Abstract
Tephritid fruit flies belonging to the Rhagoletis pomonella sibling species complex are controversial because they have been proposed to diverge in sympatry (in the absence of geographic isolation) by shifting and adapting to new host plants. Here, we report evidence suggesting a surprising source of genetic variation contributing to sympatric host shifts for these flies. From DNA sequence data for three nuclear loci and mtDNA, we infer that an ancestral, hawthorn-infesting R. pomonella population became geographically subdivided into Mexican and North American isolates ≈1.57 million years ago. Episodes of gene flow from Mexico subsequently infused the North American population with inversion polymorphism affecting key diapause traits, forming adaptive clines. Sometime later (perhaps ±1 million years), diapause variation in the latitudinal clines appears to have aided North American flies in adapting to a variety of plants with differing fruiting times, helping to spawn several new taxa. Thus, important raw genetic material facilitating the adaptive radiation of R. pomonella originated in a different time and place than the proximate ecological host shifts triggering sympatric divergence.
The apple maggot, Rhagoletis pomonella, reflects a rift in speciation theory. Mayr (1) has championed the idea that animal divergence requires the complete geographic separation of populations. Hence, the genetic origins of species arise in allopatric demes. In contrast, Walsh (2) proposed that certain host-specific phytophagous insects could speciate in sympatry when they shift and adapt to new plants. Walsh (3) cited the ancestral hawthorn (Crataegus spp.) and historically derived apple (Malus pumila)-infesting forms of R. pomonella as an example of sympatric speciation in action. Bush (4) later hypothesized that the entire complex of six or more sibling species comprising the R. pomonella group, to which the apple maggot belongs, arose by means of sympatric host shifts. Under the sympatric model, the genetic basis for adaptation to a novel host would be expected to trace to mutations occurring within the ancestral geographic range. Here, we provide evidence for a partial reconciliation of the sympatric and allopatric views by showing that inversion polymorphism forming latitudinal clines within R. pomonella and contributing to sympatric host race formation may have had much earlier geographic roots.
Genetic studies (5, 6) have shown that apple and haw flies represent partially reproductively isolated “host races,” the hypothesized first stage of sympatric speciation. The host races display significant allele frequency, but not fixed, genetic differences for six allozymes mapping to three different regions of the genome (7, 8). (Frequency differences average from 0.10 to 0.20 at sympatric sites, but are often as high as 0.3.) Inversions subsume each of these three genomic regions on chromosomes 1-3 (8) (haploid n = 6 for Rhagoletis). Markers within the inversions show patterns of strong gametic disequilibrium (5, 8), implying little or no recombination between inverted regions. The pattern of disequilibrium also suggests that alternative rearrangements may contain beneficial, coadapted suites of genes (8).
The inversions have been associated with the depth of the facultative overwintering pupal diapause (9-12). Multiple regression of genotypes for the six allozymes demarcating the inversions explained from 15% to >40% of the variation in adult eclosion time (13). Diapause variation is important because it differentially adapts apple and haw flies to a seasonal difference in the fruiting times of their hosts (9-12). In particular, the 3- to 4-week earlier phenology of apple selects for a deeper pupal diapause (greater recalcitrance to nondiapause development) in apple flies, allowing them to better withstand longer periods of warm weather before winter. A series of rearing experiments in which conditions were manipulated to simulate the seasonal difference between apple and haw fruiting times supported the “diapause hypothesis” by inducing genetic responses in allozyme frequencies in predicted directions (9-12). Life-history adaptation to apple was therefore due, in part, to inversion frequency changes in apple flies that generated ecologically based pre- and postmating reproductive isolation from haw flies.
