Abstract
The repair of double-strand DNA breaks by homologous recombination is essential for the maintenance of genome stability. In herpes simplex virus 1, double-strand DNA breaks may arise as a consequence of replication fork collapse at sites of oxidative damage, which is known to be induced upon viral infection. Double-strand DNA breaks are also generated by cleavage of viral a sequences by endonuclease G during genome isomerization. We have reconstituted a system using purified proteins in which strand invasion is coupled with DNA synthesis. In this system, the viral single-strand DNA-binding protein promotes assimilation of single-stranded DNA into a homologous supercoiled plasmid, resulting in the formation of a displacement loop. The 3′ terminus of the invading DNA serves as a primer for long-chain DNA synthesis promoted by the viral DNA replication proteins, including the polymerase and helicase-primase. Efficient extension of the invading primer also requires a DNA-relaxing enzyme (eukaryotic topoisomerase I or DNA gyrase). The viral polymerase by itself is insufficient for DNA synthesis, and a DNA-relaxing enzyme cannot substitute for the viral helicase-primase. The viral single-strand DNA-binding protein, in addition to its role in the invasion process, is also required for long-chain DNA synthesis. Form X, a topologically distinct, positively supercoiled form of displacement-loop, does not serve as a template for DNA synthesis. These observations support a model in which recombination and replication contribute toward maintaining viral genomic stability by repairing double-strand breaks. They also account for the extensive branching observed during viral replication in vivo.
Herpes simplex virus 1 (HSV-1) is a double-stranded DNA virus with a genome of ≈152 kbp (1). Replication of the genome is mediated by seven virus-encoded factors (reviewed in ref. 2). Several cellular factors have also been implicated in particular stages of the replication cycle (reviewed in refs. 1 and 2). The absence of viral genomic ends, as well as the existence of distinct viral origins and an initiator protein (UL9), is highly suggestive of an initial θ-mode of replication that generates circular DNA products. Subsequently, it is thought that replication proceeds by a rolling circle or σ-mode to produce head-to-tail genome concatemers (reviewed in ref. 2). This strategy resembles that adopted by bacteriophage λ (3).
A serious challenge to this view of HSV-1 replication has emerged from recent work that demonstrates that genome circularization is not a requisite for lytic viral replication (4). Thus, a revised model by which HSV-1 replicates is in line with the model exhibited by T4 and T7, which involves replication of a linear genome (5, 6). In this model, replication initiates at an origin in a process that requires the action of the viral initiator protein (UL9) and the single-stranded DNA-binding protein (SSB) ICP8 (7, 8). Protein-protein interactions at the origin mediate the assembly of a replisome that consists of the virus-encoded DNA helicaseprimase (UL5 and UL52 core enzyme and UL8 loading factor) and DNA polymerase (UL30 catalytic subunit and UL42 processivity factor) (reviewed in ref. 2). The UL8 protein presumably plays a central role in coordinating this process because it interacts with UL9, the UL30 subunit of the polymerase, and also mediates the interaction of the helicase-primase with ICP8 (9-12).
Replication of a linear genome would encounter a problem as the replication fork reaches an end. In T4, this problem is overcome by using recombination-dependent replication (RDR), in which the end from one genome invades into a homologous region of another and utilizes it as a template to complete lagging-strand synthesis (5). Such a mode of replication would generate highly branched replication intermediates that are commonly seen in HSV-1-infected cells (13). Indeed, direct visualization of viral replication intermediates by electron microscopy showed that ≈50% of molecules consisted of large tangled networks of DNA. The remaining molecules also exhibited topologies consistent with those of recombination intermediates/products, including lariat structures, displacement loops (D-loops) and molecules with extensive single-stranded (ss) character, and replication bubbles (14).
During the latter stages of HSV-1 replication, the genome recombines at a high frequency that has been estimated to be 0.6%/kb of genome (15). In addition to recombination acting to replicate the ends of the linear genome, it may also be required to repair DNA damage that gives rise to double-strand breaks (DSB). DSB may arise as a consequence of replication fork collapse at sites of oxidative damage, which is known to be induced upon viral infection (16, 17). DSB also arise in replicating DNA as a result of cleavage of viral a sequences by endonuclease G that may serve in genome isomerization (18, 19). Repair of such DSB by RDR would also account for the prevalence of branched replication intermediates.
One of the viral replication proteins, the SSB, referred to as ICP8, has been shown to mediate recombination reactions. In conjunction with the helicase-primase, ICP8 performs strand exchange (20) along the lines of the bacteriophage T7 SSB and helicase-primase (21). In addition, ICP8 promotes the assimilation of ssDNA into homologous supercoiled acceptor DNA, resulting in the formation of a D-loop (22). Here we show that the product of such an invasion reaction provides a template for long-chain synthesis catalyzed by the viral replication proteins in the presence of a DNA-relaxing enzyme.
