Abstract
Cystathionine γ-synthase, the first committed enzyme of methionine biosynthesis in higher plants, is encoded by the CGS1 gene in Arabidopsis thaliana. We have shown previously that the stability of the CGS1 mRNA is negatively regulated in response to methionine application [Chiba, Y., Ishikawa, M., Kijima, F., Tyson, R. H., Kim, J., Yamamoto, A., Nambara, E., Leustek, T., Wallsgrove, R. M. & Naito, S. (1999) Science 286, 1371-1374]. To determine whether methionine itself is the effector of the CGS1 exon 1-mediated posttranscriptional regulation, we carried out transfection experiments. The results suggested that, rather than methionine, S-adenosyl-l-methionine (AdoMet), or one of its metabolites, acts as the effector of this regulation. To further identify the actual effector, we exploited the wheat germ in vitro translation system. The effects of various metabolites and analogs of AdoMet were tested by using RNA carrying a CGS1 exon 1-reporter fusion. These tests identified AdoMet as the effector of this regulation. S-adenosyl-l-ethionine, an analog of AdoMet, also had effector activity. A. thaliana mto1 mutants, which are deficient in this regulation, showed a much reduced response to AdoMet in vitro, with a leaky allele showing a less reduced response. RNA translated in vitro in the presence of AdoMet contained a 5′-truncated RNA species, similar to the one that we previously suggested was an in vivo degradation intermediate of CGS1 mRNA. Together, the results show that the basic reactions of CGS1 exon 1-mediated posttranscriptional regulation occur in the wheat germ in vitro translation system, and that AdoMet acts as the effector.
Cystathionine γ-synthase (CGS; EC 4.2.99.9) catalyzes the first committed step of methionine biosynthesis in higher plants (1) (Fig. 1), which is considered to be the key regulatory step in methionine biosynthesis (2-5). Unlike many of the key-step enzymes in metabolic pathways, CGS is not an allosteric enzyme (2). CGS is encoded in Arabidopsis thaliana by the CGS1 gene (gene ID At3g01120, GenBank accession no. AB010888) (6, 7). We have previously shown (8) that CGS1 expression involves feedback regulation at the level of mRNA stability in response to methionine application in vivo. A. thaliana mto1 mutants are deficient in this feedback regulation, and overaccumulate CGS1 mRNA, CGS protein, and soluble methionine. Seven independently isolated mto1 mutants were found to carry single-base changes within the first exon of CGS1, giving rise to amino acid sequence changes (8, 9).
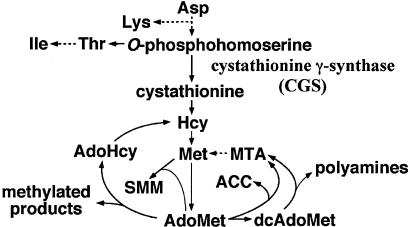
Fig. 1.
Schematic representation of the biosynthetic pathway of methionine and the metabolism of AdoMet. Dashed arrows represent multiple reactions. Hcy, l-homocysteine; AdoHcy, S-adenosyl-Hcy; MTA, 5′-deoxy-5′-(methylthio)adenosine; ACC, 1-aminocyclopropane-1-carboxylic acid; SMM, S-methyl-l-methionine; dcAdoMet, decarboxylated AdoMet.
Transient and transgenic expression experiments using CGS1 exon 1-reporter fusions (8, 10) showed that the exon 1 coding sequence of CGS1 is necessary and sufficient for its posttranscriptional regulation, in response to exogenous application of methionine. In vitro mutagenesis of CGS1 exon 1 revealed that it is its amino acid sequence that has a role in this regulation. We have identified a stretch of 11-13 amino acid residues, termed the MTO1 region, located ≈80 residues from the N terminus of CGS, and covering the mto1 mutation sites, which is important for this regulation. The MTO1 sequence is highly conserved among the CGS proteins of higher plants, but is not found elsewhere in the public nucleotide and protein databases (9). Despite the fact that the amino acid sequence of exon 1 is crucial, the exon 1 sequence has been shown to act in cis. To account for this action, we have presented a model in which the regulation occurs during the translation of CGS1 mRNA (8, 10).
In the 1980s, it was shown that CGS enzyme activity in the aquatic plant Lemna pausicostata is repressed when the plant is grown in the presence of methionine, and it has been believed that methionine feedback-regulates its own de novo biosynthesis (2, 11), although the mechanism of this regulation was unknown. We have shown that expression of CGS1 is regulated at the level of mRNA stability (8), but it still remained to be determined whether methionine itself, or one of its metabolites, is the actual effector of this regulation. Establishment of an in vitro system would not only be useful in identifying the effector, but would also greatly facilitate the elucidation of the molecular mechanisms of CGS1 regulation. According to our model that the regulation occurs during translation, the regulation could be reproduced in an in vitro translation system. We show here that the posttranscriptional regulation mediated by exon 1 of CGS1 occurs in the wheat germ in vitro translation system, and that S-adenosyl-l-methionine (AdoMet) acts as the effector.