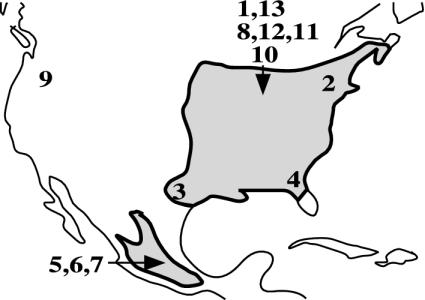
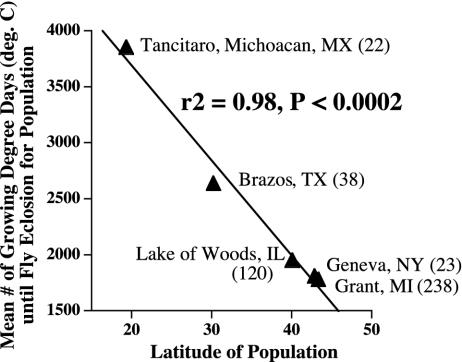
Host plant-related differentiation in R. pomonella is superimposed on a broader pattern of geographic variation (14-17). (Note: the distribution of the apple race is limited to the northeastern and midwestern United States and Canada, whereas the range of haw flies extends into Mexico; Fig. 1.) The recent shift of R. pomonella from hawthorn to the earlier-fruiting apple exposed apple flies to selection pressures similar to those experienced by haw-infesting flies in the South (i.e., warmer temperature conditions for longed periods). Mean growing degree days until eclosion, a surrogate for diapause depth (18), increase with decreasing latitude for haw-fly populations (Fig. 2). Concomitant with this phenotypic trend, allozyme alleles within the inversions that are common in the apple race at sympatric northern sites are essentially fixed in southern U.S. haw populations (14-17). Thus, the sympatrically derived apple race was extracted, in part, from preexisting adaptive inversion clines in the haw race.
Fig. 1.
Range of R. pomonella based on Bush (4) and Berlocher (17). The fly is also patchily distributed in the western United States, but these populations may be recent introductions and are not shown. Numbers indicate sampling sites in the study. (See Fig. 3 legend for site descriptions.)
Fig. 2.
Mean growing degree days (cumulative days × rearing temperature base Celsius) until adult eclosion vs. latitude for haw-fly populations from the United States and Mexico. Flies were reared for one generation in apples under standard laboratory conditions at 21°C, except for the Tancitaro, Mexico, and Urbana, IL, samples, which were reared from the field in haws at 26°C and 24°C, respectively. Numbers in parentheses are sample sizes of eclosing adults.
To understand the history of host-related diapause adaptation in R. pomonella, we sequenced three anonymous cDNA loci and mtDNA from flies sampled across the United States and a disjunct haw-infesting population in the Altiplano of Mexico (Fig. 1). The three cDNA loci P220, P2956, and P7 were chosen because they map to separate inversions on chromosomes 1, 2, and 3, respectively (7). Our rationale was that low levels of recombination within the inversions would let us resolve intact nuclear haplotypes. Comparisons of gene trees (genealogies) and coalescent events for these haplotypes among the cDNA loci and mtDNA would permit historical inferences to be made concerning the sympatric speciation process.
Materials and Methods
Fly Populations. Taxa, host plants, collecting locations, and sampling dates for flies are given in Figs. 1 and 3. Flies were collected as larvae in infested fruit and either (i) dissected from the fruit and frozen for later genetic analysis, or (ii) reared to adulthood in the laboratory.
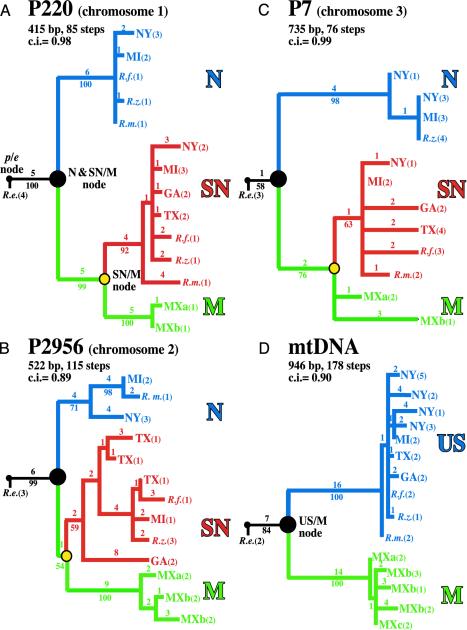
Fig. 3.