Our observations suggest a mechanism whereby RDR accounts for the extensive branching observed during viral replication in vivo (13). This interplay between recombination and replication may very well contribute toward maintaining viral genomic stability by rescuing collapsed replication forks.
Materials and Methods
Enzymes and Reagents. ICP8 (23), UL5/UL52 core enzyme, and UL8 (12) were purified as described. Their concentrations, expressed in moles of monomeric protein, were determined by using extinction coefficients of 82,720, 171,380, and 130,390 M-1 cm-1 at 280 nm, respectively, calculated from their predicted amino acid sequences (24). UL30 and UL42 were purified as described, and their concentrations, in moles of monomeric protein, were determined by using the method of Bradford using BSA as a standard (25). Calf thymus DNA topoisomerase I and proteinase K were purchased from Amersham Pharmacia and Roche Molecular Biochemicals, respectively. Bacteriophage T4 polynucleotide kinase was obtained from New England Biolabs. Escherichia coli DNA gyrase was a kind gift from Kenneth J. Marians (Memorial Sloan-Kettering Cancer Center, New York). ATP (disodium salt) and chloroquine (diphosphate salt) were purchased from Sigma. Deoxyribonucleotide triphosphate (disodium salts) and [γ-32P]ATP (4,500 Ci/mmol) were purchased from Amersham Pharmacia and ICN Biomedicals, respectively.
Nucleic Acids. Oligodeoxyribonucleotide PB11 (100-mer) (26), complementary to residues 379-478 of the minus strand of pUC18, was synthesized and gel-purified by Sigma Genosys. Its concentration was determined by using an extinction coefficient of 939,208.1 M-1 cm-1 at 260 nm. PB11 was 5′-32P-labeled with T4 polynucleotide kinase and purified by using Sephadex G-25 (fine) Quick Spin Columns (Roche Molecular Biochemicals). Form I pUC18 was prepared from E. coli JM109 by using the Promega Wizard Plus DNA purification system followed by ethanol precipitation. All DNA concentrations are expressed in moles of molecules.
D-Loop Formation. ICP8 (600 nM) was preincubated with PB11 (26.25 nM) on ice for 8 min in a buffer containing 25 mM Tris-acetate at pH 7.5, 10 mM magnesium acetate, 1 mM DTT, 1 mM ATP, and 100 μg/ml BSA. The pairing reaction was initiated by adding pUC18 form I DNA (8.75 nM), and incubation was continued for 30 min at 30°C. When stated, D-loops were purified by extraction with the Promega Wizard DNA clean-up system followed by removal of excess unannealed oligonucleotide by gel filtration through Chroma Spin + TE-1000 columns (BD Biosciences).
Recombination-Dependent DNA Synthesis Reaction. Unless otherwise stated, D-loop reactions were supplemented with a mixture to provide final concentrations of 400 nM ICP8, 60 nM UL30, 60 nM UL42, 200 nM UL5/UL52, 600 nM UL8, and 370 nM gyrase or 5 units of topoisomerase I, 500 μM each of dATP, dCTP, dGTP and TTP, and 5.8 nM DNA (with respect to plasmid of which ≈15% were D-loops). The final concentrations of buffer components were 16.7 mM Tris-acetate at pH 7.5, 6.7 mM magnesium acetate, 0.66 mM DTT, 0.66 mM ATP, and 66 μg/ml BSA. Reactions were incubated for 60 min at 30°C and quenched by the addition of termination buffer (final concentration: 50 mM EDTA/3 μg/μl proteinase K), followed by further incubation for 20 min.
Agarose Gel Electrophoresis. Reaction products were resolved either by 1D denaturing or 2D native-denaturing agarose gel electrophoresis. For 1D denaturing gels, reactions were resolved through 1% agarose containing 50 mM NaOH and 1 mM EDTA at ≈2.25 V/cm for 12 h. For 2D gels, each reaction was run in duplicate in the first dimension through 0.75% agarose-Tris acetate EDTA (pH 7.6) gels at 9 V/cm for 2 h. Under these conditions, free 100-mer migrates out of the gel. Where indicated, chloroquine was added to 25 μg/ml and electrophoresis was performed at 4.5 V/cm for 3.5 h. One of the lanes was used as a reference to visualize products resolved in the first dimension. The other lane was excised, soaked in buffer containing 50 mM NaOH and 1 mM EDTA, embedded in a second gel composed of 1% agarose in 50 mM NaOH and 1 mM EDTA, and electrophoresed at ≈2 V/cm for 7 h. After electrophoresis, the gels were dried onto DE81 chromatography paper (Whatman), analyzed, and quantitated by storage phosphor analysis with a Molecular Dynamics Storm 820.