Materials and Methods
Chemicals. The chemicals used to test the effector activity in the regulation of CGS1 expression were as follows: l-methionine (catalog no. 133-01602, Wako Chemicals, Osaka), l-homocysteine (catalog no. 085-07591, Wako Chemicals), 1-aminocyclo-propane-1-carboxylic acid (catalog no. 010-17711, Wako Chemicals), AdoMet (catalog no. A-4377, Sigma-Aldrich, St. Louis), S-adenosyl-l-homocysteine (AdoHcy; catalog no. A-9384, Sigma-Aldrich), 5′-deoxy-5′-(methylthio)adenosine (catalog no. 31122, Fluka, Buchs, SG, Switzerland), S-adenosyl-l-ethionine (AdoEtn; catalog no. A-2758, Sigma-Aldrich), adenosyl-l-ornithine (AdoOrn; catalog no. 56705, Calbiochem), S-methyl-l-methionine (SMM; catalog no. 155398, ICN), S-adenosyl-l-propionine (AdoPpn; synthesized by Eburon Organics, Rijkevorsel, Belgium), and decarboxylated AdoMet (dcAdoMet; a gift of A. Shirahata, Josai University, Saitama, Japan; ref. 12).
Genetic Materials. The A. thaliana (L.) Heynh. mutants mto1-1, mto1-2, mto1-3, mto1-4 (8, 9), and mto3-1 (13) have been described elsewhere. mto1-1 and mto3-1 mutants and their WT strains, ecotypes Columbia (Col-0) and Wassilewskija (Ws), respectively, were used in the transfection experiments. The mto3-1 mutant and Ws were obtained from M. C. Tarczynski (Pioneer Hi-Bred International, Johnston, IA).
Transfection Experiments. The tester plasmids pMI4(WT) and pMI4(mto1-1), and the control plasmid 221-LUC+ have been described (9, 14). Transfection of A. thaliana protoplasts by electroporation, treatment of the electroporated protoplasts, and reporter assays were carried out as described (9).
Construction of Plasmid DNA for in Vitro Transcription. pMI21(WT) plasmid carries the exon 1 coding region of WT CGS1 joined in-frame with the luc+ gene, which codes for the firefly (Photinus pyralis) luciferase (LUC), in the pSP64 Poly(A) vector (Promega). The exon 1 coding region, carrying XbaI and BamHI recognition sequences at its 5′ and 3′ ends, respectively, was prepared as described (9). The luc+ coding region was excised from the pSP64 luc+ plasmid (Promega) by digestion with HindIII and XbaI. The linker 5′-TCTAGGGCGGAGGTGGGAGCTC-3′ was ligated to the 3′ end of the luc+ DNA, destroying the XbaI recognition sequence. The exon 1 region and the luc+ DNA were joined with the linker 5′-GGATCCGGCGGTGGAAAGCTT-3′, and were inserted between the XbaI and SacI sites of the pSP64 Poly(A) vector. mto1-1, mto1-2, mto1-3, and mto1-4 mutant versions of the above plasmid were also constructed, and are referred to as pMI21(mto1-1), pMI21(mto1-2), pMI21(mto1-3), and pMI21(mto1-4). The pMI27 plasmid carries the sea pansy (Renilla reniformis) luciferase (RLUC) reporter gene, termed Rluc, in the pSP64 Poly(A) vector. The Rluc coding sequence was excised by digestion of the pRL-null vector (Promega) with NheI and XbaI, and was cloned into the pSP64 Poly(A) vector at its XbaI site.
In Vitro Transcription. In vitro transcription of EcoRI-linearized plasmid DNAs by SP6 polymerase was carried out by using the AmpliCap SP6 high-yield message maker kit (Epicentre Technologies, Madison, WI) in the presence of the cap analog m7G[5′]ppp[5′]GTP (Epicentre Technologies). After in vitro transcription, template DNA was digested with RNase-free DNase I, and the RNA was purified by using an RNeasy minikit (Qiagen, Valencia, CA), and was poly(A)-selected by using a GenElute mRNA miniprep kit (Sigma-Aldrich). The capped RNA transcribed from pMI21(WT) will be referred to as
Ex1(WT):Luc RNA, with the allele names used to indicate the mto1 mutant versions, and that which is transcribed from pMI27 will be referred to as Rluc RNA.
In Vitro Translation in Wheat Germ Extract. The standard reaction mixture contained 25 μl of wheat germ extract (Promega), 4 μl of 1 mM amino acid mixture lacking methionine (Promega), 1 μl of 5 μM l-methionine, 50 units of RNasin (Promega), 100 fmol of Ex1(WT):Luc RNA or one of its mto1 mutant derivatives, and 50 fmol of Rluc RNA, for a total volume of 50 μl. All reactions were carried out at 25°C for 120 min. LUC and RLUC activities were assayed by using a dual-luciferase reporter assay kit (Promega) and a Lumat LC9507 luminometer (Berthold, Nashua, NH). RNA extraction and Northern analysis were carried out as described (10) after poly(A) selection. The LUC probe was a 1.7-kb HindIII-SacI fragment of pMI21(WT). The 5′ probe that covers nucleotides 10-213 relative to the first ATG of CGS1 was prepared by PCR amplification of pMI21(WT) DNA, using the primers 5′-TCATCATTCCAGTGCCCTACCATCTTC-3′ and 5′-CTGACGGACGAAATTAGGAGGAAATC-3′. The 3′ probe that covers nucleotides 1448-1650 relative to the first ATG of luc+ was prepared by PCR amplification of pMI21(WT) DNA, using the primers 5′-GCCGTTGTTGTTTTGGAGCAC-3′ and 5′-ACGGCGATCTTTCCGCCCTTC-3′. The Rluc gene probe was a 1.0-kb NaeI-XbaI fragment of pMI27. 32P-labeled probes were prepared by using a Megaprime DNA labeling system (Amersham Pharmacia Biosciences). Hybridization signals were detected by using a BAS 1000 bioimage analyzer (Fuji).
