Abstract
Mutations in SALL1, the human homolog of the Drosophila spalt gene, result in Townes-Brocks' syndrome, which is characterized by hand/foot, anogenital, renal, and ear anomalies, including sensorineural deafness. spalt genes encode zinc finger transcription factors that are found in animals as diverse as worms, insects, and vertebrates. Here, we examine the effect of losing both of the spalt genes, spalt and spalt-related, in the fruit fly Drosophila melanogaster, and report defects similar to those in humans with Townes-Brocks' syndrome. Loss of both spalt and spalt-related function in flies yields morphological defects in the testes, genitalia, and the antenna. Furthermore, spalt/spalt-related mutant antennae show severe reductions in Johnston's organ, the major auditory organ in Drosophila. Electrophysiological analyses confirm that spalt/spalt-related mutant flies are deaf. These commonalities suggest that there is functional conservation for spalt genes between vertebrates and insects.
Keywords: Distal-less, homothorax, atonal, chordotonal organ, split hand/split foot malformation
Townes-Brocks' syndrome (TBS) is an autosomal dominant disorder that maps to the SALL1 locus at 16q12.1 (1). SALL1 is one of three known vertebrate homologs of Drosophila spalt (sal) (2, 3). The mutations in SALL1 that result in TBS typically lead to a premature stop codon after the first zinc finger (1, 4-7). Although the clinical manifestations of TBS are variable, ≈85% of patients with TBS exhibit hand/foot, anogenital, renal, and ear anomalies, including sensorineural deafness (8-13). TBS is estimated to occur in 1:250,000 liveborn (14), but may be misdiagnosed because its defects overlap with those of other genetic diseases (7, 11, 15, 16). Consistent with TBS phenotypes, human SALL1 expression has been detected in the brain, heart, ureteric bud, and developing tubules of the kidney, testes, and ovary (2, 17). In mice, sal genes are expressed in brain, testes, ovaries, kidneys, limb buds, heart, inner ear, and the lens of the eye (18, 19).
spalt genes have been identified in a variety of vertebrates and invertebrates (2, 3, 18, 20-28). In Drosophila, there are two adjacent spalt genes, spalt (sal) and spalt-related (salr). These genes use shared regulatory regions and are expressed in similar patterns throughout development. Early studies of sal single mutants indicated that sal is required for the development of the embryonic terminalia, tracheal system, and wings (20, 29-32). Mutations in sal result in partial transformations of the posterior head and anterior tail toward trunk identity (20). sal functions during tracheal development both to restrict the positions of the initial placodes and in directed migration of dorsal trunk cells (30). At larval stages, Dpp-activated sal expression in the wing pouch is activated in response to graded levels of Dpp to properly position wing pattern elements (29, 31, 33).
More recent studies of Drosophila lacking the function of both sal and salr indicate that there is significant functional redundancy between them and have revealed additional roles for these genes in the development of the larval peripheral nervous system and adult eye. During embryogenesis, sal and salr function to prevent oenocytes (cuticle secreting epidermal cells) from adopting scolopodial (peripheral neural) fates (34, 35). Later in development, lack of sal and salr activity in the developing eye leads to failure of the R7 and R8 photoreceptor differentiation (36).
Although sal and salr are expressed in the developing antenna (37), which houses the fly's auditory organ, and in the genitalia (38), there were no reported functions for them in these tissues. However, because TBS patients exhibit auditory and genital phenotypes, we asked whether sal and salr were necessary for the normal development of the antenna and genitalia in Drosophila. In addition, because TBS patients exhibit testicular defects, we also analyzed sal and salr function in Drosophila testes. We report that loss of both sal and salr leads to defects in antennal and genital cuticles and in the adult testes. Analyses of the sal and salr double null antennae indicate that these genes are required for the proper formation of the antennal auditory organ. Electrophysiological measurements of these sal and salr double null antennae confirm that sal activity is required for development of a functional auditory organ in Drosophila. This contrasts with the role of sal during differentiation of larval chordotonal organs where it plays an inhibitory role (34, 35). Together, our findings suggest that spalt genes may have some conserved roles in insects and vertebrates.
Materials and Methods
Fly Strains and Genetic Manipulations. The following fly strains were used: (i) salFCK-25/T(2:3) SM6a; TM6B (39); (ii) w; Df(2L)32FP-5 FRT40A/T(2:3) SM6a; TM6B (39); (iii) y hs-FLPase; P[y+] FRT40A;(iv) y w ey-FLPase GMR-lacZ; FRT40A; (v) sal-GAL4 (40); and (vi) UAS-GFP-nls (Bruce Edgar).
sal/salr-null clones were generated by using the FLP/FRT system (41). Animals of the genotype y hs-FLPase; P[y+] FRT40A/Df(2L)32FP-5 FRT40A were heat-shocked at 37°C for 1 h at 48-72 h after egg laying and examined either in mid-to late-third instar or as adults. sal/salr-null clones were detected by using a yellow (y) transgene on the wild-type FRT40A chromosome. y function is required to produce dark pigment in bristles and cuticle. Thus sal/salr null clones appear yellow in a dark background. sal/salr null clones also were generated by using ey-FLPase (42). The genotype of the larvae and adults examined was y w ey-FLPase GMR-lacZ; P[y+] FRT40A/Df(2L)32FP-5 FRT40A. In addition to sal/salr null clones, two additional genotypes lacking sal and salr in the antenna were examined: salFCK-25/salFCK-25 and salFCK-25/Df(2L)32FP-5 FRT40A. salFCK-25 flies have a rearrangement breakpoint upstream of salr that results in the loss of both sal and salr expression in the antenna (39). Df(2L)32FP-5 is an 85-kb deletion that removes the coding regions of both the sal and salr genes (39) but affects no other known or predicted genes. Because both salFCK-25 and Df(2L)32FP-5 FRT40A were balanced over T(2:3) SM6a; TM6B, sal mutants could be identified by lack of the Tubby phenotype.