Maximum parsimony gene trees for P220 (A), P2956 (B), P7 (C), and mtDNA (D). The position of the basal node (p/e = small filled circles) between the R. pomonella species group and its sister taxon, R. electromorpha (R.e.), was set by using R. cingulata and R. suavis as outgroups (two to nine sequences each per locus). Trees are scaled so that the longest distance from an allele to the p/e node is relatively the same across loci. Bootstrap support for nodes (10,000 replicates), sequence length (number of base pairs), branch lengths (number of steps), and consistency indexes (c.i.) are given. N, North haplotype; SN, South/North haplotype; M, Mexican haplotype; large filled circles, N and SN/M nuclear and US/M mtDNA nodes; yellow circles, SN/M nodes. Taxa, hosts, location, and collecting dates (mo/d/yr) are as follows: 1, MI = R. pomonella [apple (Malus pumila), Grant, MI, 8/15/95]; 2, NY = R. pomonella [haw (Crataegus mollis), Geneva, NY, 9/16/00]; 3, TX = R. pomonella [haw (C. mollis), Waxahachie, TX, 10/6/89]; 4, GA = R. pomonella [haw (Crataegus flava), Lizella, GA, 9/13/89]; 5, MXa = R. pomonella n.s. [haw (Crataegus mexicana), U.S. Department of Agriculture intercept, Mexico, 1989]; 6, MXb = R. pomonella n.s. [haw (C. mexicana), U.S. Department of Agriculture intercept from Mexico City, Mexico, 10/26/00]; 7, MXc = R. pomonella n.s. [haw (C. mexicana), Mexico City, Mexico, 10/9/00]; 8, R.m. = Rhagoletis mendax [blueberry (Vaccinium corymbosum), Sawyer, MI, 8/1/99]; 9, R.z. = Rhagoletis zephyria [snowberry (Symphoricarpos albus), Dixie, WA, 8/31/97]; 10, R.f. = undescribed Rhagoletis species infesting, flowering dogwood fly [(Cornus florida), South Bend, IN, 10/2/99]; 11, R.e. = R. electromorpha [gray dogwood (Cornus racemosa), Dowagiac, MI, 9/12/99]; 12, R. cingulata [black cherry (Prunus serotina), Hart, MI, 7/15/89]; 13, R. suavis [black walnut (Juglans nigra), East Lansing, MI, 7/25/89]. Numbers in parentheses after taxon names in the figure are the number of similar haplotypes sequenced from site.
DNA Sequencing. Sequence data were generated for three cDNA loci (P220, P2956, and P7) isolated from a R. pomonella EST library (7) and for mtDNA (3′ portion of COI, leucine tRNA, and COII). The three cDNA loci segregated as single-copy genes in crosses (7, 8). Genomic DNAs were PCR-amplified 35 cycles (94°C, 30 sec; 52°C, 1 min; 72°C, 1.5 min) by using locus-specific primers (7). Products were TA-cloned into pCR II vectors (Invitrogen). Four to six clones per locus per fly were sequenced in the 5′ and 3′ directions on an automated laser fluorescence sequencer (Amersham Pharmacia).
Gene Tree Construction. Maximum parsimony (MP) and maximum likelihood (ML) gene trees were constructed by using PAUP*b8 (19). For the MP analysis, deletions were treated as a fifth base pair, with indels of identical length and position recoded to count as single mutational steps. Rhagoletis electromorpha, belonging to the sister species group (Rhagoletis tabellaria)to R. pomonella (4) and the more distantly related Rhagoletis cingulata and Rhagoletis suavis (4), were used to root trees. The molecular clock was tested for each locus for R. pomonella and R. electromorpha sequences by comparing log likelihood scores enforcing-versus-relaxing the clock hypothesis for the best supported DNA substitution model identified by using MODELTEST (20). Intragenic recombination was tested by using the method of Hudson and Kaplan (21).
Estimating Divergence Dates. ML estimates for the time of divergence (tdiv) of the U.S. and disjunct Mexican Highland populations and the effective size (Neanc) of the ancestral U.S./Mexican population were simultaneously determined from our sequence data in a manner similar to that described in Takahata et al. (22) and Yang (23). Rate variation among loci was accounted for by calibrating the three nuclear genes relative to mtDNA under the assumption of an insect mtDNA clock (1.15 × 10-8 substitutions per bp/year) (24). Below we present two major competing hypotheses, lineage sorting and introgression, potentially responsible for the topologies of gene trees. These models have different ramifications for the estimation of tdiv and Neanc. Under lineage sorting, the ML estimate for U.S./Mexican subdivision is based on the depths of the nodal partitions between SN and M haplotypes among cDNA loci (see Fig. 3 and below for haplotype designations). In contrast, U.S./Mexican divergence time under introgression is related to the nodal partitions between N and SN/M nuclear haplotypes. ML estimates for gene-flow events from Mexico into the United States were derived separately for each locus (inversion) based on the nodal depth of M and SN haplotypes.