Primer Extension Studies. Poly(A) RNA from an in vitro translation reaction lacking Rluc RNA was denatured for 5 min at 67°C, and was immediately placed on ice. The primer extension reaction was carried out by using Thermoscript RNase H- reverse transcriptase (Invitrogen), and 5′ end-labeled primer TO4 (5′-TGAACAGTGAGGCTCCCAT-3′, 1 × 106 cpm) for 60 min at 50°C. The reaction mixture was formulated as recommended by the supplier. After the phenol-chloroform extraction, the products were separated on a 6% polyacrylamide/7 M urea gel, and the signals were detected by using a bioimage analyzer.
Results
AdoMet Down-Regulates CGS1 Expression in Vivo. We have reported (8) that CGS1 expression is feedback-regulated at the level of mRNA stability in response to methionine application, and that the exon 1 coding region of CGS1 is necessary and sufficient for the posttranscriptional regulation in vivo (8, 10). In these experiments, however, the applied methionine may have affected CGS1 expression after it was metabolized. Because AdoMet is the immediate metabolite of methionine (Fig. 1), we tested the effects of AdoMet on CGS1 exon 1-mediated down-regulation by using a transient expression system.
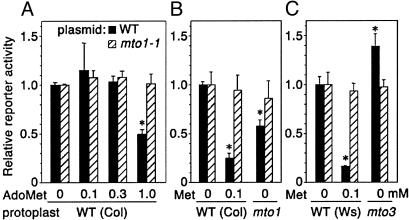
The plasmids pMI4(WT) and pMI4(mto1-1) carry the WT and mto1-1 mutant exon 1 coding sequence of CGS1, respectively, joined in-frame to the Escherichia coli β-glucuronidase (GUS) reporter gene, and placed under the control of a cauliflower mosaic virus 35S RNA promoter (8). The plasmid DNA was introduced into WT A. thaliana protoplasts by electroporation, and GUS reporter activity was analyzed relative to the LUC activity from a cotransfected control plasmid lacking the CGS1 exon 1. For pMI4(WT), addition of 1 mM AdoMet resulted in reduced reporter activity (Fig. 2A). AdoMet application did not appreciably affect the reporter activity in tests containing pMI4(mto1-1) (Fig. 2 A). These results suggested that, rather than methionine itself, AdoMet or one of its metabolites acts as the effector of CGS1 regulation.
Fig. 2.
Effects of AdoMet application and mto mutations in transient expression systems. Protoplasts were electroporated with pMI4(WT) (filled bars) or pMI4(mto1-1) (hatched bars) plasmid DNA, and were incubated for 48 h. GUS activity was normalized with LUC activity from the cotransfected 221-LUC+ plasmid, which does not carry the CGS1 exon 1. Reporter activity relative to that of untreated WT protoplasts was calculated. Averages ± SD of triplicate experiments are shown. *, values that were significantly different from those of untreated WT protoplasts (P < 0.05 by t test). (A) Effect of AdoMet treatment of Col-0 protoplasts. The electroporated protoplasts were treated with 0.1-1 mM AdoMet as indicated. (B) Effect of mto1-1 mutation. DNAs were electroporated into protoplasts of the mto1-1 mutant (mto1) and its parental WT Col-0. In the case of WT protoplasts, the effect of 0.1 mM methionine treatment was also analyzed. (C) Treatment was as in B, except that protoplasts from the mto3-1 mutant (mto3) and its parental WT Ws were used.
In another experiment designed to test the effect of AdoMet in vivo, we used protoplasts from the mto1-1 and mto3-1 mutants. The mto1-1 mutant has a defect in the negative feedback regulation of CGS1 (8), whereas the mto3-1 mutant has a defect in an AdoMet synthetase (13). Both mutants overaccumulate methionine, but their accumulation of AdoMet differs. Compared with the WT, the mto1-1 mutant has three times the level of endogenous AdoMet (15), whereas mto3-1 has ≈70% (13). The reporter activity from pMI4(WT) transfected into mto1-1 mutant protoplasts was significantly lower than that in protoplasts from its WT strain Col-0 (Fig. 2B). In contrast, in mto3-1 mutant protoplasts, the reporter activity was significantly higher than in protoplasts from its WT strain Ws (Fig. 2C). In both cases, reporter activity was not appreciably affected when pMI4(mto1-1) was used (Fig. 2 B and C). The reporter activity of pMI4(WT) responded to methionine application in both Col-0 and Ws WT protoplasts, indicating that the inability of the mto3-1 mutant to down-regulate the reporter activity is not due to the difference in ecotype. These data support the idea that, rather than methionine, AdoMet or one of its metabolites is the effector.