Immunohistochemistry, Cuticle Preparations, Testes Phenotypic Analysis, and Histology. Antibody staining and immunohistochemistry was carried out as described (43). The antibodies used were: rat anti-Sal (39) and Cy5 conjugated goat anti-HRP (Molecular Probes). Secondary antibody coupled to Cy3 was obtained from Jackson ImmunoResearch Laboratories. Rhodamine-conjugated phalloidin was obtained from Molecular Probes. Because of limited availability of the Sal antibody and the inconsistency of antibody penetration in whole mount pupal tissue, sal-GAL4 was used in combination with UAS-GFP-nls to visualize sal expression in pupal antennae. Adult antenna and genitalia were dissected in 70% ethanol and mounted in Hoyer's medium as described (44). Ten males carrying Df(2L)32FP-5 clones were selected based on the presence of genital defects. Testes were dissected in PBS 8 days after hatching. Their siblings in which no clones could be induced (Ubi-GFP-nls FRT40A/CyO and Df(2L)32FP-5 FRT40A/CyO) were used as controls. Images of spalt mutant and normal testes were collected from the same well to control for camera settings. Paraffin sections and hematoxylin and eosin staining of adult Oregon R and salFCK-25/Df(2L)32FP-5 FRT40A were carried out as described by Downs et al. (45). Imaging was carried out on a Bio-Rad MRC1024 confocal microscope and a Zeiss Axioplan microscope equipped with an Axiocam.
Electrophysiology. Electrophysiology was performed as described (46). Briefly, auditory stimuli consisted of computer-generated pulse song as well as 160- and 250-Hz sine songs. The acoustic signals were delivered frontally to the fly's head through Tygon tubing from a loudspeaker. An electrolytically sharpened tungsten electrode was inserted between the first and second antennal segments to record extracellularly from the antennal nerve. A similar, reference electrode was inserted into the dorsal side of the head. The differential signal (DAM 50 differential amplifier; World Precision Instruments, Sarasota, FL) was amplified 1,000-fold and routed to an InstruNet 100B (GW Instruments) AD/DA converter connected to a Mac PowerBook 1400. We used superscope II software (GW Instruments) to generate the stimuli and to record the signals. Each trial consisted of the auditory stimulus presented 10 consecutive times. Amplitudes are an average of the 10 recordings.
Results
spalt and spalt-related Are Required for the Development of the Drosophila Auditory Organ. TBS patients exhibit profound defects in the inner, middle, and outer ear that result in both conductive and sensorineural deafness (8, 10, 11, 47). We report here that Drosophila sal mutants also exhibit complete deafness, which is most likely both conductive and sensorineural. The Drosophila auditory apparatus consists, in part, of a large chordotonal organ termed Johnston's organ (JO) housed within the second antennal segment (a2) (46, 48, 49).
The JO is composed of functional subunits termed “scolopidia” (reviewed in ref. 48). Each scolopidium consists of approximately five cells that are thought to be clonally derived from a single precursor. These cells include two or three monodendritic neurons, a scolopale cell, a cap cell, and a ligament cell. The basally located neuronal cell bodies project ciliated dendrites apically through the actin-rich scolopales of the scolopale cells. Scolopidia act as stretch receptors: they have both an apical and a basal cuticular attachment and respond to changes in their length. The apical and basal attachments are thought to be mediated by the cap and ligament cells, respectively. Mechanical deformation of the apical cuticle at the a2/a3 joint is thought to lead to a receptor potential in the ciliated dendrites. Such signals ultimately are transmitted as action potentials via the antennal nerve to the brain.
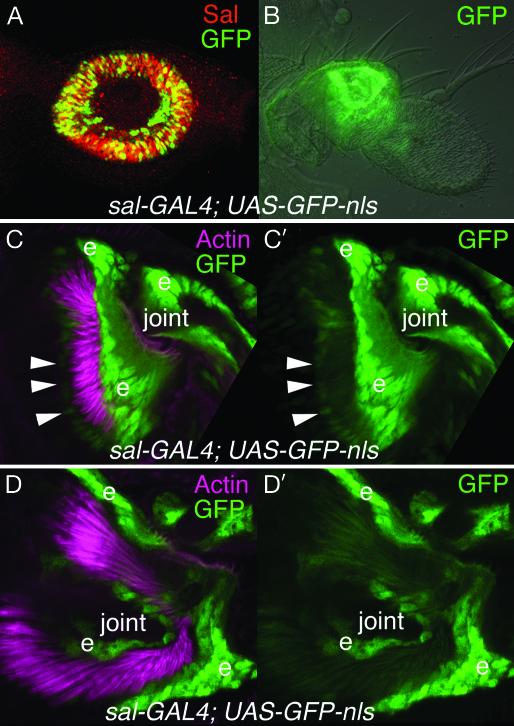
In late third-instar larvae, sal and salr expression is activated by the coexpression of Distal-less and homothorax in the antennal primordia in presumptive a2 (37-39) (Fig. 1A). At this stage, sal and salr are largely coexpressed with the chordotonal proneural gene atonal (ato) (50). ato is required for specification of scolopidial precursor cells, and ato-null flies lack the JO (46, 51). We have shown previously that ato and sal are independently regulated in the antenna (50). At pupal stages, sal expression is still detected strongly in a2 (Fig. 1B). However, the strong sal expression seems to correspond primarily with cuticle-secreting epidermal cells and cells adjacent to the scolopidial cells (Fig. 1 C, C′, D, and D′). It is likely that, by pupal stages, ato and sal expression are largely complementary instead of overlapping, suggesting that they may have different roles in constructing the JO.