Evaluating Lineage Sorting vs. Introgression Hypotheses. A likelihood ratio (LR) approach (25) was used to assess the lineage sorting vs. introgression hypotheses as causal explanations for the observed topologies of gene trees. LR tests measure the relative tenability of two competing hypotheses by comparing the ratio of their likelihoods (25). When ML1 specifies the maximum likelihood of the observed data under one hypothesis and ML2 under an alternative model, LR = ML1/ML2 (25). For non-nested hypotheses, such as lineage sorting and introgression, it has been suggested that the safest manner to assess the probability distribution for a LR test statistic is by Monte Carlo computer simulation (25). Replicate data sets are simulated under the assumptions of one hypothesis, which in our case is the null lineage sorting model. For each simulated data set, a LR is calculated anew for the competing hypotheses and the proportion of times that the observed LR value for the actual data exceeds the simulated values is taken as the significance level. The rejection level typically is set at 5% (25).
The different causes ascribed by lineage sorting and introgression for nodal partitions on gene trees (Fig. 4) make them amenable to a LR test. The ML estimate for the null lineage-sorting hypotheses (LR numerator) was found by permitting N/SN nodes to have different origin times among loci (reflecting the independent origins of inversions on chromosomes 1-3inthe common ancestor), whereas constraining the date for SN/M nuclear and US/M mtDNA nodes to be the same (reflecting the common and more recent divergence of these haplotypes resulting from the geographic isolation of Mexican Highland flies). In contrast, the ML estimate for the alternate introgression model (LR denominator) was derived holding the time of origin for N and SN/M nuclear and US/M mtDNA nodes the same across loci (= common biogeographic split of U.S./Mexican ancestor), while allowing the times for the SN/M nodes to vary (= independent gene-flow events from Mexico for each inversion).
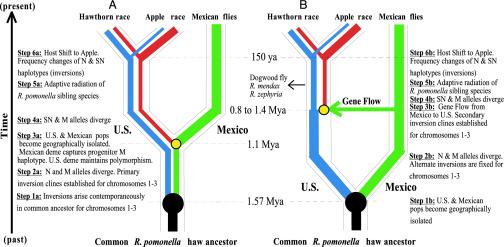
Fig. 4.
Lineage sorting (A) and introgression hypotheses (B) accounting for R. pomonella haplotypes. Thin black lines define population boundaries and sequence of divergence events. Thick colored lines represent gene trees for alleles segregating at P220, P2956, or P7 within populations. Red line, SN haplotype; blue line, N haplotype; green line, M haplotype. The generally higher frequency of SN haplotypes in the apple race at sympatric northern sites is denoted by the thicker width of the red line for the apple population.
ML estimates for gene trees and parametric bootstrapping were performed in a fashion similar to that described in Dopman et al. (26), except for the following instances. (i) ML estimates for the lineage sorting and introgression hypotheses were based on the product of likelihood scores across nuclear and mtDNA trees. (ii) To streamline the analysis, we used the mean number of substitutions separating whole classes of R. pomonella haplotypes (e.g., N vs. SN alleles), rather than including all of the sequence data for a locus. (iii) Probabilities for observing a given number of substitutions along a tree branch of a given length (in absolute time) were based on a binomial distribution by using rate constants for loci calibrated against an insect mtDNA clock (1.15 × 10-8 substitutions per bp/year). (iv) The null distribution for the LR statistic for lineage sorting was determined by simulating 10,000 trees based on ML estimates of 1.11 million years ago (Mya) for tdiv and 37,200 for Neanc under lineage sorting, and randomly chosen origin times for the inversions. (v) Mutations were arrayed on simulated trees according to a time-homogeneous Poisson process with mutations rates for the cDNA loci calibrated as above.
Results and Discussion
Nuclear and mtDNA Gene Trees. MP gene trees for P220, P2956, P7, and mtDNA are given in Fig. 3. MP and ML gene trees were virtually identical, so only the MP results are presented. None of the sequenced genes deviated significantly from a molecular clock. [P levels for P220, P2956, P7, and mtDNA and the best-fit substitution models were 0.87 (Hasegawa-Kishino-Yano or HKY model, 18 df), 0.47 (HKY+G, 18 df), 0.69 (HKY, 20 df), and 0.35 (HKY, 27 df).] We found no evidence for any recombination in the three nuclear loci or mtDNA. Hence, the gene trees can be interpreted as genealogies depicting the evolutionary relationships of haplotypes.