Establishment of the Wheat Germ in Vitro Translation System as the Experimental System. During in vivo tests of the effects of exogenously applied compounds, the efficiency of metabolite uptake into the cells can complicate interpretation of the results. We therefore exploited the in vitro translation system by using wheat germ extract to determine the actual effector of the regulation mediated by CGS1 exon 1.
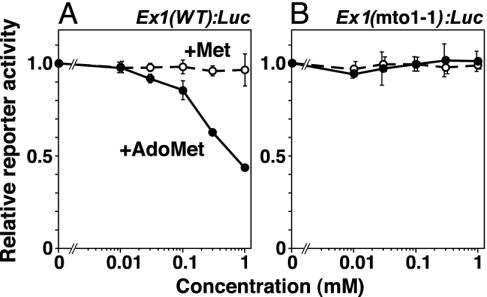
The exon 1 coding sequence of CGS1 was joined in-frame to a LUC reporter gene, transcribed in vitro in the presence of the cap analog, and this Ex1(WT):Luc RNA, or its mto1-1 mutant derivative Ex1(mto1-1):Luc, was translated in wheat germ extract. Capped Rluc RNA lacking CGS1 exon 1 was simultaneously translated, and was used as an internal control. As shown in Fig. 3A, addition of up to 1 mM methionine to the in vitro translation system did not affect the reporter activity generated by Ex1(WT):Luc RNA, whereas addition of AdoMet at concentrations >0.1 mM down-regulated the reporter activity. Neither methionine nor AdoMet affected the reporter activity when Ex1(mto1-1):Luc RNA was used (Fig. 3B). These results suggest that AdoMet or one of its metabolites acts as the effector of this down-regulation in vitro.
Fig. 3.
Effects of methionine and AdoMet on reporter activity in the in vitro translation system. Ex1(WT):Luc RNA (A) or Ex1(mto1-1):Luc RNA (B) were translated in wheat germ extract for 120 min in the presence of various concentrations of AdoMet (•) or methionine (○) as indicated. LUC activity was normalized with the control RLUC activity of the cotranslated Rluc RNA, which does not carry the CGS1 exon 1. Reporter activity relative to that in samples not treated with methionine or AdoMet was calculated, and averages ± SD of triplicate experiments are shown.
The mto1 alleles mto1-2, mto1-3, and mto1-4 (9) were also tested (Table 1), revealing that the reporter activity is unchanged or only weakly responds to AdoMet, if CGS1 exon 1 harbors any one of these mutations. The mto1-4 mutation, which has been shown to be a leaky allele in vivo (9), also showed significantly lower reporter activity than other mto1 alleles in vitro.
Table 1. Response of mto 1 alleles to AdoMet in the in vitro translation system.
| RNA | Relative reporter activity (no added AdoMet = 1) |
|---|---|
| Ex1(WT):Luc | 0.49 ± 0.03a |
| Ex1 (mto1-1):Luc | 1.02 ± 0.03c |
| Ex1 (mto1-2):Luc | 0.94 ± 0.04c |
| Ex1 (mto1-3):Luc | 0.96 ± 0.03c |
| Ex1 (mto1-4):Luc | 0.83 ± 0.03b |
Ex1:Luc RNA carrying the WT or mto 1 mutant CGS1 exon 1 was translated in wheat germ extract for 120 min in the presence of 1 mM AdoMet. LUC activity was normalized with the RLUC activity from the cotranslated Rluc RNA not carrying the CGS1 exon 1 sequence, and reporter activity relative to samples with no added AdoMet was calculated. Averages ± SD of triplicate experiments are shown. Superscript letters indicate that the values are significantly different from each other (p < 0.05 by t test).
Identification of AdoMet as the Effector of CGS1 Regulation. Because it is possible that some of the enzymes responsible for AdoMet metabolism are present in the wheat germ extract system, a number of compounds metabolically related to AdoMet were tested. When AdoMet is used in methyl transfer reactions, it is converted to AdoHcy, and AdoHcy is then recycled back to methionine through l-homocysteine (Fig. 1). Neither AdoHcy nor l-homocysteine affected the reporter activity (Fig. 4A), indicating that the metabolites of this recycling pathway are not involved in the regulation.
Fig. 4.
