Abstract
X-linked dystonia parkinsonism (XDP) is an X-linked recessive adult onset movement disorder characterized by both dystonia and parkinsonism. We report delineation of the disease gene within a 300-kb interval of Xq13.1 by allelic association. Sequencing of this region in a patient revealed five disease-specific single-nucleotide changes (here referred to as DSC) and a 48-bp deletion unique to XDP. One of the DSCs is located within an exon of a not previously described multiple transcript system that is composed of at least 16 exons. There is a minimum of three different transcription start sites that encode four different transcripts. Two of these transcripts include distal portions of the TAF1 gene (TATA-box binding protein-associated factor 1) and are alternatively spliced. Three exons overlap with ING2 (a putative tumor suppressor) and with a homologue of CIS4 (cytokine-inducible SH2 protein 4), both of which are encoded by the opposite strand. Although all DSCs are located within this multiple transcript system, only DSC3 lies within an exon. This exon is used by all alternative transcripts making a pathogenic role of DSC3 in XDP likely. The multiple transcript system is therefore referred to as DYT3 (disease locus in XDP).
Keywords: X chromosome, positional cloning, single-nucleotide polymorphism, alternative splicing, transcription factor
X-linked dystonia parkinsonism (XDP, OMIM no. 314215) is an X-linked recessive adult onset movement disorder characterized by generalized dystonia and parkinsonism in ≈50% of cases (1, 2). The disease is invariably fatal, and death occurs after a duration of up to 40 years by starvation (due to oropharyngeal dystonia) or secondary infections (mainly aspiration pneumonia). Postmortem analysis of patients' brains reveals atrophy of both caudate and putamen (3, 4). XDP originated by a genetic founder effect in the Philippine island of Panay and subsequently spread across the major Philippine islands. The disorder has also been diagnosed in Canada and in the U.S. To date, however, XDP is known only in patients of Filipino descent, suggesting genetic homogeneity of the disease.
Because the molecular pathology of XDP is likely to provide insights into both common form(s) of Parkinson's disease and dystonia, we decided to study XDP in more detail and set out to identify the underlying genetic defect. After initial assignment of the disease locus, DYT3, to the proximal long arm of the X-chromosome by linkage analysis (5-7), we took advantage of genetic homogeneity of the disorder and delineated DYT3 by association studies (8, 9) within Xq13. We then constructed a yeast artificial chromosome (10) and a bacterial artificial chromosome/P1-derived artificial chromosome/cosmid contig (11) of the critical region in Xq13.1. Association studies with short tandem repeat polymorphisms (STRPs) from the region facilitated assignment of DYT3 to an interval of ≈400 kb flanked by DXS10016 proximally and DXS559 distally (11). The entire critical interval has now been sequenced as part of the Human Genome Project (accession no. AL590763), and eight genes have been detected. They include (from proximal to distal) DXS6673E, a gene of unknown function (12); NonO (13, 14) coding for p54nrb (a nuclear RNA-binding protein); a gene coding for muscle-specific melusin (15) (that interacts with integrin); TAF1 (formerly CCG1) coding for TATA box-binding protein-associated factor (TAF)II250 (16, 17); ING2 (18), a putative tumor suppressor gene that is a member of the p33ING1 gene family; OGT, the gene for O-linked N-acetylglucosamine transferase (19-21); ACRC coding for a putative nuclear protein containing an acidic amino acid repeat (22); and CXCR3 that codes for a G-protein-coupled receptor (23). Extensive sequence analyses of both coding and noncoding regions of these genes in XDP patients did not reveal a mutation (10, 11, 22, 24), suggesting that XDP is caused by either a small structural rearrangement, a mutation in a regulatory element of a known gene, or a mutation in a hitherto unknown gene.
Materials and Methods
Patients. XDP patients were the same as those investigated before (1, 11). Controls included the 105 Filipino males studied before (1, 11) and an additional 73 male and female adult Filipinos (comprising 103 X chromosomes) with a negative family history of XDP. Blood was drawn after informed consent. Genomic DNA of patients and controls was isolated from blood samples by standard procedures.
Sequence Analysis. The 260-kb sequence (Fig. 1a) from one XDP patient was obtained by cycle sequencing of overlapping PCR products. The fragments amplified were sequenced on an ABI3700 automated sequencer with the BigDye Ver. 3.1 Cycle Sequencing kit (Applied Biosystems). Fragments were aligned by using dnastar software.