Gene Tree Topologies. A shared feature of all three nuclear gene trees was the presence of two major haplotype classes segregating in U.S. apple- and haw-fly populations (see red and blue tree branches in Figs. 3 A-C). One of the two U.S. haplotypes (designated “SN” for “South and North” in red) was more closely related to sequences from Mexican Highland haw flies (green “M” for Mexico) than the alternate haplotype class in the same U.S. host population (blue “N” for “North”). The SN haplotypes demarcated the subset of inversions generally found in higher frequency in apple than haw flies at sympatric U.S. sites. This finding was evidenced by the strong gametic disequilibrium of SN genes with corresponding allozyme alleles displaying host-related differentiation (r between P220 and NADH diaphorase-2 gene = 0.45, P < 10-3, n = 96 chromosomes; P2956/aconitase-2 gene = 0.61, P < 5 × 10-4, n = 46; P7/hydroxyacid dehydrogenase gene = 1.0, P < 10-6, n = 96). Geographic variation for the three nuclear cDNA loci also mirrored the clines observed for linked allozymes. N haplotypes were present only in R. pomonella populations from Michigan and New York, whereas SN haplotypes varied in frequency across the United States (Fig. 3). Most interestingly, all three nuclear loci showed a deep and concordant partition distinguishing N from SN and M haplotypes (compare large filled circles in Fig. 3 A-C). mtDNA had a similar partition, but strictly distinguished all U.S. from Mexican flies (Fig. 3D). We estimated that the probability for such congruence occurring by chance among nuclear and mtDNA gene trees was ≤0.025 (10,000 replicates each for computer simulations varying Neanc from 5 × 105 to 1 × 104).
Hypotheses for the Gene Trees. Lineage sorting of ancestral polymorphism and introgression are the two most likely explanations for the gene tree topologies. Under lineage sorting, the deep nodal concordance observed for P220, P2956, and P7 was caused by the chance origin of inversions on chromosomes 1-3 at a similar time in the past in the ancestral U.S./Mexican haw population (step 1a in Fig. 4A). The inversions then formed primary clines (step 2a). Some time later, as the Mexican deme became isolated, it captured progenitor M haplotypes (inversions) from the southern edge of the ancestral range (step 3a). The U.S. deme retained the ancestral polymorphism, resulting in SN alleles from the United States being more closely related to Mexican genes. Lineage sorting therefore attributes the nodal partitions between SN and M nuclear (yellow circles in Fig. 3 A-C) and U.S. and M mtDNA haplotypes to the separation of U.S. and Mexican flies, with the difference in their depths caused by coalescent time variation in the common ancestor.
In contrast, the introgression model posits that past gene flow from Mexico into the United States is responsible for the similarity of SN and M haplotypes. Under introgression, U.S. and Mexican demes became isolated first (step 1b in Fig. 4B), promoting genetic divergence and the fixation of alternate inversions in allopatry (step 2b). Note that phylogeography does not require contemporaneous origins for the inversions, as haplotypes would accumulate differences in concert from the time of isolation. Secondary gene flow from Mexico then delivered already-pronounced genetic variation for diapause traits in the guise of inversions (step 3b). The inversions were favored by selection, spreading to form adaptive clines in the U.S. haw population (step 4b). Sometime later (±1 million years), the variation in the latitudinal clines helped facilitate the historical (<150 years ago) shift from haw to apple (step 6b), the proximate basis for sympatric race formation. The introgression hypothesis therefore attributes variation in the depth of SN and M nuclear nodes to a series of gene-flow events from Mexico occurring between ≈0.8 and 1.4 Mya. The more basal concordance of cDNA and mtDNA loci (large filled circles in Fig. 3 A-C) reflects the earlier geographic separation of U.S. and Mexican demes ≈1.57 Mya (= ML estimate for tdiv under the introgression model, with the corresponding value for Neanc = 2).