Results
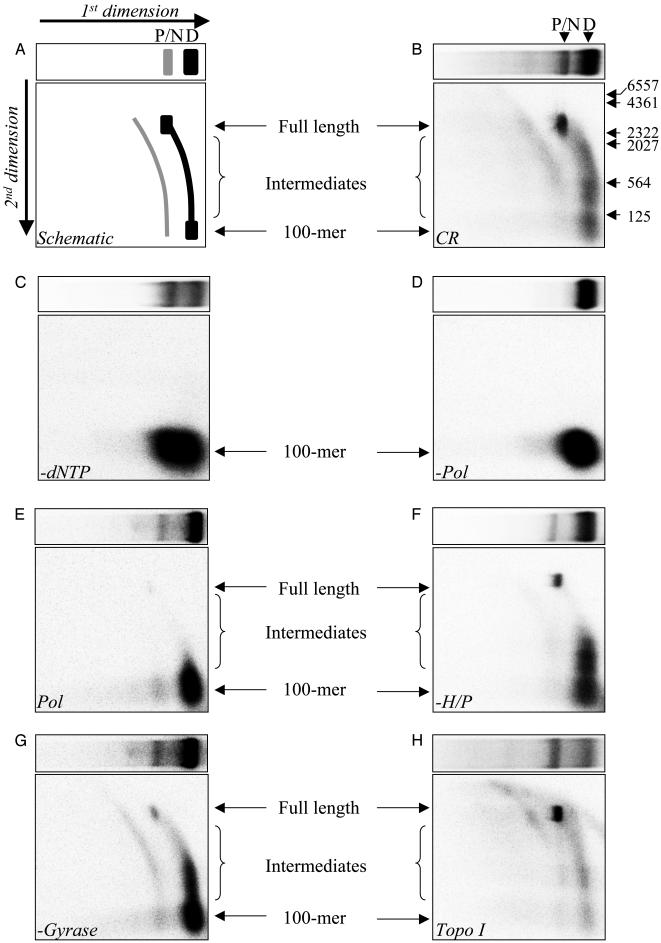
Reconstitution of Recombination-Dependent DNA Synthesis. Given our previous observation that ICP8 can promote the complete assimilation of a 100-mer into a homologous form I acceptor plasmid (22), we examined the ability of the invading 3′ terminus to serve as a primer for DNA synthesis with the HSV-1 replication proteins. Thus, D-loops were formed by preincubating ICP8 with 5′-32P-labeled 100-mer, followed by addition of homologous pUC18 (2,686 bp) to initiate the pairing reaction. In the complete reaction (CR), D-loops were supplemented with the heterodimeric polymerase (UL30 and UL42), the heterotrimeric helicase-primase (UL5, UL52, and UL8), a relaxing enzyme (E. coli gyrase or calf thymus topoisomerase I), and dNTP. Extension of the 5′-32P-labeled 100-mer in the D-loop was examined by 2D native-denaturing agarose gel electrophoresis of deproteinized reaction products as described in Materials and Methods. In this technique, as schematized in Fig. 1A, products were resolved under nondenaturing conditions in the first dimension (Upper) to separate D-loops (black band) from extension products and nicked D-loops (gray band). In the second dimension (Lower), denaturing conditions resolved extension products based on length, creating a streak originating from the 100-mer associated with D-loops (black streak) or nicked D-loops (gray streak). Complete extension of the 100-mer in the D-loop would create a species of 2,686 nt.
Fig. 1.
Recombination-dependent DNA synthesis. D-loops were formed, and the ability of the invading strand to prime DNA synthesis was examined as described in Materials and Methods. (A) Schematic depiction of the band pattern obtained by resolving the CR by 2D native-denaturing agarose gel electrophoresis. The arrows indicate the direction of migration in each dimension. The species are shaded to distinguish between primer extension originating from D-loops (black) and nicked D-loops (gray). Shown are storage phosphor images of first (Upper) and second (Lower) dimensions for each. (B) CR with gyrase. (C) CR without dNTP. (D) CR without UL30/UL42 (Pol). (E) Primer extension with UL30/UL42 (Pol) alone. (F) CR without UL5/UL52/UL8 (H/P). (G) CR without gyrase/topoisomerase I. (H) CR with topoisomerase I instead of gyrase. The positions of D-loops (D), products (P), nicked D-loops (N), 100-mer, full-length extension products (2,686 nt), intermediates, and markers (HindIII-digested λ DNA) are as indicated.
Fig. 1B shows the products of the CR using gyrase as the relaxing enzyme. Electrophoresis in the first dimension resolved two main bands: these correspond to D-loops (faster) and species comigrating with nicked/relaxed plasmid (slower), respectively, as determined by their relative mobility with respect to forms I, II, and IV (data not shown and ref. 22). In the second dimension, a streak originating from the 100-mer assimilated in the D-loop that culminates in a species corresponding to full-length product (2,686 nt) was observed. This band corresponds to the nicked/relaxed species resolved in the first dimension. Consequently, the majority of species at that position are full-length extension products (2,686 nt). Approximately 25% of the 100-mer in the D-loop was fully extended after 60 min of incubation. Intermediate extension products were also generated. A second minor streak originating from 100-mer associated with nicked D-loops, culminating in full-length products, was also observed.