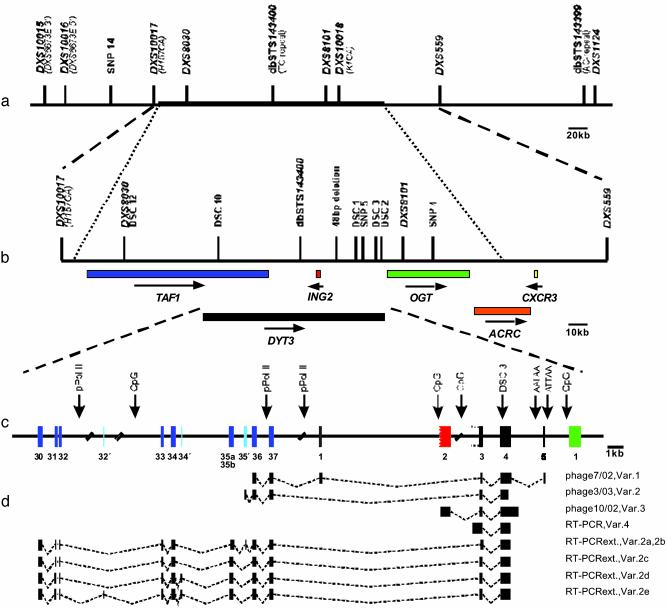
Fig. 1.
Genomic delineation and analysis of DYT3.(a) Contig of the initial XDP critical region spanning ≈400 kb (11). The thick upper line indicates the 260 kb sequenced in an XDP patient. (b) The genes assigned to the newly defined critical interval of 300 kb are given in color. Blue, TAF1 (17) (TATA box-binding protein associated factor 1); red, ING2 (18) (inhibitor of growth family member 2); green, OGT (19 -21) (O-linked-N-acetylglucosamine transferase); yellow, CXCR3 (23) [chemokine (C-X-C motif) receptor 3]; black, DYT3 (gene mutated in XDP); orange, ACRC (22) (acidic repeat-containing gene). Orientation of transcription is indicated by arrows. (c) Genomic structure of DYT3. Exons derived from TAF1 (blue) are numbered according to TAF1 nomenclature. Novel exons identified within TAF1 are marked light blue. The first facultative exon of DYT3 is numbered 1, followed by exons 2-5. Exon 2 is highlighted red, because ING2 covers part of its opposite strand. The first exon of OGT is given in green. Potentially relevant CpG islands, putative Pol II promoters, and polyadenylation sites are indicated. (d) Alternative transcripts 1, 2, and 3 were discovered in phage (phage 7/02, phage 3/03, and phage 10/02, respectively). Transcript 2 was extended by RT-PCR by using primer 189510R derived from exon 4 in combination with various TAF1-specific primers. These experiments resulted in the identification of splice variants a-e of transcript 2.
STRPs. STRPs dbSTS143400 and dbSTS143399 were detected by PCR on the basis of analysis of the published sequence of Xq13.1 (AL590763 and AL135749.3). Primers and amplification conditions are given in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org. dbSTS143400 is a TC repeat and dbSTS143399, an AC repeat. The remaining STRPs were the same as used previously (11).
Single-Nucleotide Polymorphisms (SNPs), Disease-Specific Single-Nucleotide Changes (DSCs), and 48-bp Deletion. SNPs confined to patients and DSCs were discovered by comparing the DNA sequence from a patient to sequence AL590763 by using dnastar software. Primers and amplification conditions are given in Table 4, which is published as supporting information on the PNAS web site. The characteristics of two additional SNPs identified in the region are given in Table 5, which is published as supporting information on the PNAS web site.
RT-PCR. For the identification of a transcript spanning the DSC3 region, polyA+RNA from lymphoblastoid cells of a patient and a control was transcribed in cDNA by using SuperScript II (GIBCO/BRL). RT-PCR was performed under standard conditions for 35 cycles by using primers 187750F and 189510R (Table 6, which is published as supporting information on the PNAS web site). The resulting 782-bp fragment was separated on an agarose gel, purified by using the QIAquick Gel Extraction kit (Qiagen, Chatsworth, CA), and sequenced as described above. To confirm results, RT-PCR was repeated on commercially available polyA+RNA from nucleus caudatus and human fetal brain (CLONTECH).