Testing the Competing Hypotheses. Because lineage sorting and introgression attribute different causes for nodal partitions on gene trees (Fig. 4), the two hypotheses could be evaluated by using a LR approach (Materials and Methods). Computer simulations supported the introgression model. The probability was P ≤ 0.006 of generating gene trees (10,000 replicates) under the assumptions of the null lineage sorting model as consistent with the alternative introgression model as our actual data. A variant introgression hypothesis of gene flow in the reverse direction from the United States into Mexico is unlikely. This model has problems similar to lineage sorting, requiring the contemporaneous origin of inversions in the U.S. deme soon after population subdivision. Moreover, if gene flow had occurred from the United States, then it erased all traces of prior nuclear differentiation in Mexico. In contrast, the signature of gene flow from Mexico is evident in the extant U.S. inversion clines; SN alleles, the descendents of introgressed genes from Mexico, are found at highest frequency in the southern United States (13-16).
Gene duplication and loss is another conceivable explanation for the data. Under this hypothesis, SN and N haplotypes represent paralogous loci, with one or the other of the genes having become deleted from different chromosomes. When the Mexican isolate formed, it captured only “N” deletion chromosomes, accounting for the greater similarity of SN and M nuclear haplotypes. The duplication hypothesis has a difficult time accounting for the deep nodal concordance of nuclear and mtDNA gene trees, however, unless a series of genomewide duplication events occurred just before the U.S./Mexican subdivision. The duplication model also leaves unsolved how paralogous loci became associated with different inversions. Finally, genetic crosses have given no evidence for any null or duplicated loci that would likely be observed if the duplication hypothesis were true.
Introgression, Diapause, and the Apple Race. Our results imply that inversions have introgressed in the past from Mexico into the United States. We have separately observed that these inversions are associated with diapause traits involved in the sympatric host shift of R. pomonella to apple (8). This observation suggests that gene flow introduced ready-made diapause variation in the form of inversion polymorphism that was acted on much later by selection during sympatric host shifts. However, it does not rule out the possibility that the observed diapause variation accumulated because of mutations after introgression.
Two considerations support the hypothesis that the inversions carried diapause variation at the time of their introgression. First, the phenotypic, geographic, and genealogical pattern for haplotypes is consistent with the introgression hypothesis. The evolutionarily most closely related SN and M haplotypes are associated with prolonged diapause in U.S. and Mexican populations for all three rearranged chromosomes (Fig. 1). One might expect a more equitable distribution of phenotypic effects across inversion haplotypes if diapause variation was all of recent (post gene flow) origin. Second, theory predicts that the half-life for the decay of disequilibrium between a neutral locus not tightly linked to a gene under strong epistatic selection and an inversion is on the order of the reciprocal of the double-crossing over and gene conversion rate in heterokaryotypes (27, 28). Thus, given (i) ML estimates for inversion introgression times from ≈0.8 to 1.4 Mya, (ii) that SN haplotypes are not the direct targets of selection, and (iii) that gene flux rates between inversions are on the order of 10-4 to 10-5 per meiosis, which appear typical in Drosophila (27), current levels of disequilibrium for the introduced haplotypes should be well below 0.1. Consequently, if SN haplotypes were not embedded within epistatic gene complexes at the time of introgression, then the elevated levels of disequilibrium seen across the three rearranged chromosomal regions (8) are likely due to genetic hitchhiking of older, neutral haplotypes with a series of more recent, selectively driven mutations. Such selection should reduce haplotype variation within the inversions, but no obvious trend exists for shallow coalescence times for nuclear haplotypes (Fig. 3). The balance of evidence therefore suggests that the inversions contained variation for diapause depth at their time of introduction from Mexico.
Divergence Time for Populations. Under the introgression hypothesis, we estimate that Mexican and U.S. flies initially separated ≈1.57 Mya. This date is consistent with the biogeography of many temperate plants having disjunct eastern U.S./Mexican distributions, including the Rhagoletis hosts Crataegus and Cornus (29, 30). The timeframe and concordance of multiple taxa suggest a possible role for climatic and biotic changes associated with Pleistocene glaciation in the isolation of U.S. and Mexican flies. In this regard, we have defined the extant U.S. and Mexican haw populations in terms to their current geography, but their ranges have undoubtedly changed through time. To achieve full understanding of sympatric speciation in Rhagoletis will therefore paradoxically require elucidating the dynamic biogeographic history of these flies and equating it with possible secondary contact and gene-flow events between populations, as has been done for other organisms (31).
