Abstract
Mycobacterium tuberculosis is a high GC Gram-positive member of the actinobacteria. The mycobacterial cell wall is composed of a complex assortment of lipids and is the interface between the bacterium and its environment. The biosynthesis of fatty acids plays an essential role in the formation of cell wall components, in particular mycolic acids, which have been targeted by many of the drugs used to treat M. tuberculosis infection. M. tuberculosis has ≈250 genes involved in fatty acid metabolism, a much higher proportion than in any other organism. In silico methods have been used to compare the genome of M. tuberculosis CDC1551 to a database of 58 complete bacterial genomes. The resulting alignments were scanned for genes specifically involved in fatty acid biosynthetic pathway I. Phylogenetic analysis of these alignments was used to investigate horizontal gene transfer, gene duplication, and adaptive evolution. It was found that of the eight gene families examined, five of the phylogenies reconstructed suggest that the actinobacteria have a closer relationship with the α-proteobacteria than expected. This is either due to either an ancient transfer of genes or deep paralogy and subsequent retention of the genes in unrelated lineages. Additionally, adaptive evolution and gene duplication have been an influence in the evolution of the pathway. This study provides a key insight into how M. tuberculosis has developed its unique fatty acid synthetic abilities.
The Mycobacterium genus comprises >70 species and includes the human pathogens Mycobacterium tuberculosis and Mycobacterium leprae. M. tuberculosis is a member of the actinobacteria and is a Gram-positive pathogenic bacterium. It is estimated that over the next 25 years, 40 million people will die from tuberculosis (1). The completion of many bacterial genome-sequencing projects has aided the study of these diseases in the hope of finding their mechanisms of evading host immune systems and to try to explicate their metabolic pathways. The complete genomes of the laboratory strain M. tuberculosis H37Rv (2) and a recent clinical isolate M. tuberculosis CDC1551 (3) have been sequenced. These two genomes display >99% identity at the nucleotide level, and the majority of differences between the two genomes are single-nucleotide changes, of which there are ≈1,000 (4). It has been shown that the actinobacteria form a monophyletic group on the prokaryote phylogenetic tree, and that they group beside the Thermus/Deinococcus and Cyanobacteria clade (5). The α-, β-, and γ-proteobacteria form a monophyletic grouping also. There is no suggestion of any specific relationship between these two groups.
Comparative genomic methods have allowed for the investigation of regularities and differences observed across species at the DNA level (6). Bacteria increase their DNA content by horizontal gene transfer (HGT) and gene duplication, yet there is a selective pressure to minimize genome size to promote efficiency or competitiveness during replication (7). Therefore gene loss by either large deletion events or point or frameshift mutations serves to counterbalance the increase in gene number and removes harmful mutations. HGT brings new genes into the genome that are either homologous to existing genes or entirely new sequence families (8). The potential for acquiring and replacing an existing gene generally decreases with the phylogenetic distance between the donor and recipient lineages, and an acquired gene or gene segment is more likely to be beneficial if it has some properties similar to the recipient genome (8). It is reasonable to say that we are still naïve on this point, and the incidence of HGT in microbial consortia is unknown.
It has been shown that there may be a core group of genes that remain more closely associated over a long period through evolution than the rest of the genes in a genome (9). Nevertheless, there is a growing awareness of the role of HGT in the evolution and speciation of microorganisms (10). It is now recognized that HGT has shaped the microbial genescape (11), and “it is the major, if not the sole, evolutionary source of true innovation: novel enzymatic pathways, novel membrane transporter capacities, novel energetics” (12).
Adaptive evolution (positive Darwinian selection) in protein-coding genes is thought to be an ephemeral event, frequently leading to the generation of novel function, and is generally thought to be the result of selective pressures favoring a high level of replacement substitutions (13-15). Therefore, positive selection is natural selection that favors amino acid change (16). Numerous methods have been used to detect positive selection (17-19), and there have been an abundance of cases where this effector of change has been shown to have acted on key genes, e.g., the Colubine primate lysozyme proteins (20, 21).
The M. tuberculosis genome contains ≈250 enzymes involved in fatty acid metabolism in comparison with the Escherichia coli genome that contains ≈50 such enzymes (2). It is intriguing that M. tuberculosis needs so many more of these enzymes when the genomes of these two organisms are so similar in size. Fatty acid metabolism in M. tuberculosis is vital to the survival of the bacterium in the host, because its mycolic acids, produced from elongated fatty acids, form a protective lipid layer in the cell wall, and it is this lipid layer that has proven most interesting from a drug target point of view (22). It is therefore necessary that the intricate pathways and many enzymes involved are explored to determine why fatty acid metabolism is so important to this bacterium and specifically where the genes came from.