Fig. 1.
sal expression in the developing second antennal segment. (A) Sal protein (red) and GFP (green) are coexpressed (yellow) in the presumptive second antennal segment (a2) in a third-instar larva of genotype sal-GAL4; UAS-GFP-nls. This indicates that the sal-GAL4 line approximately reproduces the sal expression pattern in the antenna. (B) In a late pupal antenna of the same genotype, GFP (green) is expressed in both epidermal cells and JO cells in a2. (C, C′, D, and D′) At earlier pupal stages, sal-GAL4 drives expression of nuclear GFP (green) strongly in epidermal cells (e) of distal a2 and proximal a3. The actin bundles of the scolopale cells have been stained with rhodamine-phalloidin (pink). The a2/a3 joint is labeled. C and C′ are lateral views of distal a2 and proximal a3. D and D′ are highe-magnification views looking distally toward the a2/a3 joint from proximal a2. GFP is weakly expressed in some of the scolopale cell nuclei (arrowheads).
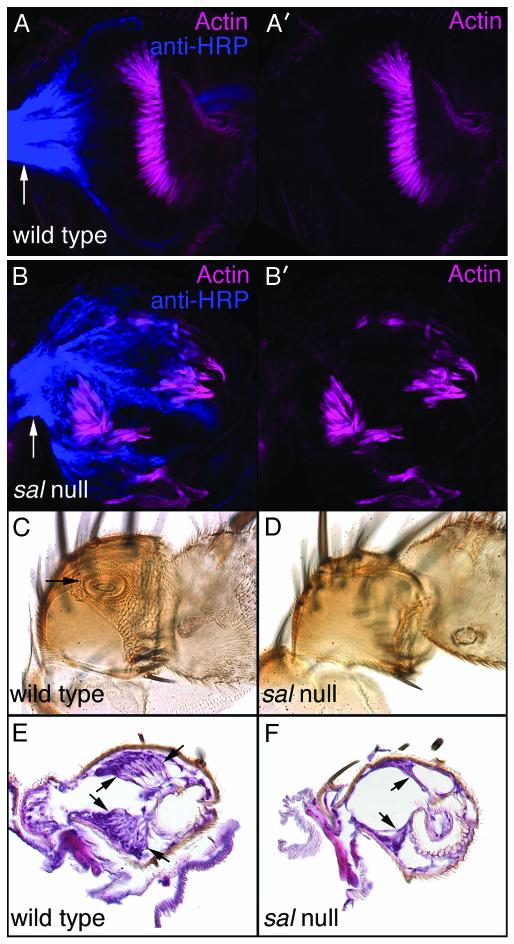
Consistent with expression in nonscolopidial cells, sal and salr mutant antennae do differentiate scolopidia (Fig. 2 A, A′, B, and B′). However, these scolopidia are malformed, reduced in number, and highly disorganized (Fig. 2 A, A′, B, and B′). They often are retracted either basally or apically as if they could not make appropriate cuticular contacts. By adult stages, most of these mutant scolopidia appear to have degenerated (see below). In a normal adult antenna, the a2/a3 joint, to which the JO is attached, is highly involuted and is seen as a circular outline via an optical section through the middle of a2 (Fig. 2C, arrow). This joint is not visible in the sal- and salr-null antennal cuticles (Fig. 2D). In addition, a2 and a3 are fused (Fig. 2D). As a consequence, a3 cannot rotate freely relative to a2. Because this rotation is necessary for conduction of acoustic vibrations absorbed by the arista into the JO (49), sal- and salr-null antennae have conductive hearing defects.
Fig. 2.
sal- and salr-null JOs develop abnormally and degenerate during pupal stages. (A and A′) Wild-type pupal antenna stained with anti-horseradish peroxidase (HRP; blue) and rhodamine-phalloidin (pink). Anti-HRP detects neuronal membranes. Phalloidin binds to actin, which is particularly prominent in the scolopales of the scolopale cells. Note the large anti-HRP-labeled axon bundle exiting a2 proximally (arrow). (B and B′) Similar staining of a salFCK-25/Df(2L)32FP-5 FRT40A antenna. Scolopidia are forming, albeit at reduced numbers, and are highly disorganized. The anti-HRP-labeled axon bundle exiting a2 proximally is indicated with an arrow. (C) Internal optical section of part of a wild-type adult antenna. The arrow indicates the a2/a3 joint to which the JO attaches. (D) Similar view of a salFCK-25/Df(2L)32FP-5 FRT40A antenna in which the specialized a2/a3 joint is lacking. (E) Histological section of part of a wild-type antenna. The arrows indicate clusters of scolopidia that comprise the JO. (F) Similar histological section of a salFCK-25/Df(2L)32FP-5 FRT40A antenna. Arrows indicate the few remaining scolopidia. In some antennae of this genotype, no scolopidia are observed.
