of a multiprotein complex (32).
There are, however, indications that the structural motifs we have described actually do play a role in the in vivo phosphorylation of β-catenin and other proteins by CK1. The most convincing clue comes from mutations found in tumors. Many mutations found in β-catenin in numerous tumors involve single base substitutions or deletions within exon 3 of β-catenin, which codes for the region of the protein that contains the targets for CK1 and GSK-3 (34-36). These findings provide strong support for the critical role played by interference with the phosphorylation and degradation of β-catenin in the evolution of many types of tumors. As expected, the majority of the mutations affect directly the serines and threonine of β-catenin that are phosphorylated by CK1 and GSK-3 (Ser-33, Ser-37, Thr-41, and Ser-45). Nevertheless, mutations of β-catenin in thyroid tumors have also been found that affect a number of other residues in the vicinity of Ser-45, the target of CK1 (45, 46). It is very significant that several mutations found in these tumors involve features that we have detected as important for CK1 recognition. Three mutations change acidic residues for lysine within the acidic cluster and one changes D58 to asparagine. The E55K mutation is identical to a change in our synthetic peptide that caused a large decrease in CK1 phosphorylation. The relevance of the S45L46S47 motif involving target Ser-45 is also strengthened by the finding of a mutation L46V and two other mutations that change Ser-47 to asparagine, which significantly reduce CK1 phosphorylation in the peptide experiments. Pro-44 and Pro-52, whose changes were found to decrease the phosphorylation in peptides, were also detected as mutants in thyroid tumors. However, there is a significant difference in the correlation between β-catenin mutations detected in thyroid tumors and CK1 recognition of mutations in Lys-49. In nine tumor samples, Lys-49 was found mutated to arginine. The corresponding change in the peptide resulted not in a decrease but rather in a higher efficiency of phosphorylation. A probable explanation for this lack of correlation in these cases may be found in the fact that in the great majority of the thyroid tumor samples analyzed by Garcia-Rostan et al. (45, 46) contain more than one mutation in the third exon of β-catenin. As a matter of fact, eight of the nine samples that contained K49R mutations also had a second change, T40I, and in three of those same samples a third mutation was also detected. Those authors found very good correlation of the T40I mutation, which may affect GSK-3 phosphorylation of Thr-41, with the appearance of poorly differentiated carcinomas. The presence of multiple mutations in these tumors suggests genetic instability, and those authors postulated that other mutations might occur in other exons of β-catenin that were not analyzed, which may explain the observation that only half of the cases that contained mutations in exon 3 of β-catenin demonstrated nuclear localization of this protein (45, 46).
Despite these caveats, the overall correlation between the features discovered through the CK1 phosphorylation of β-catenin-derived peptides and the full-length protein with mutations found in tumors argues strongly for the physiological involvement of these features in the in vivo phosphorylation of this key protein.
Another important argument for the relevance of the SLS-Xn-(E/D)n motif can be found in its predictive capacity. Among the proteins that carry this motif, the databank has provided a number of interesting possibilities. The most striking one involves APC, a protein clearly related in its function with β-catenin with which it forms multiprotein complex. It is noteworthy that the prediction of this work had already been fulfilled with respect to the APC protein because it had been shown to be phosphorylated by CK1ε (44). In this work, sites of phosphorylation were detected in two 20 amino acid repeats that activate β-catenin phosphorylation and contain SLS motifs followed by acidic clusters. A peptide reproducing the APC sequence has been shown here to be an excellent substrate of CK1 from rat liver, and its activity was shown to completely depend on the presence of two leucines in SLS motifs.
A recent report by Kang et al. (54) describes that PKA bound through presenilin to β-catenin and GSK-3 can also phosphorylate the Ser-45 of β-catenin to prime it for subsequent GSK-3 phosphorylation. In this case again, Ser-45 does not conform to the canonical consensus of PKA (RR-X-S/T-B, where R is arginine, B is a hydrophobic amino acid, and X any residue). Preliminary experiments with our β-catenin peptides and the full-length protein show that PKA can indeed phosphorylate the serine corresponding to Ser-45 in β-catenin the peptides, but at a rate that is much lower than with CK1.