There are three possibilities that may explain the large number of enzymes involved in fatty acid metabolism in M. tuberculosis: birth of genes, gene duplication (paralogy), and HGT (either recent or ancient). In bacteria such as Neisseria meningitidis, genetic exchange is so frequent that lineage boundaries are difficult to distinguish (23). In contrast, the M. tuberculosis complex has been thought until recently to owe its genome plasticity to insertion and deletion events that may have arisen due to insertion sequence-triggered events or slipped-strand mispairing during replication (24). It has been suggested, however, that M. tuberculosis has the highest number of eukaryotic-prokaryotic interkingdom gene fusions of all sequenced bacterial genomes (25), and it has even been proposed that 19 genes of eukaryotic origin have been found in the M. tuberculosis genome (26). If M. tuberculosis has indeed obtained genes from eukaryotes in the past, it would seem reasonable to speculate that the genome may also have obtained genes from other prokaryotes by HGT. For this reason, it was decided to examine some of these enzymes from the fatty acid biosynthetic pathway and determine whether indeed they were recent additions to the genome by means of HGT.
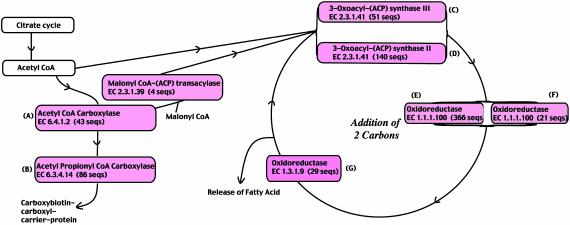
Knowledge of the biochemistry of fatty acid biosynthetic pathways I and II for M. tuberculosis is incomplete at present due in part to the difficulty in culturing this slow-growing bacterium. According to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (27), there are two discrete enzyme systems in the biosynthesis of fatty acids in mycobacteria, namely fatty acid synthase (FAS) I and II (2, 27, 28). The FAS I system consists of a single polypeptide with multiple catalytic activities that creates short precursors for elongation by other fatty acid systems (2). The FAS II system includes a host of enzymes involved in the elongation of substrates bound to an acyl-carrier protein to produce mycolic acids (2). The FAS II system for H37Rv has been well studied, because it contains the enzymes involved in the production of the cell wall mycolic acids (27, 28). Fig. 1 contains a simplified diagram of the KEGG pathway I showing all of the genes from CDC1551 involved in the biosynthesis and elongation of fatty acids. Some of the enzymes in this pathway produce mycolic acids (2), and therefore these enzymes are interesting to study, because the cell wall is the focal point of interest when it comes to interaction between the host cell and the bacterium.
Fig. 1.
Simplified diagram of the Kyoto Encyclopedia of Genes and Genomes fatty acid biosynthetic pathway I indicating the EC numbers of the enzymes involved and the number of sequences in each alignment. The colored boxes indicate that an alignment has been analyzed for this step of the pathway, and the letters of the alphabet (A-G) in order correspond to the tree diagram constructed for that alignment (see Figs. 2, 3, 4, 5 and 7-9).
The focus of this research has been to try to ascertain why the M. tuberculosis genome possesses so many genes involved in fatty acid metabolism. Another challenge is to evaluate whether the bacterium has always possessed this many genes for this function, or whether these genes were acquired from elsewhere. It is also important to determine whether gene duplication events played a significant role in expanding the repertoire of genes involved in fatty acid metabolism. Considering it has been shown that adaptive evolution can occur after duplication (29, 30), we sought to examine whether this might happen after perturbation of this pathway, following either duplication or gene acquisition.
Materials and Methods
A database of complete genome sequences for 58 organisms, including 9 Archaea, 30 Gram-negative bacteria, and 19 Gram-positive bacteria, was assembled by downloading the relevant information from www.ncbi.nlm.nih.gov, www.sanger.ac.uk, or www.tigr.org. The Kyoto Encyclopedia of Genes and Genomes database (28) was searched for the genes encoding the enzymes involved in fatty acid biosynthetic pathway I for the CDC1551 strain. BLAST (31) was used to perform a search of each CDC1551 gene against the database. Putative homologues with e values ≤10-20 were retrieved from the database and aligned by using clustalw (32). These alignments were corrected for obvious alignment ambiguity by using seal, Ver. 2.0a9 (http://evolve.zoo.ox.ac.uk/software/Se-Al/main.html). The alignments are available on request from the authors.
