Abstract
Hypermutation at the Ig loci is confined to the area between the promoter and the intronic enhancer, which includes the rearranged variable region gene segment. We identified factors that contribute to the site-specificity at the heavy chain locus. We found that distance from both the promoter and the intronic enhancer is crucial in hypermutation. The presence of the enhancer is required, and, in contrast to its definition for transcriptional activity, its effect is orientation-sensitive.
Keywords: hypermutation, transfected target sequence, termination codon, reversion
Of the mutator phenotypes in mammalian cells hypermutation in activated B lymphocytes (1–7) is one of the most intriguing ones. Unlike, for instance, in the cases of colorectal cancer cells (8) or Werner syndrome cells (9), the mutational activity at the Ig locus is site specific, i.e., it is directed exclusively to the variable (V) region and its flanking sequences of either of the Ig light (L) or heavy (H) chain. But there is no specific sequence in the V regions that attracts hypermutation (10). Because the majority of mutations are found between the promoter and the intron enhancer (11), it was concluded that these two elements are important. Indeed, the promoter apparently is required (12, 13), but it need not to be an Ig promoter (14, 15); what matters is the distance from it (13). These findings, and the observation that often there is strand polarity in the distribution of the point mutations, led to the conclusion that it is the transcriptional process that drives hypermutation.
Enhancers traditionally have been defined as elements that increase transcription in an orientation-independent way. However, since their original discovery in mammalian cells (16, 17), many other functions have been attributed to these sequences, including even repression (18). At the κ L-chain locus, the intron enhancer was found to be needed for a high rate of mutation (14), and a broad correlation between transgene expression and mutation was observed (19). But its mode of action is unknown.
The assessment of the function of the H intronic enhancer in hypermutation has posed a more difficult problem. In transfection experiments (16, 17) and in transgenic mice (20) this element is clearly needed for regular-level transcription. Furthermore, deleting this enhancer in the germ line affects VHDJH rearrangement (21).
Our finding that we can replace the Ig promoter with the thymidine kinase (tk) promoter linked to the simian virus 40 (SV40) enhancer for in vitro hypermutation (15) provided an opportunity to study the role of the intronic H enhancer in the 18–81 cell line, independently of its role to sustain transcription at the regular level. The 18–81 line faithfully reproduces features of hypermutation in vivo, such as site specificity and differentiation stage specificity (22–28). Also, as in mice (29–32) and in humans (33), the RGYW base sequence constitutes a hot-spot motif (34), although another suggested motif, the TAA sequence (29), does not (34), which may or may not reflect a difference between in vivo and in vitro. At any rate, we have termed the generator of mutations in the 18–81 cell line the GC mutator, as it clearly acts preferentially on the G within the RGYW motif (28, 34).
In this report we wanted to know whether in vitro mutability is dependent on the distance from the promoter; whether or not the major intron enhancer is needed for hypermutation at the H-chain locus and, if so, whether or not its action is distance- and orientation-independent.
MATERIALS AND METHODS
Plasmid Constructions.
All constructs are based on the plasmid ptk·VC·3′K (15). For the construction of plasmid p·E·del the 1-kb XbaI–XbaI fragment containing the large intron enhancer was excised from plasmid ptk·VC·3′K; for plasmid p·E·inv the same 1-kb XbaI–XbaI fragment was recloned into the original site, and a clone with inverted orientation was selected; for plasmid p·no·3′K the two 3′κ enhancers were excised; and to generate plasmid p·E·shift the 1-kb XbaI–XbaI fragment of plasmid p·no·3′K was excised and recloned into the XhoI site 3′ of the membrane exons. To generate plasmid p+500 the SnaI–SnaI intronic sequence fragment (475 bp) from the MICA gene (35) was excised and cloned into the EcoRV site of plasmid ptk·VC·3′K, located between the L1 and L2 exon.
Detection and Quantification of Mutants.