For extension of transcript 2, RT-PCR was performed on poly(A)+RNA from nucleus caudatus (CLONTECH). Reverse primers 187680R, 189490R, and 189510R (Table 6) were derived from exons 3 and 4. Amplification of large splice variants of transcript 2 required nested PCR that was performed in 35 cycles by using the exon 4-derived primer 189510R, followed by 30 cycles with either exon 3-derived primer 187680R or exon 4-derived nested primer 189490R. Library Screening. To identify a cDNA library containing a transcript of DYT3, four libraries were screened by PCR with primers 187750F and 189510R. The cDNA libraries were human fetal brain large insert, human fetal brain, human leukocyte, and human placenta (CLONTECH). The above-described 782-bp fragment was detected only in the human fetal brain λgt10 cDNA library. Because conventional hybridization screening of 1.5 × 106 phage clones had failed, we used a modified method for differential screening of cDNA libraries (25). Briefly, 1 × 107 plaque-forming units were divided into 28 subpools and plated on 150-mm LB-agarose plates. Lysates were generated by overlaying plates with 5 ml of SM buffer (50 mM Tris·HCl, pH 7.5/100 mM NaCl/8 mM MgSO4) for 2 h. PCR analysis on plate lysates with primers 187750F and 189510R identified the four positive subpools 5, 20, 21, and 25. The agarose plate of positive subpool 25 was cut in eight pieces and extracted separately. The subpool that gave the strongest amplification of the 782-bp fragment was used for subsequent conventional library screening. Hybridization of nitrocellulose filters (Millipore) was performed in hybridization buffer [6× SSC/5× Denhardt's/1% (wt/vol) SDS/1 M NaCl/50 mM Tris·HCl (pH 7.5)/10% dextran-sulfate/40% formamide] at 42°C. A 316-bp [α-32P]dCTP-labeled fragment generated with primers HybprobeF and 189140R (Table 6) was used as a probe. Phage inserts of positive clones were isolated by using λgt10-specific primers (CLONTECH). Sequencing of inserts was performed by using the 33P-Thermo Sequenase Radiolabeled Terminator Cycle Sequencing kit (Amersham Pharmacia) and the primers listed in Table 6. PCR analysis confirmed that splice variant 3 corresponds to subpools 21 and 25. Splice variant 1 was detected in subpools 5, 20, and 25.
All 28 primary subpools were screened by PCR for the existence of additional splice variants by using primers 120720F (located in exon 37 of TAF1) and 188885R (located proximal in exon 4, Fig. 1c). In addition to positive subpools 5, 20, and 25, a smaller fragment was amplified in subpool 9. Conventional screening of this subpool resulted in phage 3/03.
Results
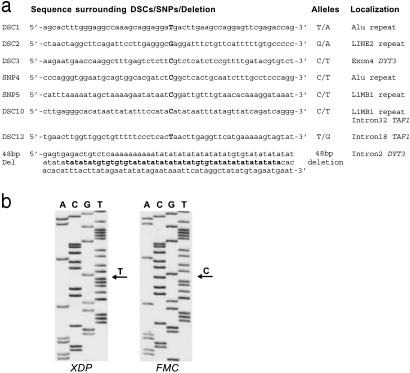
Sequence Analysis of Critical Interval in XDP Patient. We sequenced 87% (260 kb) of the 300-kb critical interval in an XDP patient (Fig. 1a). The 13% not sequenced is distal to O-linked-N-acetylglucosamine transferase, and the genes in this region (ACRC, CXCR3) had been excluded as disease genes in XDP before (ref. 22; unpublished work). Comparison to the published sequence (AL590763) revealed two SNPs (SNP 4 and 5) that were polymorphic in patients only (Fig. 1b), two SNPs that were also polymorphic in controls (Table 5), and five disease-specific single-nucleotide changes (DSC 1, 2, 3, 10, and 12) in this interval (Figs. 1b and 2a). The detection of only four SNPs within the 260 kb of the X-chromosome sequenced indicates that this region of the genome is of unusually low heterozygosity. The DSCs were found in all XDP patients (46 tested) but in none of 178 unaffected male and female Filipino controls (208 X chromosomes) without a family history of XDP. In addition to the XDP-specific DSCs, a 48-bp deletion was detected exclusively in patients.