To assess sensorineural auditory function in sal and salr mutants, we carried out both histologic analysis of the adult JO and electrophysiology of adult antennae. Most of the scolopidia that constitute the JO are missing in the adult sal- and salr-null antenna (Fig. 2 E and F). The fact that the chordotonal proneural gene atonal (ato) is expressed normally in the sal and salr null antennal discs at late third instar (50), in conjunction with our observation that some scolopidia exist in the mutant pupal and adult antennae, suggests that the chordotonal precursor cells are specified in the sal and salr mutants and are even able to differentiate, but that they are unable to organize into a cohesive system and therefore degenerate by early adult stages. This loss of adult chordotonal organs in sal and salr mutants contrasts markedly with sal function during larval chordotonal organ development. During embryogenesis, when the larval chordotonal organs are determined, sal represses chordotonal organ formation and loss of sal results in the development of ectopic scolopidia (34, 35). Thus, sal appears to have opposite effects on larval and adult chordotonal differentiation.
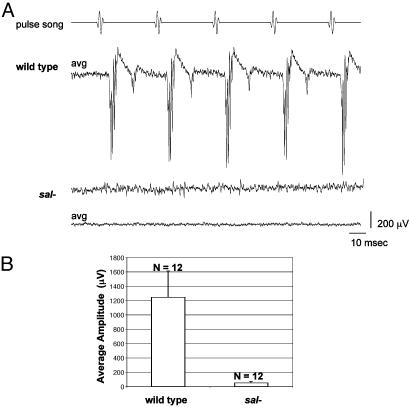
The histologic defects in the JO led us to think that sal and salr null antennae also would exhibit electrophysiologic defects. Indeed, in contrast to wild-type antennae, sal- and salr-null antennae produce no detectable electrophysiologic response to pulse songs (Fig. 3) or sine songs (not shown). We conclude that Drosophila sal and salr have important functions for both conductive and sensorineural aspects of hearing as does their human homolog SALL1.
Fig. 3.
Drosophila with sal- and salr-null antennae are completely deaf. (A) Strong sound-evoked potentials recorded from the antennal nerve of wild-type flies are completely absent in sal/salr mutants. Shown for the wild type is the averaged response (avg) to 10 presentations of the computer-generated pulse song; this average is almost indistinguishable from the response to individual presentations (not shown). For the mutant [salFCK-25/Df(2L)32FP-5 FRT40A], the response to a single presentation of the pulse song indicates the true level of unevoked background activity; averaging (10 presentations) reduces this background to reveal that there is no indication of even a subtle response. (B) Histogram of response amplitudes in sal/salr- and control flies of the Oregon R wild-type strain. The average amplitude for the control group was 1,243 μV, with a maximum of 1,622 μV, whereas the mutant flies showed an average of 52 μV, with a maximum of 85 μV. The amplitudes in sal/salr- flies represent solely background noise; there is no detectable evoked response.
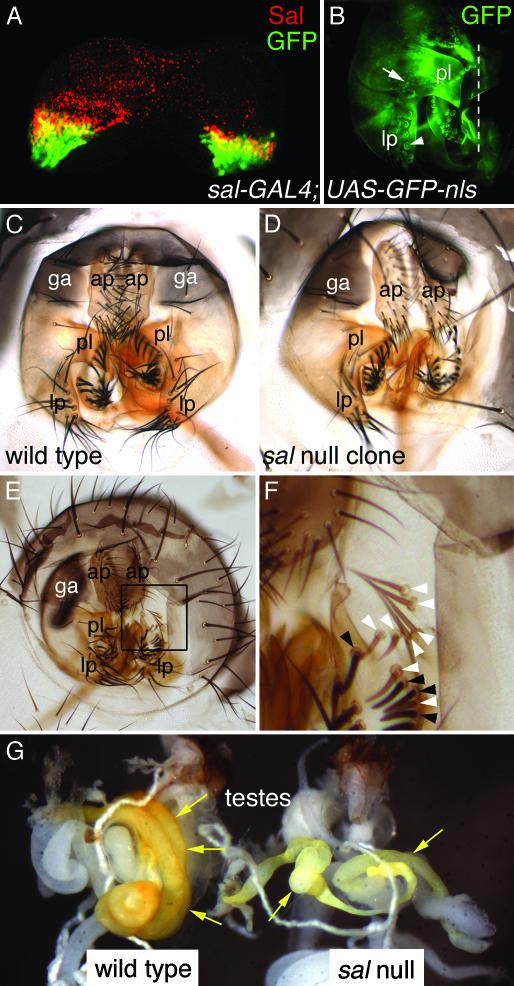
spalt and spalt-related Are Required for Drosophila Genital Development. Genital anomalies associated with TBS include hypospadias and other structural malformations (12). In Drosophila, sal and salr are differentially expressed between the male and female genital primordia (38). sal and salr expression is detected in only a few cells of the female genital disk, but is more extensive in the male genital disk. The domains of expression in male genital discs give rise to the presumptive posterior lobe, lateral plate, and genital arch that together constitute a large portion of the adult male external genitalia (Fig. 4A). Consistent with this, a sal-GAL4 enhancer trap specifically activates reporter gene expression in those structures in late pupae (Fig. 4B). The posterior lobe, lateral plate, and genital arch are lost in large somatic clones null for both sal and salr (Fig. 4 C-F). No other external genital structures were affected in the >50 clone-bearing males examined. This indicates that sal and salr play a role as significant in Drosophila genital development as does their human counterpart.