Plasmids were transfected into clone 18–81.ΔM (36). The copy number in our system is usually one, and sometimes two, as assessed with Southern blot analysis of the individual clones. The transfectants were subcloned at limiting dilution, and the subclones were grown to 105 cells, spun onto a slide, ethanol-fixed, and rehydrated in PBS + 1% BSA. The slides were stained with fluorescein isothiocyanate-conjugated goat anti-mouse IgM antibody (Fisher), washed thrice for 10 min in PBS, and μ-producing, immunofluorescent revertants were counted.
Calculation of Mutation Frequencies.
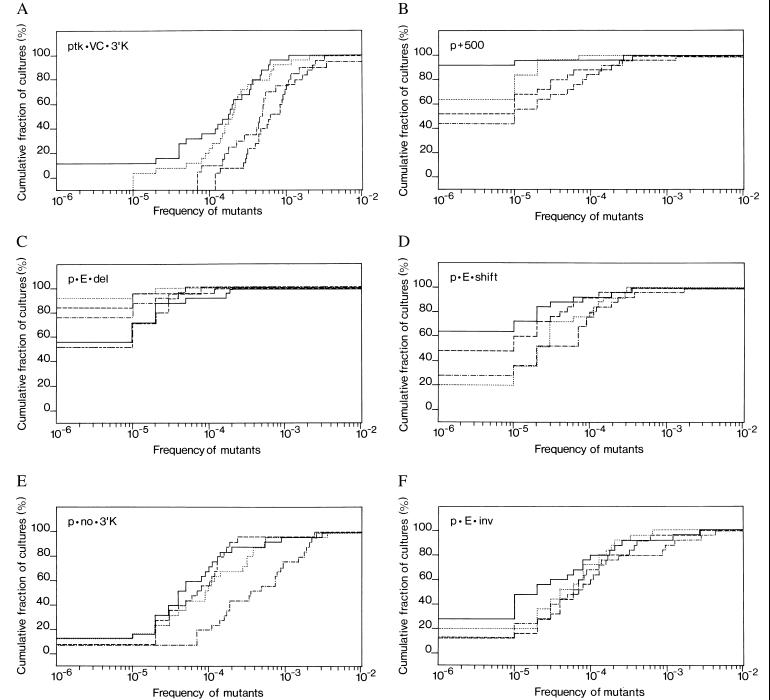
For each plasmid, the median frequency of revertants was calculated for each of the curves in the appropriate panel of Fig. 3. For those curves in which the frequency of cultures with zero revertants was greater than 50%, a value of <1 × 10−5 was used.
Figure 3.
Cumulative distribution of reversion frequencies for the plasmids tested. The plasmids used for transfection were: (A) ptk·VC·3′K, (B) p+500, (C) p·E·del, (D) p·E·shift, (E) p·no·3′K, and (F) p·E·inv. Horizontal axis: frequency of revertants (μ-producers) on a logarithmic scale. Vertical axis: fraction of cultures, in percent on a linear scale, with that frequency or less of revertants. In this graphic representation of quantitation of mutants the unit step in a curve represents a given culture, and each curve represents one transfectant, from which the cultures were derived. The 4–5 transfectants (curves) shown per construct resulted from independent transfection events. Plotting the cumulative frequencies rather than the frequencies themselves avoids the “gaps” that are caused by fluctuation and lead to a contiguous curve.
Reverse Transcription–PCR.
For beta-actin cDNA primers actin-s, AATCCTGTGGCATCCATGAAAC, and actin-as, ACGCAGCTCAGTAACAGTCCG, were used. The cDNA was obtained by a standard procedure using Superscript II (GIBCO/BRL) and then was subjected to 38 cycles in a thermal cycler, with each cycle consisting of 20 sec at 94°C, 20 sec at 62°C, and 30 sec at 72°C. The PCR product has a size of 348 bp. For the μ-chain cDNA primers V81X, GAAGAGGCTGGAGTTGGTCGCAG, and Cμ4 REV, CACACTGATGTCTGCAGGAGAGAAGC, were used. The PCR amplification reaction on cDNA was done with 40 cycles, each 1 min at 94°C, 1 min at 68°C, and 3 min at 68°C, yielding a 1.3-kb DNA product. In both cases one-tenth of the final PCR product was run on an ethidium bromide containing agarose gel.