To assess the incidence of horizontal gene transfer, robust hypotheses of phylogenetic relationships were generated by using the corrected alignments. These hypotheses were constructed with the Bayesian framework implemented in mr. bayes 2.01 (33). For the Bayesian analysis clade, probability values were generated by using the sumt command. Consensus trees were generated with PAUP* 4.0b10 (PPC) (34).
Analyses of adaptive evolution were carried out by using the likelihood ratio test (LRT) methods described by Yang (35-38). LRT is used to evaluate nested models of sequence evolution. Some models are more parameter-rich extensions of other models, and when this is the case, an LRT may be performed with twice the log-likelihood difference being compared with a χ2 distribution with the degrees of freedom equal to the difference in the number of parameters between the two models (for a more complete description, see ref. 39). The results are presented in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. These methods involve the estimation of dN:dS ratios (nonsynonymous to synonymous substitution ratio, termed ω) by using maximum likelihood estimation of parameters on a phylogenetic tree. Models that either restrict or allow ω to vary across sites (site-rate models with a variety of “classes” of sites) or across sites and lineages (branch-site models) were used. Finally, when the maximum likelihood optimization of the parameters is complete, an empirical Bayes approach is used to infer which class a site is most likely from (40). Those sites with a high probability of coming from a class of sites with a high ω are most likely to be under positive selection. The models of Yang and Nielsen (39) have been used in this paper, with the same nomenclature being preserved.
To investigate the possibility that putative HGT genes display a synonymous codon usage pattern that is unusual compared with the rest of the genome, an analysis of variation in synonymous codon usage was carried out by using gcua (40). A correspondence analysis of raw codon counts was carried out, with the first four principal axes being used to evaluate synonymous codon usage patterns. Only the first two axes are shown in Fig. 6, which is published as supporting information on the PNAS web site.
Results
Fig. 1 illustrates the enzymes involved in fatty acid biosynthetic pathway 1 and details the order chronologically in which the alignments for these enzymes will be examined.
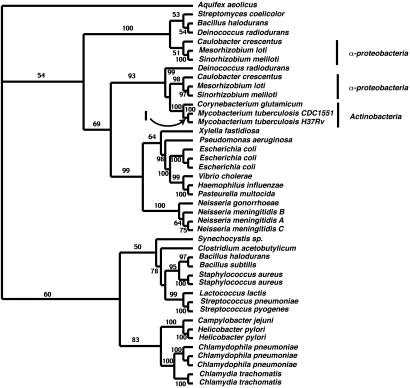
The tree illustrated in Fig. 2 was constructed from the acetyl-CoA carboxylase carboxyl transferase β-subunit. The topology of this tree is unusual and suggests either that most actinobacteria are more closely related to the proteobacteria than previously thought or they acquired genes from an ancestor of the α-proteobacteria. Most of the actinobacterial sequences form a sister-group relationship with genes from α-proteobacteria such as Sinorhizobium meliloti, Mezorhizobium loti, and Caulobacter crescentus with 99% clade credibility support. The low GC Gram-positive bacteria are located elsewhere on the tree. The exception is a sequence from Streptomyces coelicolor (an actinobacterium) that, unusually, does not group with the actinobacteria but instead forms part of a separate clade with Bacillus halodurans and Deinococcus radiodurans. Therefore in this first step of the pathway, we appear to have significant support for the ancient acquisition of this gene from the α-proteobacteria or a closer relationship than previously believed between the α-proteobacteria and the actinobacteria.
Fig. 2.
Phylogenetic tree of the acetyl-CoA carboxylase, carboxyl transferase β-subunit, corresponding to Fig. 1 A. The numbers on the branches are Bayesian clade probabilities. The arrow indicates the branch that was analyzed for signatures indicative of adaptive evolution.