Omission of dNTP from the CR prevented primer extension (Fig. 1C Lower). However, as shown in Fig. 1C Upper, during the course of the reaction, D-loops were processed into the nicked form. Extension of the primer was also dependent on the polymerase (Fig. 1D Lower). Moreover, the absence of the nicked species indicates that this activity was caused by the addition of polymerase (Fig. 1D Upper). This nicking activity was observed upon addition of polymerase alone and was attributed to a minor contaminant present in the UL30 preparation (Fig. 1E and data not shown). More importantly, addition of the polymerase alone to D-loops was not sufficient to promote long-chain synthesis, as only limited extension products were observed (Fig. 1E). Omission of the helicase-primase led to inefficient extension of the primer assimilated in the D-loop, as only 6.5% of full-length products were observed (Fig. 1F). In addition, the majority of products did not exceed ≈500 nt. Similarly, in the absence of a relaxing enzyme, only a small fraction (3%) of full-length products were observed (Fig. 1G). As in the absence of helicase-primase, the majority of products did not exceed ≈500 nt. However, extension of the 100-mer originating from the nicked D-loops was evident because this reaction is not topologically constrained. The relaxation activity of gyrase could be substituted with eukaryotic topoisomerase I, resulting in essentially complete extension of the 100-mer with fewer intermediates (Fig. 1H).
The above data indicate that the HSV-1 replication proteins use D-loops as substrates to promote long-chain primer extension in a reaction that is also dependent on a relaxation activity. Complete extension products were also observed when a 22-mer was used as the invading primer and in a continuous reaction in which D-loop formation and DNA synthesis occurred concurrently (data not shown). It should also be noted that extension of the invading primer depended on a 3′ hydroxyl group because a 2′,3′-dideoxycytidine-terminated oligonucleotide did not support DNA synthesis (data not shown).
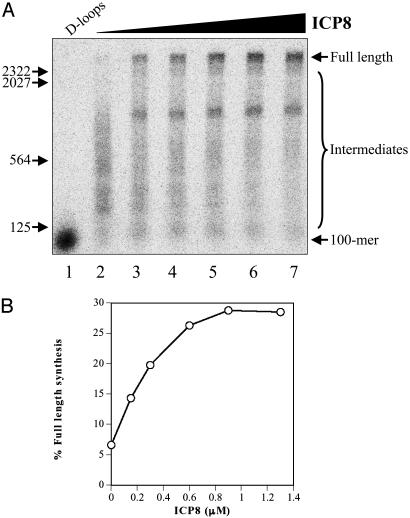
ICP8 Is Required for Long-Chain DNA Synthesis. Thus far, reactions were performed by supplementing ongoing D-loop reactions with the replication proteins. Because ICP8 is requisite for D-loop formation, its role in the DNA synthesis phase, where it may be required to function as an SSB, had to be examined using purified, deproteinized D-loops as the substrate. Consequently, purified D-loops were supplemented with replication proteins, including topoisomerase I, and increasing concentrations of ICP8. Fig. 2 shows that although limited primer extension was observed in the absence of ICP8, generation of full-length products (2,686 nt) depended on ICP8. Interestingly, increasing the concentration of ICP8 did not abolish a premature extension product (≈1,200 nt) that may be caused by pausing at a site of secondary structure (Fig. 2 A). Quantitation of the data shows that the efficiency of full-length synthesis increased with ICP8 concentration (Fig. 2B). Maximal extension of the 100-mer occurred at 900 nM ICP8,which is a 2-fold excess to what would be required to coat all of the DNA in the D-loop, based on a site size of 10 ± 1 nt per ICP8 molecule (27). The requirement for ICP8 in full-length DNA synthesis is consistent with its functioning in the capacity of an SSB.
Fig. 2.
ICP8 is required for full-length DNA synthesis. Reactions were performed as a function of ICP8 concentration with purified D-loops as described in Materials and Methods, followed by 1D denaturing agarose gel electrophoresis. (A) Storage phosphor image. Lane 1, mock-treated D-loops; lanes 2-7, extension of D-loops with 0, 150, 300, 600, 900, and 1,300 nM ICP8, respectively. (B) Quantitation of full-length synthesis products. The positions of 100-mer, full-length extension products (2,686 nt), intermediates, and markers (HindIII-digested λ DNA) are as indicated.