Effects of metabolites and analogs of AdoMet on reporter activity in vitro. (A) Effects of AdoMet metabolites. Ex1(WT):Luc RNA (filled bars) or Ex1(mto1-1):Luc RNA (hatched bars) were translated in wheat germ extract for 120 min in the presence of 1 mM of various metabolites of AdoMet. LUC activity was normalized with the control RLUC activity from the cotranslated Rluc RNA, and reporter activity relative to samples without added metabolites was calculated. Averages ± SD of triplicate experiments are shown. *, the value for Ex1(WT):Luc RNA is significantly different from that for Ex1(mto1-1):Luc RNA (P < 0.05 by t test). Abbreviations are the same as in Fig. 1. (B) Effect of AdoHcy on AdoMet-mediated down-regulation. In vitro translation was carried out in the presence of 0.3 mM AdoMet plus various concentrations of AdoHcy. (C) Effect of decarboxylated AdoMet (dcAdoMet). In vitro translation was carried out in the presence of 0.3 mM dcAdoMet. (D) Effects of analogs of AdoMet. In vitro translation was carried out in the presence of analogs at 1 mM. AdoEtn, S-adenosyl-l-ethionine; AdoPpn, S-adenosyl-l-propionine; and AdoOrn, adenosyl-l-ornithine. (E) Schematic representation of compounds used in this study that are metabolically and/or structurally related to AdoMet. The 5′-deoxyadenosine moiety (Ado) and the sulfonium sulfur (S+) are shaded. Abbreviations are the same as in A-D.
Both low-molecular-weight compounds and macromolecules act as acceptors of AdoMet-mediated methyl transfer reactions. It is possible that a product of these methyl transfer reactions acts as the effector. AdoHcy has been shown to be a potent competitive inhibitor of a number of methyltransferases that use AdoMet as the methyl donor (16). We therefore tested whether AdoHcy competes with AdoMet in the down-regulation of reporter activity. AdoMet was added to 0.3 mM, and the concentration of AdoHcy was varied between 0.3 and 2 mM. As shown in Fig. 4B, AdoHcy exhibited no competitive effect on the AdoMet-mediated down-regulation of reporter activity, even when AdoHcy was added at ≈6-fold the concentration of AdoMet. Because inhibitor constants for AdoHcy are usually about equal to, or lower than, the Km values for AdoMet (16), it is unlikely that any of the methylated products, such as a methylated protein factor, acts as the effector of this regulation.
AdoMet is used in the biosynthesis of polyamines after it is decarboxylated by AdoMet decarboxylase. In this reaction, the methylthio moiety is recycled back to methionine through 5′-deoxy-5′-(methylthio)adenosine (Fig. 1). Because 1 mM dcAdoMet was inhibitory to the in vitro translation reaction (data not shown), the effect of this compound was tested at 0.3 mM. As shown in Fig. 4C, dcAdoMet down-regulated reporter activity, but the effect was ≈30% that of AdoMet. Addition of spermidine and spermine did not appreciably affect relative reporter activities (data not shown). 5′-Deoxy-5′-(methylthio)adenosine also did not affect the reporter activity (Fig. 4A).
Other metabolites that diverge from AdoMet include 1-aminocyclopropane-1-carboxylic acid (17) and SMM (ref. 18 and Fig. 1). Neither compound appreciably affected reporter activity (Fig. 4A). These results support the idea that AdoMet, and not a metabolite of AdoMet, is the effector.
AdoEtn, an Analog of AdoMet, also Down-Regulates Reporter Activity.
To obtain insight into the structural features of the effector, the effects of AdoMet analogs were tested. AdoEtn has a sulfur-linked ethyl group in place of the methyl group of AdoMet (Fig. 4E). As shown in Fig. 4D, AdoEtn down-regulated reporter activity to a similar extent as that of AdoMet. We also tested AdoPpn and AdoOrn, which have a sulfur-linked propyl group and a carbon-linked amino group, respectively, in place of the sulfur-linked methyl group of AdoMet (Fig. 4E). Neither compound affected reporter activity (Fig. 4D).
Identification of a 5′-Truncated RNA Species After in Vitro Translation.
We have reported (8) that when methionine is applied to WT A. thaliana calli, accumulation of CGS1 mRNA is reduced, and a related short RNA species that is truncated at its 5′ end appears. We have suggested that this 5′-truncated RNA is a degradation intermediate of CGS1 mRNA. A similar RNA species was also detected in methionine-treated transgenic A. thaliana carrying essentially the same construct as pMI4(WT) (10), which indicates that the exon 1 coding sequence of CGS1 alone is sufficient to produce the 5′-truncated RNA.
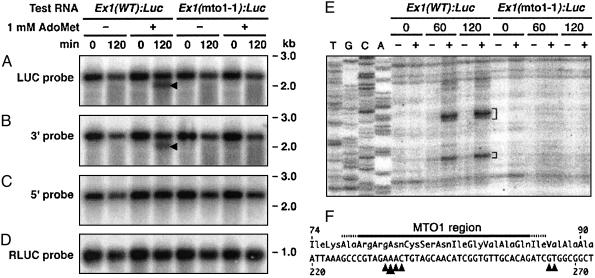
We tested whether the 5′-truncated RNA is produced in our in vitro system. RNA was analyzed after translation in wheat germ extract in the presence or absence of 1 mM AdoMet. Northern hybridization was carried out after poly(A) selection. As shown in Fig. 5A, the luc+ probe detected an RNA species ≈300 bases shorter than the full-length RNA when Ex1(WT):Luc RNA was translated in the presence of AdoMet. This short RNA band was also detected by the probe covering the 3′ region of the luc+ reporter gene (Fig. 5B), but not with the probe covering the 5′ region of the CGS1 exon 1 coding sequence (Fig. 5C), indicating that it is truncated at its 5′ end. The amounts of the full-length RNA remaining after the in vitro reaction were not significantly different between the presence and absence of AdoMet. Ex1(mto1-1):Luc RNA (Fig. 5A) and cotranslated Rluc RNA (Fig. 5D) did not produce a truncated RNA species. Northern hybridization of total RNA produced similar results to those of poly(A)-selected RNA (data not shown).