Fig. 4.
sal and salr expression and function in the male genitalia and the testes. (A) Sal protein (red) and GFP (green) are coexpressed (yellow) in the male genital disk of a third-instar larva of genotype sal-GAL4;UAS-GFP-nls. This indicates that the sal-GAL4 line reproduces the sal expression pattern in the genital disk. There is GFP in some cells where Sal protein is not detected. This may be caused by perdurance of GFP. (B) Left half of male pupal genitals of the same genotype in which GFP is expressed in the lateral plate, posterior lobe, and genital arch. The arrow points to the nucleus of a GFP-expressing cell. The arrowhead indicates a bristle socket with background autofluorescence. The dotted line indicates the ventral midline. The sal/salr expression domain in the female genital disk (not shown) is much smaller than that in males. This finding suggests that sal and salr may be differentially regulated between the sexes and that this could contribute to Drosophila sexual dimorphism. (C) Wild-type adult male anal plates and genitalia. (D) Adult male anal plates and genitalia from an animal of genotype y hs-FLPase; P[y+] FRT40A/Df(2L)32FP-5 FRT40A in which large sal-null clones were induced. The right lateral plate and posterior lobe are absent, as is a portion of the genital arch. (E) Adult male anal plates and genitalia from another animal of genotype y hs-FLPase; P[y+] FRT40A/Df(2L)32FP-5 FRT40A in which sal/salr null clones were induced. The right genital arch and posterior lobe are absent, as is a portion of the lateral plate. (F) Higher-magnification view of the area boxed in E. Beneath where the right posterior lobe would have been in a wild-type animal, sal/salr null clones can be detected by virtue of their y- bristles (white arrowheads). This tissue appears normal, and thus probably does not require sal or salr function. Adjacent wild-type tissue has dark bristles (black arrowheads). (G) Testes (arrows) from wild-type (Left) and sal mutant [Right; y hs-FLPase; FRT40A/Df(2L)32FP-5 FRT40A] animals in which clones had been induced. The testes from the sal mutant are smaller, lighter in color, and abnormally coiled. ap, anal plate; ga, genital arch; lp, lateral plate; pl, posterior lobe (pl).
spalt and spalt-related Are Required for Drosophila Testes Development. sal expression has been detected in both mouse and human testes (17-19). However, we have not been able to detect sal expression in the Drosophila testes. Nonetheless, we find that flies with clones null for both sal genes exhibit morphological defects in the testes (Fig. 4G). Mutant testes are improperly coiled, reduced in size, and lighter in color (Fig. 4G). Because these phenotypes were produced by clones, testes completely null for sal and salr might exhibit more severe defects. The lack of detectable sal expression in developing Drosophila testes could reflect a timing or other detection problem or may indicate that the effects of sal on testes development are not tissue autonomous. Together, the Drosophila testes phenotypes in conjunction with the observed expression of vertebrate sal gene expression in the testes, suggest the sal genes may have essential roles for testes development in both flies and vertebrates. The testes and genitalia of vertebrates and flies generally are not considered homologous. It therefore is unclear whether the roles of sal in their development represents conservation or convergence. However, because the last common ancestor of vertebrates and flies probably reproduced sexually, it should perhaps not surprise us to find developmental genetic parallels in the development of these organs.
The differential expression of sal between male and female genital discs and the male-specific role of sal in development of external genitalia and gonads indicate that sal plays a critical role in sexual dimorphism in Drosophila. Therefore, it will be important to determine how sex-determining genes such as Sex lethal, transformer, and doublesex interact with sal and salr.
Discussion
In light of a recent report that Drosophila sal and salr mutants exhibit eye defects (36), we were intrigued that a patient of age 44 with sudden optic neuropathy was subsequently diagnosed with TBS because of both physical deformities typical of the syndrome and a heterozygous mutation in SALL1 (52). Together with the reported expression of sal in the mouse eye (19), this leads us to think it likely that sal is involved in vertebrate eye development. We suggest that Drosophila sal and salr mutant phenotypes may provide clues to other overlooked or less penetrant defects associated with TBS. That sal functions in both Drosophila and human limb development, in conjunction with its putative function in vertebrate eye development, and our finding that the auditory organ, genitalia, and testes are affected by loss of sal function in both humans and flies indicates that sal functions may be evolutionarily conserved between insects and vertebrates. Studying sal in Drosophila therefore may lead to insights into the roles of sal genes in vertebrate development.