Implications of Introgression for the R. pomonella Group. If true, introgression from Mexico could also have implications for the radiation of the R. pomonella complex. Each sibling species in the group infests a different, nonoverlapping set of host plants (4, 17). Like the host races, these species differ in diapause traits differentially adapting them to variation in host fruiting time (17). They also differ from each other in allozyme frequencies, but to a much greater extent than the host races. Many of these differences involve loci mapping to the same inverted regions of chromosomes 1-3 differentiating the host races (17). Indeed, R. mendax, R. zephyria, and the undescribed flowering dogwood fly all possessed different combinations of N and SN haplotypes in common with apple and haw flies for P220, P2956, and P7 (Fig. 3 A-C), implying that they share inversion polymorphism. Thus, not only the host races, but more distinct host-related forms of R. pomonella appear to have evolved from prestanding genetic variation (inversion clines) created by past adaptive introgression from Mexico. However, in the case of the sibling species, we cannot discount a possible role for recent gene flow contributing to the shared polymorphism.
Conclusions
Our findings suggest that allopatry and secondary introgression may sometimes act in conjunction with, rather than distinction from, host shifts to facilitate sympatric speciation in R. pomonella. Walsh (3) and Bush (4) may therefore have been correct in postulating that sympatric host shifts represent the proximate ecological basis for R. pomonella divergence. But Mayr (1) could also be right in that initial allopatry provided a genetic architecture in the form of inversion polymorphism conducive for sympatric speciation. Moreover, the view of both Anderson (32) and Stebbins (33) that hybridization is often a catalyst for the genesis of new ecotypes and species could also be germane to Rhagoletis. However, introgression did not immediately precipitate the divergence of sympatric Rhagoletis populations. Rather, a significant time lag occurred. The initial impact was likely the favorable spread of the inversions, with the creation of secondary clines adapting the U.S. haw population to latitudinal differences in the environment. This spread provided a large genetic base for diapause that was later capitalized on for shifts to novel plants with differing fruiting phenologies. We do not mean to imply that all of the genes responsible for sympatric host shifts arose in allopatry or are contained within inversions. For example, premating isolation stemming from differences in host-plant preference also plays a critical role in R. pomonella divergence (34), and could have a simpler allelic history than the diapause-related inversions we have investigated here. Regardless, before gene flow, no host shifting and no speciation appear to have occurred between Mexican and U.S. isolates (haws are and were the common hosts for both populations). It was only after gene flow and when diapause traits later became associated with variation in host preference that R. pomonella apparently radiated sympatrically to fill open plant niches. Questions remain concerning why mtDNA haplotypes did not introgress with nuclear alleles (perhaps gene flow is male-mediated) and the current taxonomic status of Mexican flies. Nevertheless, our results evoke a surprising “PostModern” synthesis of processes and personalities, adding to a growing literature (35-38) implying that the origins of animal species can be as dynamic and rich (reticulate) as those for plants.
Acknowledgments
We thank E. Dyreson, H. Hollocher, L. Knowles, D. Lodge, S. Lyons, W. Maddison, B. McPheron, S. Muse, R. Nielsen, D. Posada, D. Schwarz, J. Thorne, U. Stolz, J. Wakeley, and X. Xie. This work was supported by grants from the National Science Foundation (Integrated Research Challenges and Graduate Research Traineeship) and the U.S. Department of Agriculture (to J.L.F. and S.H.B.) and by the State of Indiana 21 Century Fund (to J.L.F.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Mya, million years ago; MP, maximum parsimony; ML, maximum likelihood; LR, likelihood ratio.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY152494 -AY152510 (P220), AY152511-AY152526 (P2956), AY152527-AY152542 (P7), and AY152477-AY152493 (mtDNA)].
References
- 1.Mayr, E. (1963) Animal Species and Evolution (Belknap, Cambridge, MA).
- 2.Walsh, B. J. (1864) Proc. Entomol. Soc. Philadelphia 3, 403-430. [Google Scholar]
- 3.Walsh, B. J. (1867) J. Horticulture 2, 338-343. [Google Scholar]
- 4.Bush, G. L. (1966) The Taxonomy, Cytology and Evolution of the Genus Rhagoletis in North America (Mus. Comp. Zool., Cambridge, MA).