Fig. 5.
Analysis of RNA after in vitro translation. (A-D) Ex1(WT):Luc RNA or Ex1(mto1-1):Luc RNA were translated in wheat germ extract in the presence (+) or absence (-) of 1 mM AdoMet. Poly(A) RNA was extracted immediately after (0 min) or 120 min after the start of in vitro translation, and was fractionated in 1% agarose gels containing formaldehyde. Northern hybridizations were carried out by using a 32P-labeled luc+ gene probe (A), a probe covering the 3′ region of the luc+ coding sequence (B), or a probe covering the 5′ region of the CGS1 exon 1 coding sequence (C). Northern hybridization was also carried out by using an RLUC probe to detect the cotranslated Rluc RNA (D). Positions of size markers are indicated to the right. Arrowheads in A and B indicate the positions of the 5′-truncated RNA. Representative results of triplicate experiments are shown. (E) Detection of the 5′ ends of truncated RNA species by primer extension. Poly(A) RNA was prepared as in A, and primer extension reactions were carried out by using TO4 primer. Lanes T, G, C, and A are the sequence ladder resulting from the same primer. Brackets indicate the 5′ ends of the truncated RNA. (F) The 5′ ends were mapped on the nucleotide (numbered from the first ATG) and the amino acid sequence. Arrowheads indicate positions of the 5′ ends with the large arrowhead pointing to the most prominent 5′ end. The MTO1 region (9) is marked above the amino acid sequence. Similar results were obtained in quadruplicate experiments.
To map the 5′ end of the truncated RNA, we carried out primer extension studies. When Ex1(WT):Luc RNA was translated in the presence of AdoMet, the 5′ end of the truncated RNA resolved into two regions 234-237 and 265-266 bases from the first ATG, with position 235 most prominent (Fig. 5 E and F). These 5′ ends were located within or very close to the MTO1 region (9). Primer extension studies using poly(A) RNA from methionine-treated WT calli showed that at least the most prominent 5′ end of the truncated RNA in vitro was also detected in vivo, suggesting that the truncated RNA produced in these two systems are essentially the same (see Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site,www.pnas.org.).
Discussion
Identification of AdoMet as the Natural Effector of the Posttranscriptional Regulation Mediated by CGS1 Exon 1. Transient expression experiments suggested that, rather than methionine, AdoMet or one of its metabolites is the effector of CGS1 regulation in vivo. The results corroborate previous findings (13, 19) with the mto3 mutants and cosuppression lines of the AdoMet synthetase gene in A. thaliana (20). In these systems, the steady-state level of CGS1 mRNA was the same as, or greater than, that in WT, despite the fact that these plants overaccumulate methionine. In AdoMet feeding experiments, 1 mM AdoMet was less effective in down-regulating expression than was 0.1 mM methionine (compare Fig. 2 A and B). This may be due, however, to AdoMet being taken up less efficiently into cells or being more labile in the experimental medium than in methionine. Although AdoMet transporters have been characterized from Saccharomyces cerevisiae (21), homologous transporters do not appear to exist in the A. thaliana genome.
In this article, a wheat germ in vitro translation system was used to identify the effector of the regulation mediated by CGS1 exon 1. A number of different metabolites related to AdoMet were tested, and AdoMet was found to be the most effective in blocking the reporter activity. dcAdoMet was also effective, although weakly. However, because the concentration of dcAdoMet in vivo has been reported to be ≈2% of that of AdoMet (22), it is unlikely that this metabolite exerts a greater effect than AdoMet in vivo. Our results establish that AdoMet is the natural effector of the posttranscriptional regulation mediated by the exon 1 of CGS1, and suggest that in the in vivo experiments (8-10), the down-regulation was induced after methionine was converted to AdoMet by AdoMet synthetase.
AdoEtn, an analog of AdoMet, was shown to be as effective as AdoMet in down-regulating reporter activity. AdoMet has an α-aminobutyric acid moiety, a 5′-deoxyadenosine moiety, and a methyl group linked to the sulfonium sulfur (Fig. 4E). AdoEtn differs from AdoMet by containing a sulfur-linked ethyl group (Fig. 4E). Therefore, the regulation mechanism tolerates either a methyl or an ethyl group. However, a propyl group is not tolerated at this position, because AdoPpn (Fig. 4E) was not effective. AdoHcy lacks the sulfur-linked alkyl group (Fig. 4E) and was also ineffective, suggesting that the sulfonium group is a necessary component of effector. The fact that AdoOrn (Fig. 4E) had no effect supports this notion. Although SMM is a sulfonium compound (Fig. 4E), it had no effect on reporter activity, demonstrating the importance of the 5′-deoxyadenosine moiety. The carboxylic acid group is necessary, but is less important than the sulfonium and 5′-deoxyadenosine moieties, as dcAdoMet was a weak effector.