RESULTS AND DISCUSSION
For the studies in vitro, we introduced an indicator Ig μ H-chain gene into the mutator-positive cell line 18–81 by stable transfection (28); as a result of an amber termination codon (TAG), translation is terminated prematurely, and μ H-chains are not expressed (Fig. 1). On reversion of the termination codon in the transfected gene, expression of full-length μ protein chains can be detected by immunofluorescence; as the host cell line has lost both endogenous μ chain alleles, any μ chain synthesized must have been encoded by the transfected μ chain gene. In all plasmids, the Ig promoter is replaced by the tk promoter. For full hypermutation, the plasmids also contain a 3′ κ enhancer (15).
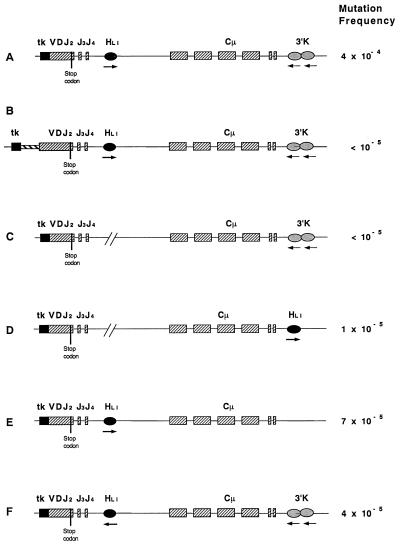
Figure 1.
Plasmids to monitor mutator activity. The VDJ exon encoding the V region is functionally rearranged but no μ chain is expressed because of a TAG termination codon within the D segment. The 3′ κ enhancer is added to the minimal μ gene, and the Ig promoter has been replaced by the tk promoter. tk, tk promoter; VDJ2, V region segment; HLI, large intron enhancer, Cμ, constant region exons; 3′K, 3′ κ enhancer.
Distance from the Promoter.
If the promoter is the 5′ boundary for hypermutation and transcription is important for the process, then increasing the distance from the promoter without changing the distance from a putative 3′ element may result in a lower mutation frequency. Previously, we found that moving the hot-spot motif upstream, and thus nearer to the promoter, but further away from the intronic H enhancer resulted in a decreased mutation frequency (34). However, such a result also could be interpreted that the RGYW motif is a minimal motif only.
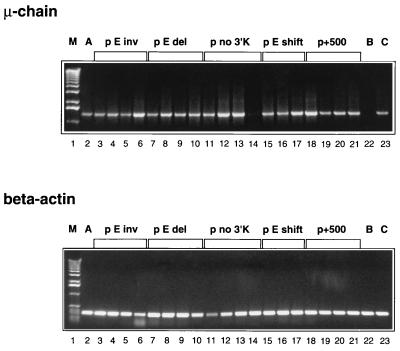
We inserted into the standard plasmid ptk·VC·3′K (driven by a tk promoter/SV40 enhancer) (Fig. 1A) between the L1 and L2 exon a 500-bp stuffer to generate plasmid p+500 (Fig. 1B), in which the V region is thus moved further away from the promoter. As for all other plasmids described below, cells of the line 18–81 were transfected, 4–5 independent transfectants were subcloned, and the frequency of revertants expressing μ H-chain was determined for 25 subclones for each transfectant. In all revertants the fluorescence intensity was similar (not shown); and the steady-state mRNA expression levels in the transfectants were similar, as determined by reverse transcription–PCR analysis, regardless of the particular type of construct (Fig. 2). A subclone with an endogenous μ gene containing a stop codon had a comparable state level (Fig. 2, lane 23), whereas no μcDNA was amplified in the clone that was used for transfections and lacked a μ gene (Fig. 2, lane 22). The mutation results were plotted as cumulative distributions of revertant frequencies in Fig. 3. From the plots, the median revertant frequencies for each plasmid were estimated, and the median of the plasmid medians are given in Fig. 3 and listed in Fig. 1 as the “mutation frequency.” Standard plasmid ptk·VC·3′K (Fig. 1A) is the reference plasmid, which previously showed full hypermutability at 4 × 10−4 (15); we have repeated the mutation frequency measurement for the present study with a new transfectant, and found it to be 3 × 10−4 (Fig. 3A). We thus average it at 4 × 10−4. In comparison, plasmid p+500 (Fig. 1B) mutated at a frequency of approximately two orders of magnitude lower (Fig. 3B). Thus we conclude that the GC mutator is sensitive to the distance from the promoter. That distance from the promoter is an important factor in hypermutation has been shown in vivo (13), which adds another striking similarity between the in vivo process and the effect of the GC mutator, as defined in vitro.