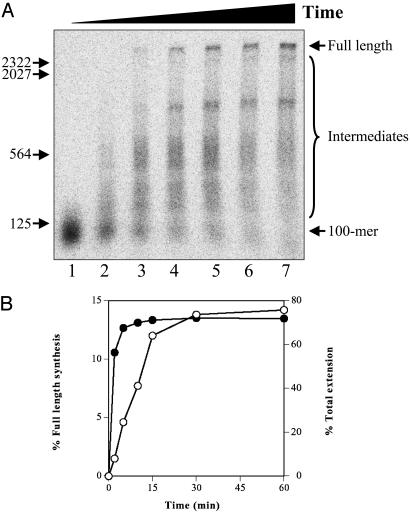
Kinetics of Recombination-Dependent DNA Synthesis. Fig. 3 shows a time course of DNA synthesis on purified D-loops supplemented with the replication proteins, including topoisomerase I and ICP8. Full-length products (2,686 nt) were observed as early as 5 min after initiation of the reaction (Fig. 3A, lane 3). Based on these data, a lower limit for the rate of DNA synthesis was calculated to be ≈10 nt/s. DNA synthesis paused at several discrete sites (intermediates), notably the ≈1,200-nt species observed in Fig. 2A. The rate of full-length product synthesis was linear in the first 15 min of the reaction. It should be noted that although ≈70% of the primer in the D-loop was rapidly (within ≈5 min) extended, only ≈15% was converted to full-length product (Fig. 3B).
Fig. 3.
Kinetics of recombination-dependent DNA synthesis. Reactions were performed as a function of time with purified D-loops as described in Materials and Methods, followed by 1D denaturing agarose gel electrophoresis. (A) Storage phosphor image. Lanes 1-7: 0, 2, 5, 10, 15, 30, and 60 min, respectively. (B) Quantitation of full-length synthesis products (open circles) and total extension products (filled circles). The positions of 100-mer, full-length extension products (2,686 nt), intermediates, and markers (HindIII-digested λ DNA) are as indicated.
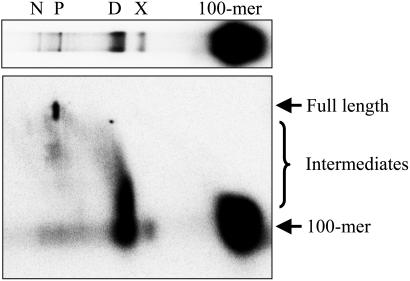
Form X Is Not a Template for DNA Synthesis. We had previously shown that strand assimilation promoted by ICP8 gives rise to a distinct topological form of D-loop, referred to as form X, that is resolved upon electrophoresis in the presence of the intercalating agent chloroquine (22). Relative to D-loops, form X exhibits a faster electrophoretic mobility, indicating that it is more positively supercoiled (22). To examine whether form X is a substrate for DNA synthesis, reaction products were resolved on a 2D native chloroquine-denaturing agarose gel. Resolution in the first dimension separated D-loops from form X, nicked D-loops, and extension products (Fig. 4). Subsequent resolution of the products by denaturing electrophoresis in the second dimension shows that the 100-mer originating from form X was not extended, whereas that from the D-loop reached full length (2,686 nt) (Fig. 4).
Fig. 4.
Form X is not a substrate for DNA synthesis. D-loops were formed, and the ability of the invading strand to prime DNA synthesis was examined as described in Materials and Methods. Reaction products were resolved by 2D native chloroquine-denaturing agarose gel electrophoresis. Storage phosphor images of first (Upper) and second (Lower) dimensions. The positions of D-loops (D), form X (X), products (P), nicked D-loops (N), 100-mer, full-length extension products (2,686 nt), intermediates, and markers (HindIII-digested λ DNA) are as indicated.
Discussion
As presented in the introduction, scenarios exist during HSV-1 replication that generate DSB, which would necessitate repair by RDR. Here we have reconstituted recombination-dependent DNA synthesis mediated by HSV-1 enzymes. To our knowledge, this is the first report of such a reaction reconstituted with eukaryotic viral proteins. We have used 2D native-denaturing agarose gel electrophoresis, a technique adapted from Xu and Marians (28), to visualize extension of a primer that was assimilated into a supercoiled plasmid template. In our system, strand assimilation is promoted by the viral SSB, ICP8, as described (22). D-loops generated in this reaction are similar to those formed by the action of RecA and related recombinases (e.g., Rad51 and T4 UvsX) (29). However, unlike these recombinases, ICP8 acts via an annealing mechanism (42). To exclude the possibility of protein-independent strand assimilation, we chose reaction conditions (importantly 10 mM Mg2+ and 30°C) under which D-loop formation is ICP8-dependent. D-loops formed by the action of ICP8 nucleate the assembly of a replication fork that involves the viral replication proteins, including the polymerase catalytic subunit (UL30), processivity factor (UL42), helicase-primase core enzyme (UL5/UL52), loading factor (UL8), SSB (ICP8), and a relaxing enzyme (E. coli gyrase or eukaryotic topoisomerase I).