It was recently reported (23) that in a wheat germ in vitro-coupled transcription/translation system, methionine down-regulated the accumulation of transcripts from the full-length CGS1 cDNA, although production of the 5′-truncated RNA species was not discussed. Regions of CGS1 other than exon 1 might carry additional regulatory functions, however, the reported response should represent a minor component in vivo as it does not corroborate the CGS1 mRNA levels in the methionine overaccumulation lines discussed above (13, 19, 20). In our in vitro system, addition of 1 mM methionine had no effect on the RNA profile (data not shown).
Use of the Wheat Germ in Vitro Translation System to Study the Mechanism of CGS1 Regulation. Two lines of evidence suggest that the reactions that we observed in the wheat germ in vitro translation system reflect the posttranscriptional regulation of CGS1 in vivo. First, the response to AdoMet was strongly reduced when the CGS1 exon 1 carried one of the mto1 mutant alleles. In addition, Ex1(mto1-4):Luc RNA showed significantly lower reporter activity than those RNAs that carry the other mto1 alleles (Table 1). The mto1-4 mutant has been shown to be a leaky allele in vivo (9). This finding means that the phenotypic differences identified genetically in vivo, and the level of response in vitro, are interrelated. Second, analysis of Ex1(WT):Luc RNA after in vitro translation in the presence of AdoMet, revealed the presence of a 5′-truncated RNA species, which we have suggested to be a degradation intermediate (8, 10). In addition, the 5′ end of the truncated RNA species produced in vivo and in vitro were essentially the same. These data suggest that the same RNA degradation event that occurs in vivo also occurs in vitro.
In contrast, the amounts of full-length Ex1(WT):Luc RNA that remained after in vitro translation in the presence and absence of AdoMet did not match the difference in reporter activities. This result suggests that the down-regulation of reporter activity is not a direct consequence of mRNA degradation. It is possible that the down-regulation of reporter activity that we observed in vitro could have mainly reflected a translational repression. In some eukaryotic genes, translational regulation by a peptide encoded by an upstream ORF is known (24). Although the regulation is not linked to mRNA degradation, the mechanism shares similarity to the CGS1 system in that a short stretch of amino acid sequence acts in cis. Unlike the CGS1 system, the regulation by an upstream ORF is usually exerted at the termination of translation. However, in the case of the arginine attenuator peptide in arg-2 of Neurospora crassa and its S. cerevisiae counterpart, which encode an enzyme for arginine biosynthesis, the stop codon is dispensable, and in vitro translation systems reproduced the response (25, 26). Among the genes whose expression is regulated at the level of mRNA stability, there are cases where the stability determinants are located in their coding sequences (27-31). One such gene is c-myc, for which in vitro studies have been applied. Studies using rabbit reticulocyte lysate from which tRNA was depleted by nuclease treatment have shown that c-myc translation pauses at one of the rare codons, and suggested that this triggers c-myc mRNA degradation (28). However, it is unlikely that rare codons are involved in the regulation of CGS1, because among the 40 in vivo- and in vitro-generated mutants that we have obtained with lesions within the CGS1 exon 1 (8, 9), there was no correlation between codon usage and phenotype (data not shown), and the in vitro translation system used in this article contained ample amounts of added tRNA.
The data presented in this article suggest that the basic reactions of CGS1 regulation occur in the wheat germ in vitro translation system. This system allows us to examine the molecular mechanisms of posttranscriptional regulation of CGS1, by providing a biochemical strategy to complement the genetic advantages of the A. thaliana system, which has proven amenable in study of the control of mRNA stability (8, 32).
Supplementary Material
Acknowledgments
We thank M. C. Tarczynski and A. Shirahata for materials; P. J. Green, T. Leustek, M. C. Tarczynski, M. Ishikawa, and D. B. Goto for critical readings; I. Lambein for careful reading of the manuscript; S. Kudo and S. Moriyama for technical assistance; and K. Fujiwara for general assistance. We used the Radioisotope Laboratory of the Graduate School of Agriculture, Hokkaido University. This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [12138201 and 13440233 (to S.N.) and 14035208 (to H.O.)]; Japan Society for the Promotion of Science Grant RFTF97L00601 (to S.N.); and Japan Society for the Promotion of Science Fellows Grant 06660 (to Y.C.). Y.C. and M.Y. are supported by the Japan Society for the Promotion of Science.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CGS, cystathionine γ-synthase; AdoMet, S-adenosyl-l-methionine; AdoHcy, S-adenosyl-l-homocysteine; dcAdoMet, decarboxylated AdoMet; AdoEtn, S-adenosyl-l-ethionine; SMM, S-methyl-l-methionine; AdoPpn, S-adenosyl-l-propionine; AdoOrn, adenosyl-l-ornithine; Col-0, Columbia WT; Ws, Wassilewskija WT; LUC, Photinus pyralis luciferase; RLUC, Renilla reniformis luciferase; GUS, β-glucuronidase.
References
- 1.Matthews, B. F. (1999) in Plant Amino Acids: Biochemistry and Biotechnology, ed. Singh, B. K. (Dekker, New York), pp. 205-225.