Figure 2.
Steady-state levels of μ mRNA in cells transfected with various constructs, as determined by reverse transcription–PCR. (Upper) Expression of mRNA encoding μ chain in clones transfected with various constructs, marked at the top of the photographs. M, marker. A, clone transfected with ptk·VC·3′K, which has been derived and evaluated for this study, in addition to the three evaluated previously (15). B, untransfected 18–81 subclone 4.3 lacking Cμ. C, subclone with endogenous μ gene containing a stop codon. (Lower) Expression of mRNA encoding β actin in clones transfected with various constructs. Three or four independent transfectants for each construct (except A and C) shown.
Intronic H Enhancer as a cis Element for Hypermutation.
As we can replace the Ig promoter by the tk promoter (15), just as in vivo (14), the specificity-determining element(s) may be located in the major intron. It is probable, but not certain, that this element is located 5′ to the Sμ switch region, because switched cells continue to mutate. In analogy to the large intron κ enhancer this points to the H enhancer. To assess the role of the major intron enhancer, we generated four different plasmids, in which we changed the context of the enhancer. For the first one we introduced into the standard plasmid a 1-kb XbaI–XbaI fragment deletion, which contains all elements of the H enhancer, to produce plasmid p·E·del (Fig. 1C). This deletion caused the mutation frequency to drop by two orders of magnitude (Fig. 3C). Therefore, the intronic H enhancer is necessary for hypermutation at the H locus.
The question arises whether the effect of the enhancer is dissociated from its conventional role as a transcriptional enhancer. Because in our constructs the tk promoter is linked to the SV40 enhancer, lack of the intronic H enhancer is not likely to decrease regular-level transcription. Indeed, the steady-state mRNA level (Fig. 2), and, therefore, likely the transcription rate, seems to be no different for the constructs with or without the H enhancer. Importantly, the revertant clones showed the same fluorescence intensity for all the constructs. Thus, although transcription may be a prerequisite for hypermutation to occur (19), the intronic H enhancer functions in hypermutation apparently not or not exclusively via enhancing transcription.
Position of the H Enhancer.
Given that the H enhancer is so important, is its function position-independent, as is the case for transcriptional enhancement? To answer this question, we moved, in plasmid p·E·shift, the H enhancer 3′ to the C region, replacing there the 3′K enhancer (Fig. 1D). [We could not leave the 3′K enhancer in its place, because in such a construct the close proximity of the two enhancers can cause antihypermutation (15).] The mutation frequency in construct p·E·shift was decreased to 1 × 10−5 (Fig. 3D). This decrease corresponds to a drop by one order of magnitude when compared with either the standard plasmid, which contains the 3′κ enhancer (with 4 × 10−4), or to a 7-fold decrease, when compared with the one of plasmid p·no·3′K (Fig. 1E), at 7 × 10−5 (Fig. 3E). In turn, the frequency of plasmid p·no·3′K is almost the same as the 6.7 × 10−5 for the plasmid pVC·SV40, which has the Ig promoter, the large intron enhancer and the SV40 enhancer 3′ to the Cμ (15). Although we have not studied yet how the SV40 enhancer at the promoter site influences the mutation rate, we have shown previously that an additional enhancer (of any kind) is required for full hypermutation of the minimal μ transgene, which is fully transcribed without such an enhancer. The fact that p·no·3′K and pVC·SV40 have the same number of enhancers and close mutation rates indicates the reliability of our method. At any rate, moving the H intronic enhancer from its original place causes the mutation frequency to drop 7-fold. We conclude that mutability is also a function of the distance from the H enhancer.