- 5.Feder, J. F., Chilcote, C. A. & Bush, G. L. (1988) Nature 336, 61-64. [Google Scholar]
- 6.McPheron, B. A., Smith, D. C. & Berlocher, S. H. (1988) Nature 336, 64-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roethele, J. B., Romero-Severson, J. & Feder, J. L. (2001) Ann. Entomol. Soc. Am. 94, 936-947. [Google Scholar]
- 8.Feder, J. L., Roethele, J. B., Filchak, K. E., Niedbalski, J. & Romero-Severson, J., Genetics 163, 939-953. [DOI] [PMC free article] [PubMed]
- 9.Feder, J. L., Roethele, J. B., Wlazlo, B. & Berlocher, S. H. (1997) Proc. Natl. Acad. Sci. USA 94, 11417-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feder, J. L., Stolz, U., Lewis, K. M., Perry, W., Roethele, J. B. & Rogers, A. (1997) Evolution (Lawrence, Kans.) 51, 1862-1876. [DOI] [PubMed] [Google Scholar]
- 11.Feder, J. L. & Filchak, K. E. (1999) Entomol. Exp. Appl. 91, 211-225. [Google Scholar]
- 12.Filchak, K. E., Roethele, J. B. & Feder, J. L. (2000) Nature 407, 739-742. [DOI] [PubMed] [Google Scholar]
- 13.Feder, J. L., Hunt, T. A. & Bush, G. L. (1993) Entomol. Exp. Appl. 69, 117-135. [Google Scholar]
- 14.Feder, J. L., Chilcote, C. A. & Bush, G. L. (1990) Evolution (Lawrence, Kans.) 44, 570-594. [DOI] [PubMed] [Google Scholar]
- 15.Feder, J. L. & Bush, G. L. (1989) Heredity 63, 245-266. [Google Scholar]
- 16.Berlocher, S. H. & McPheron, B. A. (1996) Heredity 77, 83-99. [Google Scholar]
- 17.Berlocher, S. H. (2000) Evolution (Lawrence, Kans.) 54, 543-557. [DOI] [PubMed] [Google Scholar]
- 18.Tauber, M. J. & Tauber, C. A. (1976) Annu. Rev. Entomol. 21, 81-107. [Google Scholar]
- 19.Swofford, D. L. (2000) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b8.
- 20.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 21.Hudson, R. R. & Kaplan, N. L. (1985) Genetics 111, 147-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahata, N., Satta, Y. & Klein, J. (1995) Theor. Popul. Biol. 48, 198-221. [DOI] [PubMed] [Google Scholar]
- 23.Yang, Z. (1997) Genet. Res. 69, 111-116. [DOI] [PubMed] [Google Scholar]
- 24.Brower, A. V. Z. (1994) Proc. Natl. Acad. Sci. USA 91, 6491-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huelsenbeck, J. P. & Crandall, K. A. (1997) Annu. Rev. Ecol. Syst. 28, 437-466. [Google Scholar]
- 26.Dopman, E. B., Sword, G. A. & Hillis, D. M. (2002) Evolution (Lawrence, Kans.) 56, 731-740. [DOI] [PubMed] [Google Scholar]
- 27.Ishii, K. & Charlesworth, B. (1977) Genet. Res. Camb. 30, 93-106. [Google Scholar]
- 28.Nei, M. & Li, W.-H. (1980) Genet. Res. 35, 65-83. [DOI] [PubMed] [Google Scholar]
- 29.Graham, A. (1999) Am. J. Bot. 86, 32-38. [PubMed] [Google Scholar]
- 30.Xiang, Q., Brunsfeld, S. J., Soltis, D. E. & Soltis, P. S. (1996) Syst. Bot. 21, 515-534. [Google Scholar]
- 31.Hewitt, G. (2000) Nature 405, 907-913. [DOI] [PubMed] [Google Scholar]
- 32.Anderson, E. (1949) Introgressive Hybridization (Wiley, New York).
- 33.Stebbins, G. L. (1959) Proc. Am. Philos. Soc. 103, 231-251. [Google Scholar]
- 34.Feder, J. L., Opp, S., Wlazlo, B., Reynolds, K., Go, W. & Spisak, S. (1994) Proc. Natl. Acad. Sci. USA 91, 7990-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold, M. L. (1992) Annu. Rev. Ecol. Syst. 23, 237-261. [Google Scholar]
- 36.Dowling, T. E. & Secor, C. L. (1997) Annu. Rev. Ecol. Syst. 28, 593-618. [Google Scholar]
- 37.Grant, P. R. & Grant, B. R. (2002) Science 296, 707-711. [DOI] [PubMed] [Google Scholar]
- 38.Vollmer, S. & Palumbi, S. R. (2002) Science 296, 2023-2025. [DOI] [PubMed] [Google Scholar]