Our data show that a mixture of the above replication proteins was capable of efficient extension of the invading primer; 75% of the 100-mer was used in the reaction, of which ≈15% reached full length (2,686 nt). Several intermediates, notably one at ≈1,200 nt, were detected, even in the presence of excess SSB. They may be attributed to pausing and dissociation of the replisome at specific sites in the template that impede its progress, as seen in T4 (30). The lower limit for the rate of DNA synthesis in our system is ≈10 nt/s at 30°C. This is within the same magnitude as the rate of DNA unwinding for the helicaseprimase in the presence of ICP8 (≈24-60 nt/s) (31) and to the estimated rate of replication fork progression in vivo for pseudorabies virus (≈50 nt/s) (32), that were determined at 37°C.
The coupling of strand invasion with DNA synthesis has been demonstrated in other systems. In T4, after inactivation of origin-dependent initiation, recombination provides a major pathway for initiating DNA synthesis from invading 3′ termini (reviewed in ref. 33). In this process, the two recombination proteins UvsX and UvsY, together with the SSB (gp32), catalyze strand invasion. The invading 3′ terminus in the resulting D-loop is used to prime leading-strand DNA synthesis by the phage replisome [polymerase (gp43), clamp (gp45), clamp loader (gp44 and gp62), helicase (gp41), loading protein (gp59), and SSB (gp32)]. Addition of the primase (gp61) leads to primer synthesis and lagging-strand synthesis, thereby re-creating a semiconservative replication fork. Aside from its role in initiating replication, RDR is also thought to play an important role in repairing DSB (reviewed in ref. 34). A hallmark of RDR in T4 is the formation of highly branched replication intermediates.
In E. coli, homologous recombination is emerging to be the dominant pathway for repair of DSB in an error-free manner (35, 36). Recently, a direct coupling of recombination and replication was elegantly demonstrated using purified E. coli proteins (37). In that study, Xu and Marians reconstituted the initial steps of the DSB pathway. In this system, RecBCD enzyme acts on χ-containing linear duplex donor DNA to provide 3′ terminal ssDNA for strand invasion into a homologous supercoiled plasmid catalyzed by RecA and SSB. The 3′ end of the invading primer provides a platform for the assembly of a bona fide replication fork consisting of the “restart” primosomal proteins (PriA, PriB, PriC, DnaT, DnaB, DnaC, and DnaG), RecO, RecR, polymerase III holoenzyme, and gyrase, which mediate leading- and lagging-strand synthesis.
Our data demonstrate that, after a strand invasion event, the viral replication proteins promote efficient long-chain synthesis that is representative of leading-strand synthesis. As yet, we have not been able to demonstrate lagging-strand synthesis. As expected, DNA synthesis in our system was absolutely dependent on the viral polymerase and dNTP. The polymerase by itself promoted only limited primer extension. Efficient full-length synthesis depended on the viral SSB (ICP8), helicase-primase, and a relaxing enzyme. We examined the role of ICP8 in the DNA synthesis phase of the reaction using purified D-loops. Omission of ICP8 only gave rise to intermediate extension products. Full-length product synthesis required coating concentrations of ICP8. In this regard, ICP8 presumably acts as an SSB. Consistent with this notion, it was previously shown that coating concentrations of ICP8 are required for full-length replication of primed M13 ssDNA (38). Helicase-primase is expected to play a pivotal role by loading at the replication fork and unwinding the duplex to provide the template for the polymerase. Indeed, in the absence of helicase-primase, there was an ≈4-fold reduction in the amount of full-length product and an accumulation of intermediates. The dependence of long-chain synthesis on helicase action observed here is in contrast to DnaB-independent elongation described for E. coli (28, 37). Helicase-primase-driven extension would generate positive linkages as a result of the unwinding of the template (39). An enzymatic activity that relaxes these positive supercoils would therefore be required for fork progression. Omission of this relaxation activity led to an ≈8-fold drop in full-length synthesis. Thus, full-length product synthesis was seen to depend on gyrase or eukaryotic topoisomerase I, both of which are known to remove positive linkages (39, 40). In our in vitro assays, either enzyme performs this function, although we would expect that topoisomerase I would fulfill this role in vivo.
We observed that extension occurred from primers assimilated into D-loops as well as nicked D-loops. Nicked D-loops arose during the course of the reaction from a minor endonuclease contaminant in the UL30 preparation. Extension of the 100-mer originating from the nicked species was independent of a relaxation activity, because these structures are not topologically constrained, but depend on the helicase-primase. Our data indicate that form X, a positively supercoiled form of D-loop, fails to participate in the elongation reaction. This finding is consistent with the reaction described for E. coli (28).