It seems significant that the proteins NF-AT4, β-catenin, and APC, which contain the noncanonical sequence motif described in this work, are involved in the regulation of the access of a transcription factor to the nucleus to ultimately affect cell division. Through different mechanisms, masking a nuclear localization signal in the case of NF-AT4 or triggering proteolytic recognition for β-catenin, concerted phosphorylation involving CK1 and GSK-3 act to oppose the access to the nucleus of a transcription factor with the resulting block to proliferation. Pursuing the analogy further, the cAMP-dependent PKA also appears to participate in the priming phosphorylation of the NF-AT family of factors (30) and in the phosphorylation of Ser-45 of β-catenin bound to presenilin (54). In both cases, PKA acts to prime further phosphorylation by GSK-3. Recently, these same three kinases, PKA, CK1 and GSK-3, have been elegantly shown to cooperate through hierarchical phosphorylation of Cubitus interruptus with the resulting degradation of this developmental transcription factor (55). In this case, however, the phosphorylations strictly obey the known canonical sequence motifs.
Concerted action of several kinases on a common substrate, as in this example, suggests the possible existence of polykinase complexes that may facilitate their collective function. In the post-genomic era, the important task of collating and cataloguing the 518 different genes of the human “kinome” according to sequence similarity (13) must be followed by the much more difficult task of ordering the enzymes according to their cellular function. As part of this effort, special relevance should be given to mechanisms of signal integration through the concerted action of protein kinases on transcription factor proteins.
Supplementary Material
Acknowledgments
The work in the laboratory of J.E.A. was supported by Project 1030462 of Fondo Nacional de Investigaciín Cientiífica y Tecnolígica and a grant from the Wellcome Trust. The work in the laboratory of L.A.P. was supported by grants from the Ministero dell'Istruzione, dell'Università e della Ricerca (COFIN 2001 and 2002).
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
Abbreviations: CK, casein kinase; NF-AT4, nuclear factor for activated T cells 4; GSK-3, glycogen synthase-3; PKA, protein kinase A; APC, adenomatous polyposis coli.
References
- 1.Appella, E. & Anderson, C. W. (2001) Eur. J. Biochem. 268, 2764-2772. [DOI] [PubMed] [Google Scholar]
- 2.Flotow, H., Graves, P. R., Wang, A., Fiol, C. J., Roeske, R. W. & Roach, P. J. (1990) J. Biol. Chem. 265, 14264-14269. [PubMed] [Google Scholar]
- 3.Meggio, F., Perich, J. W., Reynolds, E. C. & Pinna, L. A. (1991) FEBS Lett. 283, 303-306. [DOI] [PubMed] [Google Scholar]
- 4.Harwood, A. J. (2001) Cell 105, 821-824. [DOI] [PubMed] [Google Scholar]
- 5.Dajani, R., Fraser, E., Roe, S. M., Young, N., Good, V., Dale, T. C. & Pearl, L. H. (2001) Cell 105, 721-732. [DOI] [PubMed] [Google Scholar]
- 6.ter Haar, E., Coll, J. T., Austen, D. A., Hsiao, H.-M., Swenson, L. & Jain, J. (2001) Nat. Struct. Biol. 8, 593-596. [DOI] [PubMed] [Google Scholar]