Orientation of the H Enhancer.
As mentioned above, the effect on transcription is independent of the orientation of the enhancer in respect to the promoter. But the requirement for the H enhancer in hypermutation apparently is not caused by its effect on transcription. Thus it is possible that hypermutation is sensitive to the enhancer orientation. If one views the promoter and enhancer not only as apparent, but also as functional boundaries for hypermutation, one may expect the enhancer to confer orientation to the hypermutation process. To test this we inverted the XbaI–XbaI fragment including the H enhancer of the standard construct to generate p·E·inv (Fig. 1F). For this plasmid the mutation frequency is an order of magnitude lower than the one in the standard plasmid (4 × 10−5; Fig. 3F). Because the steady-state mRNA level (Fig. 2), and, therefore, likely the transcription rate, is no different from the construct with the H enhancer in the original orientation, this finding confirms the notion that the H enhancer functions in hypermutation not or at least not exclusively via enhancing transcription. Barring the possibility that we have disrupted a cis-acting sequence or separated two such sequences it also indicates that the H enhancer gives, in a yet unspecified way, a direction to hypermutation.
Conclusion.
In conclusion, distance from the promoter is crucial in hypermutation at the H locus. The presence of the H enhancer is required; but its effect is orientation- and distance-sensitive, which ipso facto contrasts with its conventional definition. Our findings also bring us one step further in our goal to assemble the minimal cis-acting elements in a cassette, which on transfection into the 18–81 cell line allows hypermutation of any gene that it contains.
Acknowledgments
We thank Charles Steinberg for editing the manuscript and Siamak Bahram for providing the MICA gene construct. The Basel Institute for Immunology was founded and is sponsored by F.-Hoffmann-La Roche, Basel, Switzerland. This work was supported by National Institutes of Health Grant 1R01 GM37699 and by funds from the Markey Trust to M.W.
ABBREVIATIONS
- V
variable
- L
light
- H
heavy
- tk
thymidine kinase
- SV40
simian virus 40
References
- 1.Scharff M D, Poltoratsky V, Green N S. J Exp Med. 1997;185:185–188. doi: 10.1084/jem.185.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neuberger M S, Milstein C. Curr Opin Immunol. 1995;7:248–254. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 3.Jolly C J, Wagner S D, Rada C, Klix N, Milstein C, Neuberger M S. Semin Immunol. 1996;8:159–161. doi: 10.1006/smim.1996.0020. [DOI] [PubMed] [Google Scholar]
- 4.Reynaud C A, Quint L, Bertocci B, Weill J C. Semin Immunol. 1996;8:125–129. doi: 10.1006/smim.1996.0016. [DOI] [PubMed] [Google Scholar]
- 5.Storb U, Peters A, Klotz E, Rogerson B, Hackett J., Jr Semin Immunol. 1996;8:131–140. doi: 10.1006/smim.1996.0017. [DOI] [PubMed] [Google Scholar]
- 6.Tumas-Brundage K, Vora K A, Giusti A M, Manser T. Semin Immunol. 1996;8:141–150. doi: 10.1006/smim.1996.0018. [DOI] [PubMed] [Google Scholar]
- 7.Wabl M, Steinberg C. Curr Opin Immunol. 1996;8:89–92. doi: 10.1016/s0952-7915(96)80110-5. [DOI] [PubMed] [Google Scholar]
- 8.Dunlop M G. Curr Opin Genet Dev. 1996;6:76–81. doi: 10.1016/s0959-437x(96)90014-3. [DOI] [PubMed] [Google Scholar]