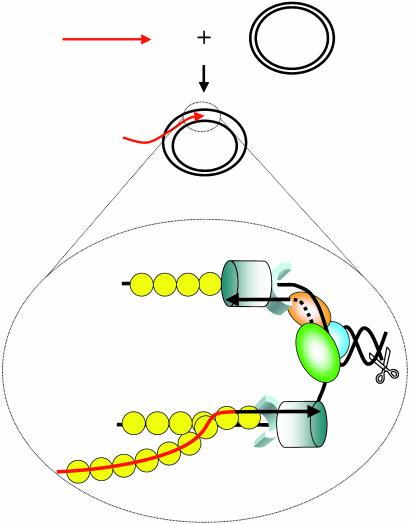
The model emerging from the findings presented in this paper is portrayed in Fig. 5. As shown, DSB are processed to provide 3′-terminal ssDNA fragments that invade into homologous acceptor regions. Processing may involve exonucleolytic degradation by the viral UL12 protein, DNA unwinding (UL9 helicase or helicase-primase), or helix destabilization by ICP8 (reviewed in ref. 2). In the in vitro system described here, the invading 100-mer mimics the outcome of DSB processing. In vivo, homologous sequences that are required for the pairing step may be provided either by the presence of repeated sequences in the viral genome or by duplication of genomic sequences after replication. ICP8-promoted D-loops provide a structure for the assembly of a replisome consisting of the polymerase (UL30 and UL42), helicase-primase (UL5/UL52 and UL8), SSB (ICP8), and a relaxing enzyme. Although the invading primer would be used for leading-strand synthesis, the primase would theoretically synthesize an RNA primer for lagging-strand synthesis on the opposite strand. The model predicts that the fork consists of two molecules of the viral polymerase that promote leading- and lagging-strand synthesis, respectively. Coordinated leading- and lagging-strand synthesis by the HSV-1 replisome has been demonstrated by Elias and colleagues (41). RDR along the lines proposed here would account for the generation of highly branched structures that exist late during viral replication. Finally, it is possible that RDR is a mechanism by which viral replication initiates independent of origin or UL9 function as seen in T4 (34).
Fig. 5.
Model for recombination-dependent replication in HSV-1. (Upper) The assimilation of a ssDNA (red arrow) into a homologous duplex (black circle) by ICP8. (Lower) The detailed events that culminate in the assembly of a replication fork after ICP8 (yellow circles)-mediated strand assimilation. The three ovals (orange, green, and blue) represent the heterotrimeric helicaseprimase (UL5/UL52/UL8). The cylinder and attached semicircle represent the polymerase (UL30) and its processivity factor (UL42), respectively. The pair of scissors represents the action of a relaxing enzyme. The solid and dashed lines represent DNA and RNA synthesis, respectively. The 3′ primer end is depicted by an arrowhead.
Acknowledgments
This work was supported by National Institutes of Health Grant GM62643 and Florida Biomedical Research Program Grant BM 022 (to P.E.B.).
Abbreviations: CR, complete reaction; D-loop, displacement loop; DSB, double-strand breaks; HSV-1, herpes simplex virus 1; RDR, recombination-dependent replication; SSB, single-stranded DNA-binding protein; ss, single-stranded.
References
- 1.Lehman, I. R. & Boehmer, P. E. (1999) J. Biol. Chem. 274, 28059-28062. [DOI] [PubMed] [Google Scholar]
- 2.Boehmer, P. E. & Nimonkar, A. V. (2003) IUBMB Life 55, 13-22. [DOI] [PubMed] [Google Scholar]
- 3.Enquist, L. W. & Skalka, A. (1973) J. Mol. Biol. 75, 185-212. [DOI] [PubMed] [Google Scholar]
- 4.Jackson, S. A. & DeLuca, N. A. (2003) Proc. Natl. Acad. Sci. USA 100, 7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luder, A. & Mosig, G. (1982) Proc. Natl. Acad. Sci. USA 79, 1101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson, C. C. (1983) Cell 33, 315-317. [DOI] [PubMed] [Google Scholar]
- 7.Aslani, A., Olsson, M. & Elias, P. (2002) J. Biol. Chem. 277, 41204-41212. [DOI] [PubMed] [Google Scholar]
- 8.Makhov, A. M., Lee, S. S., Lehman, I. R. & Griffith, J. D. (2003) Proc. Natl. Acad. Sci. USA 100, 898-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLean, G. W., Abbotts, A. P., Parry, M. E., Marsden, H. S. & Stow, N. D. (1994) J. Gen. Virol. 75, 2699-2706. [DOI] [PubMed] [Google Scholar]