Given that this dataset of 43 sequences is sufficiently small to perform a number of likelihood ratio tests for heterogeneity of selective pressures across sites and lineages, we performed a number of analyses for evidence of adaptive evolution. For the branch-site models, we marked the lineage leading to the two M. tuberculosis sequences. We tested a variety of models against null hypotheses of simpler models and found that two models were interesting insofar as they were significantly better fit to the data than the simpler models. These were models B and 7. In both cases, these models suggest that the majority of sites are under strong purifying selection. In the case of model 7, which is a site-rate model, it suggests that all sites in the alignment are under strong purifying selection. However, when the branch leading to the M. tuberculosis sequences is marked as a “foreground” branch, the likelihood is significantly greater (P < 0.005) than the null model (model 3, k = 2), and as many as 20.5% of sites on this branch are hypothesized to be under strong positive selection for change (ω = 82.8). Using Bayesian estimation, eight sites are likely to be in the class of sites with an ω>1 in the foreground lineage (P > 0.95).
Moving to the next enzyme in the pathway, there are two genes in the CDC1551 genome that perform this enzymatic function, MT2576 and MT3384. A diagram of this tree is too large to include here and is published as supporting information on the PNAS web site. Fig. 7, which is published as supporting information on the PNAS web site, indicates yet another example of either an ancient horizontal gene transfer or perhaps deep paralogy and differential retention of paralogues. There are four regions of the tree in which we find actinobacteria. The top of the tree houses a diverse group of low GC Gram-positive bacteria. Three of the clades containing actinobacterial sequences are closely related to proteobacterial sequences, specifically the α-proteobacteria. The fourth clade of actinobacteria have a sister grouping with a sequence from Halobacterium (an Archaeon). The Halobacterium gene groups with the actinobacteria with 100% clade credibility support and may suggest an interdomain horizontal gene transfer event. It is unlikely that this represents differential retention of ancient paralogues.
The third enzyme involved in this pathway is Malonyl CoA-(acp) transacylase. Unusually, this alignment was actinobacteria specific, containing only one gene from M. leprae, one from H37Rv, one from CDC1551, and one from S. coelicolor. Therefore it was not possible to construct a useful tree. However, this is an interesting finding, because it appears that although other bacterial species possess the gene for this enzyme, their genes are not homologous to the actinobacterial genes.
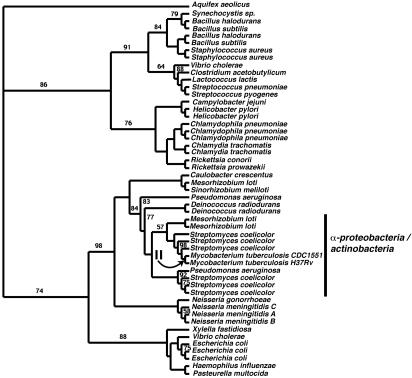
The next part of the pathway relates to the cyclic elongation of the fatty acids. The enzyme EC 2.3.1.41 in CDC1551, is encoded by four genes: MT0557, MT2305, MT2306, and MT3021.1. The first of these genes, MT0557, is annotated as a 3-oxoacyl (acp) synthase II. The alignment produced for this family contains 51 sequences and appears to be distinct from the alignments containing the other three genes. The tree (Fig. 3) produced from this alignment provides evidence that once again the actinobacterial homologues of this gene are more closely related to the proteobacteria than would have been expected. The actinobacterial clades reside within a large group of proteobacteria. The grouping of the actinobacteria beside M. loti is supported by 57% clade credibility. The S. coelicolor genes form a clade with Pseudomonas aeruginosa, a γ-proteobacteria, and this clade is supported by a 100% clade credibility value. This alignment was small enough to perform analyses of adaptive evolution. We used site-rate and branch-site models with the lineage leading to the two Mycobacterium sequences marked as a foreground branch. We did not detect signatures indicative of adaptive evolution anywhere in this dataset.
Fig. 3.
Phylogenetic tree inferred from the 3-oxoacyl-(acp) synthase III genes. This tree corresponds to Fig. 1C. Numbers on internal branches indicate clade credibility values for those internal branches where the values were <100. All unlabeled branches correspond to credibility values of 100. The arrow indicates the branch that was analyzed for signatures indicative of adaptive evolution.