- 7.Frame, S., Cohen, P. & Biondi, R. M. (2001) Mol. Cell 7, 1321-1327. [DOI] [PubMed] [Google Scholar]
- 8.Pinna, L. A. & Ruzzene, M. (1996) Biochim. Biophys. Acta 1314, 191-225. [DOI] [PubMed] [Google Scholar]
- 9.Allende, J. E. & Allende, C. C. (1995) FASEB J. 9, 313-323. [DOI] [PubMed] [Google Scholar]
- 10.Meggio, F. & Pinna, L. A. (2003) FASEB J. 17, 349-368. [DOI] [PubMed] [Google Scholar]
- 11.Guerra, B. & Issinger, O.-G. (1999) Electrophoresis 20, 391-408. [DOI] [PubMed] [Google Scholar]
- 12.Litchfield, D. W. (2003) Biochem. J. 369, 1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. (2002) Science 298, 1912-1934. [DOI] [PubMed] [Google Scholar]
- 14.Gross, S. & Anderson, R. (1998) Cell. Signalling 10, 699-711. [DOI] [PubMed] [Google Scholar]
- 15.Burzio, V., Antonelli, M., Allende, C. C. & Allende, J. E. (2002) J. Cell. Biochem. 86, 805-814. [DOI] [PubMed] [Google Scholar]
- 16.Kloss, B., Prince, J., Saez, L., Blau, J., Rothenfluh, A., Westley, C. & Young, M. (1998) Cell 94, 97-107. [DOI] [PubMed] [Google Scholar]
- 17.Liu, F., Virshup, D. M., Nairn, A. C. & Greengard, P. (2002) J. Biol. Chem. 277, 45393-45399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwab, C., De Maggio, A., Ghoshal, N., Binder, L., Kuret, J. & McGeer, P. (2000) Neurobiol. Aging 21, 503-510. [DOI] [PubMed] [Google Scholar]
- 19.Tobin, A. B., Totty, N. F., Sterlin, A. E. & Nahorski, S. R. (1997) J. Biol. Chem. 272, 20844-20849. [DOI] [PubMed] [Google Scholar]
- 20.Zhu, J., Shibasaki, F., Price, R., Guillemot, J. C., Yano, T., Dötsch, V., Wagner, G., Ferrara, P. & McKeon, F. (1998) Cell 93, 851-681. [DOI] [PubMed] [Google Scholar]
- 21.Pulgar, V., Marin, O., Meggio, F., Allende, C. C., Allende, J. E. & Pinna, L. A. (1999) Eur. J. Biochem. 260, 520-526. [DOI] [PubMed] [Google Scholar]
- 22.Graves, P. R. & Roach, P. J. (1995) J. Biol. Chem. 270, 21689-21694. [DOI] [PubMed] [Google Scholar]
- 23.Cegielska, A., Gietzen, K. F., Rivers, A. & Virshup, D. M. (1998) J. Biol. Chem. 273, 1357-1364. [DOI] [PubMed] [Google Scholar]
- 24.Marin, O., Meggio, F., Sarno, S., Andretta. M. & Pinna, L. A. (1994) Eur. J. Biochem. 223, 647-653. [DOI] [PubMed] [Google Scholar]
- 25.Crabtree, G. R. (1999) Cell 96, 611-614. [DOI] [PubMed] [Google Scholar]
- 26.Neilson, J., Stankunas, K. & Crabtree, G. R. (2001) Curr. Opin. Immunol. 13, 346-350. [DOI] [PubMed] [Google Scholar]
- 27.Gomez del Arco, P., Martinez-Martinez, S., Maldonado, J. L., Ortega-Perez, I. & Redondo, J. M. (2000) J. Biol. Chem. 275, 13872-13878. [DOI] [PubMed] [Google Scholar]
- 28.Porter, C. M., Havens, M. A. & Clipstone, N. A. (2002) J. Biol. Chem. 275, 3543-3551. [DOI] [PubMed] [Google Scholar]
- 29.Chow, C. W., Dong, C., Flavell, R. A. & Davis, R. J. (2000) Mol. Cell. Biol. 20, 5227-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheridan, C. M., Heist, E. K., Beals, C. R., Crabtree, G. R. & Gardner, P. (2002) J. Biol. Chem. 277, 48664-48676. [DOI] [PubMed] [Google Scholar]
- 31.Marin, O., Burzio, V., Boschetti, M., Meggio, F., Allende, C. C., Allende, J. E. & Pinna, L. A. (2002) Biochemistry 41, 618-627. [DOI] [PubMed] [Google Scholar]
- 32.Polasis, P. (2002) Curr. Biol. 12, R499-R501. [DOI] [PubMed] [Google Scholar]