- 10.Marsden, H. S., McLean, G. W., Barnard, E. C., Francis, G. J., MacEachran, K., Murphy, M., McVey, G., Cross, A., Abbotts, A. P. & Stow, N. D. (1997) J. Virol. 71, 6390-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkenberg, M., Bushnell, D. A., Elias, P. & Lehman, I. R. (1997) J. Biol. Chem. 272, 22766-22770. [DOI] [PubMed] [Google Scholar]
- 12.Tanguy Le Gac, N., Villani, G., Hoffmann, J. S. & Boehmer, P. E. (1996) J. Biol. Chem. 271, 21645-21651. [DOI] [PubMed] [Google Scholar]
- 13.Severini, A., Scraba, D. G. & Tyrrell, D. L. (1996) J. Virol. 70, 3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob, R. J. & Roizman, B. (1977) J. Virol. 23, 394-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smiley, J. R., Wagner, M. J., Summers, W. P. & Summers, W. C. (1980) Virology 102, 83-93. [DOI] [PubMed] [Google Scholar]
- 16.Valyi-Nagy, T., Olson, S. J., Valyi-Nagy, K., Montine, T. J. & Dermody, T. S. (2000) Virology 278, 309-321. [DOI] [PubMed] [Google Scholar]
- 17.Milatovic, D., Zhang, Y., Olson, S. J., Montine, K. S., Roberts, L. J., II, Morrow, J. D., Montine, T. J., Dermody, T. S. & Valyi-Nagy, T. (2002) J. Neurovirol. 8, 295-305. [DOI] [PubMed] [Google Scholar]
- 18.Wohlrab, F., Chatterjee, S. & Wells, R. D. (1991) Proc. Natl. Acad. Sci. USA 88, 6432-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, K. J., Zemelman, B. V. & Lehman, I. R. (2002) J. Biol. Chem. 277, 21071-21079. [DOI] [PubMed] [Google Scholar]
- 20.Nimonkar, A. V. & Boehmer, P. E. (2002) J. Biol. Chem. 277, 15182-15189. [DOI] [PubMed] [Google Scholar]
- 21.Kong, D. & Richardson, C. C. (1996) EMBO J. 15, 2010-2019. [PMC free article] [PubMed] [Google Scholar]
- 22.Nimonkar, A. V. & Boehmer, P. E. (2003) J. Biol. Chem. 278, 9678-9682. [DOI] [PubMed] [Google Scholar]
- 23.Boehmer, P. E. & Lehman, I. R. (1993) Proc. Natl. Acad. Sci. USA 90, 8444-8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill, S. C. & von Hippel, P. H. (1989) Anal. Biochem. 182, 319-326. [DOI] [PubMed] [Google Scholar]
- 25.Boehmer, P. E. (1996) Methods Enzymol. 275, 16-35. [DOI] [PubMed] [Google Scholar]
- 26.Boehmer, P. E., Dodson, M. S. & Lehman, I. R. (1993) J. Biol. Chem. 268, 1220-1225. [PubMed] [Google Scholar]
- 27.Gourves, A. S., Tanguy Le Gac, N., Villani, G., Boehmer, P. E. & Johnson, N. P. (2000) J. Biol. Chem. 275, 10864-10869. [DOI] [PubMed] [Google Scholar]
- 28.Xu, L. & Marians, K. J. (2002) J. Biol. Chem. 277, 14321-14328. [DOI] [PubMed] [Google Scholar]
- 29.Bianco, P. R., Tracy, R. B. & Kowalczykowski, S. C. (1998) Front. Biosci. 3, D570-D603. [DOI] [PubMed] [Google Scholar]
- 30.Formosa, T. & Alberts, B. M. (1986) Cell 47, 793-806. [DOI] [PubMed] [Google Scholar]
- 31.Falkenberg, M., Lehman, I. R. & Elias, P. (1998) J. Biol. Chem. 273, 32154-32157. [DOI] [PubMed] [Google Scholar]
- 32.Ben-Porat, T., Blankenship, M. L., DeMarchi, J. M. & Kaplan, A. S. (1977) J. Virol. 22, 734-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bleuit, J., Xu, H., Ma, Y., Wang, T., Liu, J. & Morrical, S. (2001) Proc. Natl. Acad. Sci. USA 98, 8298-8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, E. S., Kutter, E., Mosig, G., Arisaka, F., Kunisawa, T. & Ruger, W. (2003) Microbiol. Mol. Biol. Rev. 67, 86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox, M. M. (2002) Mutat. Res. 510, 107-120. [DOI] [PubMed] [Google Scholar]
- 36.Marians, K. J. (2000) Curr. Opin. Genet. Dev. 10, 151-156. [DOI] [PubMed] [Google Scholar]
- 37.Xu, L. & Marians, K. J. (2003) Mol. Cell 11, 817-826. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez, T. R. & Lehman, I. R. (1994) J. Biol. Chem. 265, 11227-11232. [PubMed] [Google Scholar]
- 39.Hiasa, H. & Marians, K. J. (1996) J. Biol. Chem. 271, 21529-21535. [DOI] [PubMed] [Google Scholar]
- 40.Gellert, M. (1981) Annu. Rev. Biochem. 50, 879-910. [DOI] [PubMed] [Google Scholar]
- 41.Falkenberg, M., Lehman, I. R. & Elias, P. (2000) Proc. Natl. Acad. Sci. USA 97, 3896-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimonkar, A. V. & Boehmer, P. E. (2003) Nucleic Acids Res. 31, in press. [DOI] [PMC free article] [PubMed]