The other genes corresponding to the enzyme EC 2.3.1.41 are MT2305, MT2306, and MT3021.1, and one alignment was generated that contained these genes and their homologues. One of the most interesting observations in Fig. 8, which is published as supporting information on the PNAS web site, was that there was a large amount of Mycobacterium-specific gene duplication. The region of the tree containing these duplications shows a number of clades in which we exclusively find Mycobacterium species, and there is evidence of several gene duplication events. It is interesting to note that, even though the other actinobacteria do feature in this region of the tree, their homologs have not duplicated to the same extent as M. tuberculosis. Besides this very large duplication of mycobacterial genes, there are three lesser clades of actinobacteria. The clade at the top of the tree groups the mycobacteria with an M. loti gene from the α-proteobacteria, suggesting that this may be a horizontal gene transfer event or evidence for deep paralogy. It is interesting that this clade is grouped in the middle of two clades of genes from Bacillus subtilis, a low GC Gram-positive bacterium. Toward the bottom of the tree, there is a clade of actinobacteria containing a gene duplication of the mycobacteria and multiple homologs from S. coelicolor. The closest clade to this contains α-proteobacteria and two S. coelicolor homologs. In this instance, perhaps the α-proteobacteria acquired the genes from the actinobacteria, or again the closeness of the two groups on the tree could be due to ancient artifacts.
The next stage of the elongation cycle involves the oxidoreductases (EC 1.1.1.100). There are five genes that correspond to this EC number in CDC1551. Once again, one of the query genes produced an alignment with homologues that were dissimilar to the large alignment produced by the other four query genes. The large alignment contained four of the five genes, MT0793, MT1393, MT1530, MT3664, and all their homologues, and in total, 366 sequences were used to construct the consensus tree (Fig. 9, which is published as supporting information on the PNAS web site). It appears to have been quite a promiscuous gene, because the tree shows evidence of clades comprised of mycobacteria with proteobacteria, Gram-negative bacteria grouped with Archaea, Gram-positive bacteria with γ-proteobacteria, and almost every combination of prokaryote possible. It is possible that this gene is highly prone to HGT events. The phylogenetic groupings do not appear to conform to those seen when using the16S rRNA gene, for example.
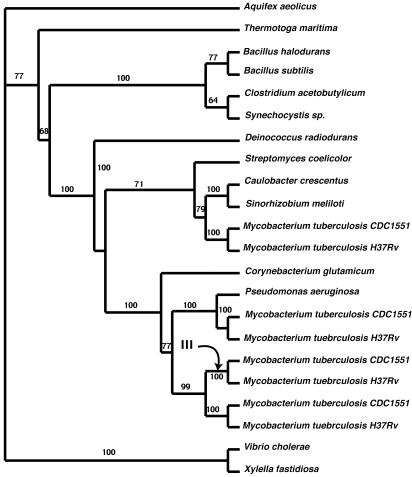
The fifth gene, (MT3606) 3-oxoacyl-(acp)-reductase, consisted of an alignment of 22 sequences. The resulting tree (Fig. 4) shows an example, once again, of a Mycobacterium-specific gene duplication. This tree is also interesting, because there may be evidence that the proteobacteria acquired genes from the actinobacteria. C. crescentus and S. meliloti genes are grouped with the actinobacteria with clade credibility support of 71%. P. aeruginosa, a γ-proteobacterium, is grouped in the middle of the actinobacteria with a clade credibility support value of 100%. An alternative but less likely explanation may be that the genes are closely related due to deep paralogy. The analyses of adaptive evolution for this dataset indicate that there are lineage-specific episodes of adaptive selection. The site-rate models all indicate there is no category of site for which there is positive selection across the tree. However, using the branch-site model, model B, there is significant support for adaptive evolution on the branch labeled in Fig. 4. In fact, it is estimated that a total of 14.6% of sites are likely to be under positive selection on this lineage (ω = 2.58, P > 0.95).
Fig. 4.
Phylogenetic tree inferred from 3-oxoacyl (acp) reductase genes. This tree corresponds to Fig. 1F. The numbers on the internal branches are Bayesian clade credibility values. The arrow indicates the branch that was analyzed for signatures indicative of adaptive evolution.
The final enzyme found in the pathway is the enoyl (acp) reductase (MT1531). The tree in Fig. 5 is distinctive in that is does not appear to show any evidence of HGT. There is minimal evidence of paralogy in this gene family, and the shape of the tree does not indicate any specific relationship between the α-proteobacteria and the actinobacteria, despite the presence of sequences from both sets of organisms on the tree. This is the only case in the pathway where there is a monophyletic clade of actinobacteria that group separately from the other α-proteobacteria, as we might expect. The actinobacteria group together with a clade credibility support value of 100%, and there is very high support (99% clade credibility support) for the grouping of the α-proteobacteria with other proteobacteria. This enzyme shows no evidence of positive selection by using either site-rate or branch-site models.