Although human SALL1 and Drosophila sal mutants exhibit some strikingly similar phenotypes, there are fundamental differences in the nature of the mutations leading to the defects. The Drosophila phenotypes result from being null for at least one sal gene, whereas TBS is caused by a heterozygous mutation in a single human sal, SALL1, that leads to production of a truncated protein. The two Drosophila genes are adjacent, similarly expressed, share enhancers, exhibit significant redundancy, and probably result from a relatively recent duplication event (39). The four known human genes reside on separate chromosomes and their mouse and chick homologs exhibit overlapping, but still distinct, expression patterns. Therefore, they are less likely to be as functionally redundant. However, it remains unclear whether the effects of SALL1 mutations in TBS patients result from haploinsufficiency or whether the truncated SALL1 proteins act dominantly and negatively to interfere with normal SALL1 function (and possibly the functions of other SALL genes). Sall1 heterozygous null mice are viable and appear normal, whereas homozygous null mice die perinatally with only severe kidney defects (53). These results lead us to think that the mutations in TBS are dominant negative. However, the expression patterns of mouse Sall1 and human SALL1 are not identical, so they may not serve identical functions, and the effects of Sall1 mutations might not be comparable between mice and humans.
The primary set of defects associated with TBS overlaps substantially with those of several other genetic disorders (7, 11, 15, 16). This could indicate that there is an overlapping set of regulatory genes at work in the human developmental genetic pathways used to construct the ear, appendage, renal, and anogenital organ systems. Yet, in some cases, the genes disrupted in patients with these other disorders have not yet been identified. Further analyses of spalt function and of the genetic hierarchies that pattern each of the affected tissues in Drosophila are likely to provide insights both into the ontogeny of analogous tissues in vertebrates and into these human disorders.
The observations that humans and Drosophila with sal mutations exhibit both conductive and sensorineural deafness has implications for our thinking about the evolution of auditory systems. We previously have shown that sal (38) and ato (50) are activated by Distal-less (Dll) during antennal development. The data presented here, when integrated with existing information, allow us to build a model in which Dll regulates both ato and sal and all three are required for the normal development of a functional Drosophila auditory system (Fig. 5A). Interestingly, the homologs of Dll and ato as well as sal are required for the development of a normal vertebrate auditory system. A second human syndrome that includes limb and auditory defects, split hand/split foot malformation has been linked to vertebrate homologs of Dll, the Dlx genes (54-56), and mice null for the ato homolog Math1 lack inner ear hair cells (57). Thus, despite the fact that sal/salr/SALL1, ato/Math1, and Dll/Dlx have pleiotropic effects in both Drosophila and vertebrates, their expression in the developing ear appears to represent a unique and tissue-specific constellation. If the regulatory interactions among Dll, ato, and sal homologs prove to be the same in vertebrates as in Drosophila (Fig. 5B), it would provide strong support for a view that these genes were used in the development of an ancestral auditory system and that the existence of such a system predated the arthropod/vertebrate divergence. One prediction of this model is that the developing auditory organs of other animals also would require this genetic cascade.
Fig. 5.
Structures affected in both Drosophila and humans with sal mutations and the genetic hierarchies governing ear development. (A) Dll regulates both sal and ato during development of the Drosophila auditory apparatus (38, 48). (B) Dll, sal, and ato homologs, the Dlx, Sall, and Ath genes, respectively, are involved in vertebrate ear development (8, 11-13, 54-57). If their genetic relationships prove similar, it would support the idea that a common vertebrate/invertebrate ancestor possessed a primitive ear whose development was governed by these genes.
Acknowledgments
We thank Jeanne Bourdeau-Heller and Karen Downs for assistance with histology, Shane Andrews for assistance with fly husbandry, and Carol Dizack for artwork. We are grateful to Sean Carroll for access to his confocal microscope, Rosa Barrio for sal mutants and antibody, Steve Cohen for the sal-GAL4 stock, and the Bloomington Drosophila Stock Center for other Drosophila lines used in these experiments. P.D.S.D. was supported by National Institutes of Health Predoctoral Training Grant T32-HD07477 and a Cremer fellowship. G.B.-F. is the recipient of a Young Investigator Award in Molecular Studies of Evolution from the Sloan Foundation and the National Science Foundation. This work also was supported in part by National Institutes of Health Grants DC04848-01 (to D.F.E.) and GM59871-01 (to G.B.-F.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TBS, Townes-Brocks' syndrome; JO, Johnston's organ.