- 2.Thompson, G. A., Datko, A. H., Mudd, S. H. & Giovanelli, J. (1982) Plant Physiol. 69, 1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giovanelli, J., Mudd, S. H. & Datko, A. H. (1989) Plant Physiol. 90, 1584-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gakière, B., Ravanel, S., Droux, M., Douce, R. & Job, D. (2000) C. R. Acad. Sci. Ser. III 323, 841-851. [DOI] [PubMed] [Google Scholar]
- 5.Kim, J. & Leustek, T. (2000) Plant Sci. (Shannon, Irel.) 151, 9-18. [Google Scholar]
- 6.Kim, J. & Leustek, T. (1996) Plant Mol. Biol. 32, 1117-1124. [DOI] [PubMed] [Google Scholar]
- 7.Kim, J., Chiba, Y., Yamamoto, A., Naito, S. & Leustek, T. (1999) Plant Physiol. 120, 635-636. [Google Scholar]
- 8.Chiba, Y., Ishikawa, M., Kijima, F., Tyson, R. H., Kim, J., Yamamoto, A., Nambara, E., Leustek, T., Wallsgrove, R. M. & Naito, S. (1999) Science 286, 1371-1374. [DOI] [PubMed] [Google Scholar]
- 9.Ominato, K., Akita, H., Suzuki, A., Kijima, F., Yoshino, T., Yoshino, M., Chiba, Y., Onouchi, H. & Naito, S. (2002) J. Biol. Chem. 277, 36380-36386. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki, A., Shirata, Y., Ishida, H., Chiba, Y., Onouchi, H. & Naito, S. (2001) Plant Cell Physiol. 42, 1174-1180. [DOI] [PubMed] [Google Scholar]
- 11.Giovanelli, J., Mudd, S. H. & Datko, A. (1985) Plant Physiol. 77, 450-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samejima, K., Nakazawa, Y. & Matsunaga, I. (1978) Chem. Pharm. Bull. 26, 1480-1485. [Google Scholar]
- 13.Shen, B., Li, C. & Tarczynski, M. C. (2002) Plant J. 29, 371-380. [DOI] [PubMed] [Google Scholar]
- 14.Matsuo, N., Minami, M., Maeda, T. & Hiratsuka, K. (2001) Plant Biotechnol. 18, 71-75. [Google Scholar]
- 15.Bartlem, D., Lambein, I., Okamoto, T., Itaya, A., Uda, Y., Kijima, F., Tamaki, Y., Nambara, E. & Naito, S. (2000) Plant Physiol. 123, 101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueland, P. M. (1982) Pharmacol. Rev. 34, 223-253. [PubMed] [Google Scholar]
- 17.Kende, H. (1993) Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 283-307. [Google Scholar]
- 18.Ranocha, P., McNeil, S. D., Ziemark, M. J., Li, C., Tarczynski, M. C. & Hanson, A. D. (2001) Plant J. 25, 575-584. [DOI] [PubMed] [Google Scholar]
- 19.Goto, D. B., Ogi, M., Kijima, F., Kumagai, T., van Werven, F., Onouchi, H. & Naito, S. (2002) Genes Genet. Syst. 77, 89-95. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J., Lee, M., Chalam, R., Martin, M. N., Leustek, T. & Boerjan, W. (2002) Plant Physiol. 128, 95-107. [PMC free article] [PubMed] [Google Scholar]
- 21.Rouillon, A., Surdin-Kerjan, Y. & Thomas, D. (1999) J. Biol. Chem. 274, 28096-28105. [DOI] [PubMed] [Google Scholar]
- 22.Hanfrey, C., Franceschetti, M., Mayer, M. J., Illingworth, C. & Michael, A. J. (2002) J. Biol. Chem. 277, 44131-44139. [DOI] [PubMed] [Google Scholar]
- 23.Kreft, O., Hoefgen, R. & Hesse, H. (2003) Plant Physiol. 131, 1843-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris, D. R. & Geballe, A. P. (2000) Mol. Cell. Biol. 20, 8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, Z., Fang, P. & Sachs, M. S. (1998) Mol. Cell. Biol. 18, 7528-7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, Z., Gaba, A. & Sachs, M. S. (1999) J. Biol. Chem. 274, 37565-37574. [DOI] [PubMed] [Google Scholar]
- 27.Gay, D. A., Sisodia, S. S. & Cleveland, D. W. (1989) Proc. Natl. Acad. Sci. USA 86, 5763-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemm, I. & Ross, J. (2002) Mol. Cell. Biol. 22, 3959-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caponigro, G., Muhlrad, D. & Parker, R. (1993) Mol. Cell. Biol. 13, 5141-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellington, C. L., Greenberg, M. E. & Belasco J. G. (1993) Mol. Cell. Biol. 13, 5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tierney, M. J. & Medcalf, R. L. (2001) J. Biol. Chem. 276, 13675-13684. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, M. A., Pérez-Amador, M. A., Lidder, P. & Green, P. J. (2000) Proc. Natl. Acad. Sci. USA 97, 13991-13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.