Fig. 5.
Phylogenetic tree inferred from enoyl (acp) reductase genes. This tree corresponds to Fig. 1G. The numbers on the internal branches are Bayesian clade credibility values. The arrow indicates the branch that was analyzed for signatures indicative of adaptive evolution.
It has been shown in E. coli that genes thought to have been acquired recently by the genome often display a codon preference different than most other genes in the genome (41). To examine whether these horizontally transferred genes display an unusual codon usage, an analysis of variation in synonymous codon usage in M. tuberculosis CDC1551 was carried out. Correspondence analysis has already been performed for the H37Rv strain (42), and our results concur with the authors' findings. The results are shown in Fig. 6. The first axis explains 14% of the total inertia of the data, and the second axis explains a further 8%. As can be seen, most of the variation in axis 1 is due to a small number of outlier genes. It transpires that these genes mostly belong to either the PE or PPE families of proteins, as was found in the analysis of the H37Rv strain (42). The genes from fatty acid pathway I do not display an unusual synonymous codon usage. The codon usage of these genes is very similar to most genes in the M. tuberculosis CDC1551 genome.
Discussion
Fatty acid biosynthesis is likely a very ancient pathway. The relative ubiquity of fatty acid synthesis across all of the major groups of organisms provides reasonable support for this hypothesis. It might seem likely that in a similar way to translation or replication, a mechanism of carrying out this function would have been invented early in evolution, and the contemporary operation of this pathway would have remained largely unchanged. However, from the evidence of this study, we can see that in the actinobacteria in general and in M. tuberculosis in particular, this pathway has been changed significantly. Fatty acid biosynthesis is an essential part of the M. tuberculosis lifestyle with the genome containing ≈250 fatty acid metabolizing enzymes.
In this study, we have shown that five protein families from the fatty acid biosynthetic pathway group the actinobacteria with the proteobacteria and specifically the α-proteobacteria, suggesting HGT. Another step in the pathway appears to be a de novo invention by the actinobacteria, although greater sampling of genomes will help clarify this situation. A further two gene families have extensive levels of gene duplication. Evidence for adaptive evolution has been found in two gene families, and only one step in the pathway (Fig. 5) shows no evidence for gene acquisition, duplication, or positive selection.
Many of the fatty acid biosynthesis genes in this study seem to have been acquired from, or are more closely related than expected to, the α-proteobacteria (i.e., the rhizosphere, bacteria, and relatives). On the one hand, this seems to be an unusual situation, given the likelihood that this pathway existed in early prokaryotes and is likely to have been fully functional possessing most, if not all, of the genetic components to adequately synthesize fatty acids. On the other hand, the closeness of the relationship between the mycobacterial genes and the α-proteobacterial genes seems reasonable when one considers that the majority of the members of the Mycobacterium genus are soil-dwelling bacteria (43), and that most members of the α-proteobacteria are also soil dwellers, suggesting there has been opportunity for these organisms to exchange genetic material. It is more than likely that M. tuberculosis and the other actinobacteria had native copies of these enzymes, and they have been replaced by HGT from proteobacteria.
The Malonyl CoA transacylase enzyme is almost a bottleneck in one section of the cycle and is unique in this pathway, because its gene is actinobacterium-specific. This enzyme function exists in other bacterial species, but the genes responsible are not homologous to the ones from the actinobacteria. Given its central role in the pathway, it is most unusual to think that it was invented late in evolution, and that this invention has replaced the gene that was responsible for catalyzing this reaction. It is also curious that there is no homolog in the Corynebacterium glutamicum genome. If we consider this fact, along with the fact that two other enzymatic functions in this pathway (EC 2.3.1.41 and EC 1.1.1.100) are carried out by separate groups of nonhomologous proteins, it seems likely that this pathway is evolving in many ways apart from point mutation, and that turnover and replacement of genes and gene families is characteristic of the evolution of the pathway.
The position of all of these fatty acid synthesis genes in the M. tuberculosis CDC1551 genome varies. Obviously the MT2303, MT2305, and MT2306 genes lie very close to each other on the genome, as do the MT1530 and MT1531 genes. It is possible that these two groups of genes were ancient HGT events of either single genes or an operon from another source. It must be noted, however, that the results for MT2303 indicate that it is a Mycobacterium-specific gene. The MT1531 gene also does not appear to be the result of gene transfer according to our findings.