- 33.Hino, S., Michiue, T., Asashima, M. & Kikuchi, A. (2003) J. Biol. Chem. 278, 14066-14073. [DOI] [PubMed] [Google Scholar]
- 34.Morin, P. J., Sparks, A. B., Korinek, V., Barker, N., Clevers, H., Vogelstein, B. & Kinzler, K. W. (1997) Science 275, 1787-1790. [DOI] [PubMed] [Google Scholar]
- 35.Abraham, S. C., Montgomery, E. A., Giardello, F. M. & Wu, T.-T. (2001) Am. J. Pathol. 158, 1073-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, R., Zhai, Y., Feavon, E. R. & Cho, K. R. (2001) Cancer Res. 61, 8247-8255. [PubMed] [Google Scholar]
- 37.Peters, J. M., McKay, R. M., McKay, J. P. & Graff, J. M. (1999) Nature 401, 345-350. [DOI] [PubMed] [Google Scholar]
- 38.Sakanaka, C., Leong, P., Xu, L., Harrison, S. & Williams, L. (1999) Proc. Natl. Acad. Sci. USA 96, 12548-12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKay, R. M., Peters, J. M. & Graff, J. M. (2001) Dev. Biol. 235, 388-396. [DOI] [PubMed] [Google Scholar]
- 40.Liu, C., Li, Y., Semenov, M., Han, C., Baeg, G-H., Tan, Y., Zhang, Z., Lin, X. & He, X. (2002) Cell 108, 837-847. [DOI] [PubMed] [Google Scholar]
- 41.Sakanaka, C. (2002) J. Biochem. 132, 697-703. [DOI] [PubMed] [Google Scholar]
- 42.Yanagawa, S.-I., Matsuda, Y., Lee, J.-S., Matsubayashi, H., Sese, S., Kadowaki, T. & Ishimoto, A. (2002) EMBO J. 21, 1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amit, S., Hatzubai, A., Birman, Y., Andersen, J. S., Ben-Shushan, E., Mann, M., Ben-Neriah, Y. & Alkalay, I. (2002) Genes Dev. 16, 1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubinfield, B., Tice, D. A. & Polakis, P. (2001) J. Biol. Chem. 276, 39037-39045. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Rostan, G., Tallini, G., Herrero, A., D'Aquila, T. G., Cargangiu, M. L. & Rimm, D. L. (1999) Cancer Res. 59, 1811-1815. [PubMed] [Google Scholar]
- 46.Garcia-Rostan, G., Camp, R. L., Herrero, A., Cargangiu, M. L., Rimm, D. L. & Tallini, G. (2001) Am. J. Pathol. 158, 987-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meggio, F., Donella, D. A. & Pinna, L. A. (1981) J. Biol. Chem. 256, 11958-11961. [PubMed] [Google Scholar]
- 48.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 51-59. [DOI] [PubMed] [Google Scholar]
- 49.Marin, O., Meggio, F., Sarno, S. & Pinna, L. A. (1997) Biochemistry 36, 7192-7198. [DOI] [PubMed] [Google Scholar]
- 50.Meggio, F., Donella-Deana, A. & Pinna, L. A. (1979) FEBS Lett. 106, 76-80. [DOI] [PubMed] [Google Scholar]
- 51.Songyang, Z., Ping Lu, K., Kwon, Y. T., Tsai, L.-H., Filhol, O., Cochet, C., Brickley, D. A., Soderling, T. R., Bartleson, Ch., Graves, D. J., et al. (1996) Mol. Cell. Biol. 16, 6486-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Longenecker, K. L., Roach, P. J. & Hurley, T. D. (1996) J. Mol. Biol. 257, 618-631. [DOI] [PubMed] [Google Scholar]
- 53.Ren, P. & Ponder, J. W. (2002) J. Comput. Chem. 23, 1497-1506. [DOI] [PubMed] [Google Scholar]
- 54.Kang, D. E., Soriano, S., Xia, X., Eberhard, C. G., Strooper, B. D., Zheng, H. & Koo, E. H. (2002) Cell 110, 751-762. [DOI] [PubMed] [Google Scholar]
- 55.Price, M. A. & Kalderon, D. (2002) Cell 108, 823-835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.