- 9.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 10.Yelamos J, Klix N, Goyenechea B, Lozano F, Chui Y L, Gonzalez-Fernandez A, Pannell R, Neuberger M S, Milstein C. Nature (London) 1995;376:225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- 11.Gearhart P J. In: Fundamental Immunology. Paul W E, editor. New York: Raven; 1993. pp. 865–885. [Google Scholar]
- 12.Peters A, Storb U. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 13.Tumas-Brundage K, Manser T. J Exp Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betz A G, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger M S. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 15.Bachl J, Wabl M. Immunogenetics. 1996;45:59–64. doi: 10.1007/s002510050167. [DOI] [PubMed] [Google Scholar]
- 16.Gillies S D, Morrison S L, Oi V T, Tonegawa S. Cell. 1993;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 17.Banerji J, Olson L, Schaffner W. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 18.Zong R T, Scheuermann R H. J Biol Chem. 1995;270:24010–24018. doi: 10.1074/jbc.270.41.24010. [DOI] [PubMed] [Google Scholar]
- 19.Goyenechea B, Klix N, Yelamos J, Williams G T, Riddell A, Neuberger M S, Milstein C. EMBO J. 1997;16:3987–3994. doi: 10.1093/emboj/16.13.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosschedl R, Baltimore D. Cell. 1985;41:885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- 21.Serwe M, Sablitzky F. EMBO J. 1993;12:2321–2327. doi: 10.1002/j.1460-2075.1993.tb05886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wabl M, Burrows P D, von Gabain A, Steinberg C. Proc Natl Acad Sci USA. 1985;82:479–482. doi: 10.1073/pnas.82.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer J, Jäck H-M, Ellis N, Wabl M. Proc Natl Acad Sci USA. 1986;83:6950–6953. doi: 10.1073/pnas.83.18.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jäck H-M, Wabl M. Proc Natl Acad Sci USA. 1987;84:4934–4938. doi: 10.1073/pnas.84.14.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wabl M, Jäck H-M, Meyer J, Beck-Engeser G, von Borstel R C, Steinberg C M. Immunol Rev. 1987;96:91–107. doi: 10.1111/j.1600-065x.1987.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhu M, Rabinowitz J L, Green N S, Kobrin B J, Scharff M D. Proc Natl Acad Sci USA. 1995;92:2810–2814. doi: 10.1073/pnas.92.7.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green N S, Rabinowitz J L, Zhu M, Kobrin B J, Scharff M D. Proc Natl Acad Sci USA. 1995;92:6304–6308. doi: 10.1073/pnas.92.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachl J, Wabl M. Proc Natl Acad Sci USA. 1996;93:851–855. doi: 10.1073/pnas.93.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogozin I B, Kolchanov N A. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 30.Betz A G, Neuberger M S, Milstein C. Immunol Today. 1993;14:405–411. doi: 10.1016/0167-5699(93)90144-a. [DOI] [PubMed] [Google Scholar]
- 31.Wagner S D, Milstein C, Neuberger M S. Nature (London) 1995;376:732. doi: 10.1038/376732a0. [DOI] [PubMed] [Google Scholar]
- 32.Goyenechea B, Milstein C. Proc Natl Acad Sci USA. 1996;93:13979–13984. doi: 10.1073/pnas.93.24.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorner T, Brezinschek H P, Brezinschek R I, Foster S J, Domiati-Saad R, Lipsky P E. J Immunol. 1997;158:2779–2789. [PubMed] [Google Scholar]
- 34.Bachl, J., Steinberg, C. & Wabl, M. (1998) Eur. J. Immunol., in press. [DOI] [PubMed]
- 35.Bahram S, Mizuki N, Inoko H, Spies T. Immunogenetics. 1996;44:80–81. doi: 10.1007/BF02602661. [DOI] [PubMed] [Google Scholar]
- 36.Jäck H-M, McDowell M, Steinberg C M, Wabl M. Proc Natl Acad Sci USA. 1988;85:1581–1585. doi: 10.1073/pnas.85.5.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]