We have examined a number of genes in the pathway for evidence of lineage-specific adaptive evolution and have confirmed the presence of selective pressures for amino acid change in two of the gene families in this pathway. In other words, the strains of M. tuberculosis that possessed specific changes in these genes were at an advantage compared with strains that did not have these changes.
The first gene in which we see adaptive evolution is that in which there is only one member of the gene family in M. tuberculosis. This is the acetyl-CoA carobxylase, which represents the first step in the pathway. The genes are very similar to α-proteobacterial genes, appear to have been acquired by the actinobacteria, and have been extensively modified in the mycobacteria. In the case of one of the oxidoreductase genes (Figs. 1F and 4), we see evidence of gene family expansion by duplication followed by positive selection in one of the genes. This is likely to be a kind of “tweaking” of the mechanism of fatty acid synthesis, perhaps for improved functioning after major alterations of the pathway.
The codon usage pattern for the genes involved in fatty acid biosynthesis is typical of the remainder of the genes in the genome. It was found that those genes that had the least similarity of codon usage to the majority of tuberculosis genes were the PE and PPE family of proteins, as was previously found to be the case in the H37Rv strain (42). The absence of any appreciable difference in synonymous codon usage between putative recent acquisitions and the rest of the genome can be for either of two reasons. The incoming genes could have had a codon usage pattern very similar to the average pattern in the CDC1551 strain, or perhaps enough time has elapsed since acquisition of these genes that mutational pressure has caused these genes to ameliorate toward the typical CDC1551 codon usage pattern.
In summary, this ancient pathway has been extensively modified in M. tuberculosis, with HGT, duplication, and adaptive evolution contributing to these modifications. These results should help explain why M. tuberculosis is so accomplished at working with fatty acids.
Supplementary Material
Acknowledgments
Thanks to Steve Gordon for advice and valued opinions. This work was supported by the Health Research Board, Higher Education Authority Program for Research in Third Level Institutes Cycles II and III, Enterprise Ireland, and National University of Ireland Maynooth (scholarship to R.J.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: HGT, horizontal gene transfer.
References
- 1.World Health Organization (2001) Stop TB Annual Report WHO/CDS/STB/2002.17 (W.H.O., Geneva).
- 2.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, et al. (1998) Nature 393, 537-544. [DOI] [PubMed] [Google Scholar]
- 3.Valway, S. E., Sanchez, M. P., Shinnick, T. F., Orme, I., Agerton, T., Hoy, D., Jones, J. S., Westmoreland, H. & Onorato, I. M. (1998) N. Engl. J. Med. 338, 633-639. [DOI] [PubMed] [Google Scholar]
- 4.Betts, J. C., Dodson, P., Quan, S., Lewis, A. P., Thomas, P. J., Duncan, K. & McAdam, R. A. (2000) Microbiology 146, 3205-3216. [DOI] [PubMed] [Google Scholar]
- 5.Wolf, Y. I., Rogozin, I. B., Grishin, N. V. & Koonin, E. V. (2002) Trends Genet. 18, 472-479. [DOI] [PubMed] [Google Scholar]
- 6.Lillo, F., Basile, S. & Mantegna, R. N. (2002) Bioinformatics 18, 971-979. [DOI] [PubMed] [Google Scholar]
- 7.Mira, A., Ochman, H. & Moran, N. A. (2001) Trends Genet. 17, 589-596. [DOI] [PubMed] [Google Scholar]
- 8.Ochman, H. (2001) Curr. Opin. Genet. Dev. 11, 616-619. [DOI] [PubMed] [Google Scholar]