References
- 1.Kohlhase, J., Wischermann, A., Reichenbach, H., Froster, U. & Engel, W. (1998) Nat. Genet. 18, 81-83. [DOI] [PubMed] [Google Scholar]
- 2.Kohlhase, J., Schuh, R., Dowe, G., Kuhnlein, R. P., Jackle, H., Schroeder, B., Schulz-Schaeffer, W., Kretzschmar, H. A., Kohler, A., Muller, U., et al. (1996) Genomics 38, 291-298. [DOI] [PubMed] [Google Scholar]
- 3.Kohlhase, J., Hausmann, S., Stojmenovic, G., Dixkens, C., Bink, K., Schulz-Schaeffer, W., Altmann, M. & Engel, W. (1999) Genomics 62, 216-222. [DOI] [PubMed] [Google Scholar]
- 4.Kohlhase, J., Taschner, P. E., Burfeind, P., Pasche, B., Newman, B., Blanck, C., Breuning, M. H., ten Kate, L. P., Maaswinkel-Mooy, P., Mitulla, B., et al. (1999) Am. J. Hum. Genet. 64, 435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marlin, S., Blanchard, S., Slim, R., Lacombe, D., Denoyelle, F., Alessandri, J., Calzolari, E., Drouin-Garraud, V., Ferraz, F., Fourmaintraux, A., et al. (1999) Hum. Mutat. 14, 377-386. [DOI] [PubMed] [Google Scholar]
- 6.Kohlhase, J. (2000) Hum. Mutat. 16, 460-466. [DOI] [PubMed] [Google Scholar]
- 7.Keegan, C. E., Mulliken, J. B., Wu, B. L. & Korf, B. R. (2001) Genet. Med. 3, 310-313. [DOI] [PubMed] [Google Scholar]
- 8.Townes, P. L. & Brocks, E. R. (1972) J. Pediatr. 81, 321-326. [DOI] [PubMed] [Google Scholar]
- 9.Kurnit, D. M., Steele, M. W., Pinsky, L. & Dibbins, A. (1978) J. Pediatr. 93, 270-273. [DOI] [PubMed] [Google Scholar]
- 10.Monteiro de Pina-Neta, J. (1984) Am. J. Med. Genet. 18, 147-152. [DOI] [PubMed] [Google Scholar]
- 11.Powell, C. M. & Michaelis, R. C. (1999) J. Med. Genet. 36, 89-93. [PMC free article] [PubMed] [Google Scholar]
- 12.Salerno, A., Kohlhase, J. & Kaplan, B. S. (2000) Pediatr. Nephrol. 14, 25-28. [DOI] [PubMed] [Google Scholar]
- 13.Surka, W. S., Kohlhase, J., Neunert, C. E., Schneider, D. S. & Proud, V. K. (2001) Am. J. Med. Genet. 102, 250-257. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Frias, M. L., Bermejo Sanchez, E., Arroyo Carrera, I., Perez Fernandez, J. L., Pardo Romero, M., Buron Martinez, E. & Hernandez Ramon, F. (1999) Anal. Esp. Pediatr. 50, 57-60. [PubMed] [Google Scholar]
- 15.Ferraz, F. G., Nunes, L., Ferraz, M. E., Sousa, J. P., Santos, M., Carvalho, C. & Maroteaux, P. (1989) Ann. Genet. 32, 120-123. [PubMed] [Google Scholar]
- 16.Konig, R., Schick, U. & Fuchs, S. (1990) Eur. J. Pediatr. 150, 100-103. [DOI] [PubMed] [Google Scholar]
- 17.Ma, Y., Singer, D. B., Gozman, A., Ford, D., Chai, L., Steinhoff, M. M., Hansen, K. & Maizel, A. L. (2001) Pediatr. Nephrol. 16, 701-709. [DOI] [PubMed] [Google Scholar]
- 18.Ott, T., Kaestner, K. H., Monaghan, A. P. & Schutz, G. (1996) Mech. Dev. 56, 117-128. [DOI] [PubMed] [Google Scholar]
- 19.Buck, A., Kispert, A. & Kohlhase, J. (2001) Mech. Dev. 104, 143-146. [DOI] [PubMed] [Google Scholar]
- 20.Kuhnlein, R., Frommer, G., Friedrich, M., Gonzalez-Gaitan, M., Weber, A., Wagner-Bernholz, J., Gehring, W., Jackle, H. & Schuh, R. (1994) EMBO J. 13, 168-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basson, M. & Horvitz, H. R. (1996) Genes Dev. 10, 1953-1965. [DOI] [PubMed] [Google Scholar]
- 22.Hollemann, T., Schuh, R., Pieler, T. & Stick, R. (1996) Mech. Dev. 55, 19-32. [DOI] [PubMed] [Google Scholar]
- 23.Koster, R., Stick, R., Loosli, F. & Wittbrodt, J. (1997) Mech. Dev. 124, 3147-3156. [DOI] [PubMed] [Google Scholar]
- 24.Onuma, Y., Nishinakamura, R., Takahashi, S., Yokota, T. & Asashima, M. (1999) Biochem. Biophys. Res. Commun. 264, 151-156. [DOI] [PubMed] [Google Scholar]
- 25.Buck, A., Archangelo, L., Dixkens, C. & Kohlhase, J. (2000) Cytogenet. Cell Genet. 89, 150-153. [DOI] [PubMed] [Google Scholar]
- 26.Farrell, E. R. & Munsterberg, A. E. (2000) Dev. Biol. 225, 447-458. [DOI] [PubMed] [Google Scholar]
- 27.Kohlhase, J., Altmann, M., Archangelo, L., Dixkens, C. & Engel, W. (2000) Mamm. Genome 11, 64-68. [DOI] [PubMed] [Google Scholar]
- 28.Farrell, E. R., Tosh, G., Church, E. & Munsterberg, A. E. (2001) Mech. Dev. 102, 227-230. [DOI] [PubMed] [Google Scholar]
- 29.de Celis, J., Barrio, R. & Kafatos, F. C. (1996) Nature 381, 421-424. [DOI] [PubMed] [Google Scholar]