- 9.Daubin, V. A., Gouy, M. & Perriere, G. (2002) Genome Res. 12, 1080-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucher, Y. & Doolittle, W. F. (2000) Mol. Microbiol. 37, 703-716. [DOI] [PubMed] [Google Scholar]
- 11.Ragan, M. A. (2001) Curr. Opin. Genet. Dev. 11, 620-626. [DOI] [PubMed] [Google Scholar]
- 12.Woese, C. R. (2000) Proc. Natl. Acad. Sci. USA 97, 8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart, C. B., Schilling, J. W. & Wilson, A. C. (1987) Nature 330, 401-404. [DOI] [PubMed] [Google Scholar]
- 14.Irwin, D. M. (1995) J. Mol. Evol. 41, 299-312. [PubMed] [Google Scholar]
- 15.Irwin, D. M. & Wilson, A. C. (1990) J. Biol. Chem. 265, 4944-4952. [PubMed] [Google Scholar]
- 16.Bush, R. M. (2001) Nat. Rev. Genet. 2, 387-392. [DOI] [PubMed] [Google Scholar]
- 17.Li, W.-H. (1993) J. Mol. Biol. 36, 96-99. [Google Scholar]
- 18.Yang, Z. (1994) J. Mol. Evol. 39, 306-314. [DOI] [PubMed] [Google Scholar]
- 19.McDonald, J. H. & Kreitman, M. (1991) Nature 351, 652-654. [DOI] [PubMed] [Google Scholar]
- 20.Messier, W. & Stewart, C.-B. (1997) Nature 385, 151-154. [DOI] [PubMed] [Google Scholar]
- 21.Yang, Z. (1998) Mol. Biol. Evol. 15, 568-573. [DOI] [PubMed] [Google Scholar]
- 22.Barry, C. E., III, Lee, R. E., Mdluli, K., Sampson, A. E., Schroeder, B. G., Slayden, R. A. & Yuan, Y. (1998) Prog. Lipid Res. 37, 143-179. [DOI] [PubMed] [Google Scholar]
- 23.Davis, J., Smith, A. L., Hughes, W. R. & Golomb, M. (2001) J. Bacteriol. 183, 4626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domenech, P., Barry, C. E., III, & Cole, S. T. (2001) Curr. Opin. Microbiol. 4, 28-34. [DOI] [PubMed] [Google Scholar]
- 25.Wolf, Y. I., Kondrashov, A. S. & Koonin, E. V. (2000) Genome Biol. 1, 0013.1-0013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamieldien, J., Ptitsyn, A. & Hide, W. (2002) Trends Genet. 18, 5-8. [DOI] [PubMed] [Google Scholar]
- 27.Kanehisa, M., Goto, S., Kawashima, S. & Nakaya, A. (2002) Nucleic Acids Res. 30, 42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolattukudy, P. E., Fernandes, N. D., Azad, A. K., Fitzmaurice, A.-M. & Sirakova, T. D. (1997) Mol. Microbiol. 24, 263-270. [DOI] [PubMed] [Google Scholar]
- 29.Wolfe, K. H. & Shields, D. C. (1997) Nature 387, 708-713. [DOI] [PubMed] [Google Scholar]
- 30.Hughes, A. L. (2002) Trends Genet. 18, 433-434. [DOI] [PubMed] [Google Scholar]
- 31.Altschul, S. F., Gis, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huelsenbeck, J. P. & Ronquist, F. R. (2001) Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- 34.Swofford, D. L. (1998) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 35.Yang, Z. (1997) Comp. Appl. Biosci. 13, 555-556. [DOI] [PubMed] [Google Scholar]
- 36.Goldman, N. & Yang, Z. (1994) Mol. Biol. Evol. 11, 725-736. [DOI] [PubMed] [Google Scholar]
- 37.Yang, Z. (1998) Mol. Biol. Evol. 15, 568-573. [DOI] [PubMed] [Google Scholar]
- 38.Yang, Z. & Bielawski, J. P. (2000) Trends Ecol. Evol. 15, 496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, Z. & Nielsen, R. (2002) Mol. Biol. Evol. 19, 908-917. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen, R. & Yang, Z. (1998) Genetics 134, 1271-1276. [Google Scholar]
- 40.McInerney, J. O. (1998) Bioinformatics 14, 372-373. [DOI] [PubMed] [Google Scholar]
- 41.Medigue, C., Rouxel, T., Vigier, P., Henaut, A. & Danchin, A. (1991) J. Mol. Biol. 222, 851-856. [DOI] [PubMed] [Google Scholar]
- 42.Tekaia, F., Gordon, S. V., Garnier, T., Brosch, R., Barrell, B. G. & Cole, S. T. (1999) Tubercule Lung Dis. 79, 329-342. [DOI] [PubMed] [Google Scholar]
- 43.Brosch, R., Gordon, S. V., Eiglmeier, K., Garnier, T. & Cole, S. T. (2000) Res. Microbiol. 151, 135-142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.