- 30.Kuhnlein, R. P. & Schuh, R. (1996) Development (Cambridge, U.K.) 122, 2215-2223. [DOI] [PubMed] [Google Scholar]
- 31.Sturtevant, M., Biehs, B., Marin, E. & Bier, E. (1997) Development (Cambridge, U.K.) 124, 21-32. [DOI] [PubMed] [Google Scholar]
- 32.de Celis, J. F., Barrio, R. & Kafatos, F. C. (1999) Development (Cambridge, U.K.) 126, 2653-2662. [DOI] [PubMed] [Google Scholar]
- 33.Nellen, D., Burke, R., Struhl, G. & Basler, K. (1996) Cell 85, 357-368. [DOI] [PubMed] [Google Scholar]
- 34.Elstob, P. R., Brodu, V. & Gould, A. P. (2001) Development (Cambridge, U.K.) 128, 723-732. [DOI] [PubMed] [Google Scholar]
- 35.Rusten, T. E., Cantera, R., Urban, J., Technau, G., Kafatos, F. C. & Barrio, R. (2001) Development (Cambridge, U.K.) 128, 711-722. [DOI] [PubMed] [Google Scholar]
- 36.Mollereau, B., Dominguez, M., Webel, R., Colley, N. J., Keung, B., de Celis, J. F. & Desplan, C. (2001) Nature 412, 911-913. [DOI] [PubMed] [Google Scholar]
- 37.Wagner-Bernholz, J. T., Wilson, C., Gibson, G., Schuh, R. & Gehring, W. J. (1991) Genes Dev. 5, 2467-2480. [DOI] [PubMed] [Google Scholar]
- 38.Dong, P. D. S., Chu, J. & Panganiban, G. (2000) Development (Cambridge, U.K.) 127, 209-216. [DOI] [PubMed] [Google Scholar]
- 39.Barrio, R., de Celis, J. F., Bolshakov, S. & Kafatos, F. C. (1999) Dev. Biol. 215, 33-47. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, U., Joensson, F., Speicher, S. A. & Knust, E. (1995) Genetics 139, 203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, T. & Rubin, G. (1993) Development (Cambridge, U.K.) 117, 1223-1237. [DOI] [PubMed] [Google Scholar]
- 42.Newsome, T. P., Asling, B. & Dickson, B. J. (2000) Development (Cambridge, U.K.) 127, 851-860. [DOI] [PubMed] [Google Scholar]
- 43.Halder, G., Callaerts, P., Flister, S., Walldorf, U., Kloter, U. & Gehring, W. (1998) Development (Cambridge, U.K.) 125, 2181-2191. [DOI] [PubMed] [Google Scholar]
- 44.Stern, D. L. & Sucena, E. (2000) in Preparation of Larval and Adult Cuticles for Light Microscopy, eds. Sullivan, W., Ashburner, M. & Hawley, R. S. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 601-616.
- 45.Downs, K. M., Gifford, S., Blahnik, M. & Gardner, R. L. (1998) Development (Cambridge, U.K.) 125, 4507-4520. [DOI] [PubMed] [Google Scholar]
- 46.Eberl, D., Hardy, R. & Kernan, M. (2000) J. Neurosci. 20, 5981-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossmiller, D. R. & Pasic, T. R. (1994) Otolaryngol. Head Neck Surg. 111, 175-180. [DOI] [PubMed] [Google Scholar]
- 48.Eberl, D. F. (1999) Curr. Opin. Neurobiol. 9, 389-393. [DOI] [PubMed] [Google Scholar]
- 49.Goepfert, M. & Robert, D. (2001) Nature 411, 908. [DOI] [PubMed] [Google Scholar]
- 50.Dong, P. D. S., Dicks, J. S. & Panganiban, G. (2002) Development (Cambridge, U.K.) 129, 1967-1974. [DOI] [PubMed] [Google Scholar]
- 51.Jarman, A. P., Sun, Y., Jan, L. Y. & Jan, Y. N. (1995) Development (Cambridge, U.K.) 121, 2019-2030. [DOI] [PubMed] [Google Scholar]
- 52.Blanck, C., Kohlhase, J., Engels, S., Burfeind, P., Engel, W., Bottani, A., Patel, M. S., Kroes, H. Y. & Cobben, J. M. (2000) J. Med. Genet. 37, 303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishinakamura, R., Matsumoto, Y., Nakao, K., Nakamura, K., Sato, A., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Scully, S., Lacey, D. L., et al. (2001) Development (Cambridge, U.K.) 128, 3105-3115. [DOI] [PubMed] [Google Scholar]
- 54.Crackower, M. A., Scherer, S. W., Rommens, J. M., Hui, C. C., Poorkaj, P., Soder, S., Cobben, J. M., Hudgins, L., Evans, J. P. & Tsui, L. C. (1996) Hum. Mol. Genet. 5, 571-579. [DOI] [PubMed] [Google Scholar]
- 55.Scherer, S. W., Poorkaj, P., Massa, H., Soder, S., Allen, T., Nunes, M., Geshuri, D., Wong, E., Belloni, E., Little, S., et al. (1994) Hum. Mol. Genet. 3, 1345-1354. [DOI] [PubMed] [Google Scholar]
- 56.Robledo, R. F., Rajan, L., Li, X. & Lufkin, T. (2002) Genes Dev. 16, 1089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bermingham, N. S., Hassan, B. A., Price, S. D., Vollrath, M. A., Ben-Arie, N., Eatock, R. A., Bellen, H. J., Lysakowski, A. & Zoghbi, H. Y. (1999) Science 284, 1837-1841. [DOI] [PubMed] [Google Scholar]