Fig. 2.
Analysis of polymorphic changes in DYT3 critical region. (a) DSCs 1, 2, 3, 10, and 12; SNPs 4 and 5; and the 48-bp deletion in their sequence context. (b) Sequence analysis of DSC3, the only DSC embedded in an exonic DNA sequence in an XDP patient and a Filipino male control.
Haplotype of Critical Interval in XDP. The DSCs, SNPs 4 and 5, and the 48-bp deletion discovered by sequencing together with STRP loci DXS10017, DXS8030, dbSTS143400 (TC-repeat), DXS8101, and DXS10018 established an XDP-specific haplotype that is interrupted by SNP5 located between DSC1 and DSC3 (Fig. 1b, Table 1). Because SNPs 4 and 5 are not polymorphic in controls, they obviously originated on an ancestral XDP chromosome after the mutation had been introduced into the population. In addition to the 178 male and female Filipino controls, we analyzed DSCs 1, 2, 3, 10, and 12 and the 48-bp deletion in 180 Caucasians, 80 Japanese, and 60 Cubans (total of 629 X chromosomes) and confirmed XDP specificity of the various sequence changes at the corresponding loci. In two Cuban controls, a similar but not identical rearrangement of the 48-bp deletion region was found.
Table 1. Alleles at various STRP, DSC, and SNP loci in XDP patients.
| Patient no. | DXS7119 | DXS10015 | DXS10016 | SNP14 | DXS10017 | DSC12 | DXS8030 | DSC10 | dbSTS143300 | 48 bpDel | DSC1 | SNP5 | DSC3 | DSC2 | DXS8101 | DXS10018 | SNP4 | DXS559 | dbSTS143399 | DXS1124 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 4 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 5 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 8 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 9 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 14 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 17 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 20 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 27 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 29 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 32 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 46 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 242 | 158 | 116 |
| 12 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 13 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 22 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 26 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 28 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 38 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 40 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 43 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 45 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 47 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 37 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 2 | 240 | 158 | 116 |
| 31 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 1 | 1 | 1 | 271 | 142 | 1 | 244 | 150 | 118 |
| 41 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | - | 242 | 158 | 116 |
| 1 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 10 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 11 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 16 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 18 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 19 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 21 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 25 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 33 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 34 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 36 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 35 | 396 | 138 | 130 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 39 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 118 |
| 7 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 240 | 158 | 116 |
| 23 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 240 | 158 | 116 |
| 24 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 246 | 158 | 116 |
| 42 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 248 | 150 | 118 |
| 15 | 396 | 138 | 128 | 1 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 146 | 2 | 242 | 158 | 120 |
| 3 | 400 | 142 | 128 | 2 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 6 | 400 | 142 | 128 | 2 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 44 | 400 | 142 | 128 | 2 | 294 | 1 | 244 | 1 | 194 | 1 | 1 | 2 | 1 | 1 | 271 | 142 | 2 | 242 | 158 | 116 |
| 30 | 400 | 142 | 124 | 2 | 270 | 2 | 242 | 2 | 196 | 2 | 2 | 2 | 2 | 2 | 260 | 144 | 2 | 240 | 150 | 116 |
Note the common XDP-specific haplotype between DXS10017 and DXS10018. This haplotype is interrupted by SNP5, which is not polymorphic in controls (for further details, see text). For SNP and DSC, alleles 1 are the mutated (Fig. 2) and alleles 2 are the wild-type alleles. Patient 30 turned out not to be affected by XDP. Analysis of his core family did not provide evidence of linkage of the disease locus to the X chromosome, and his ethnicity appears to be pure Chinese. Because this patient had been included in previous studies, however, he is listed in this table as well. Boldface indicates recombinations in patients.
Analysis of DNA Sequences Surrounding DSCs and SNPs. The DSCs were not located within either the structural or regulatory region of a known gene. Rather, most changes had occurred within repetitive DNA. DSC1 and SNP4 are located within Alu, and DSCs 2 and 10 and SNP5 lie within long interspersed nuclear element sequences (Fig. 2a).
Only DSC3 is located in a region of unique DNA not related to an annotated gene. Because in silico analysis detected a CIS4 (cytokine-inducible SH2 protein 4) (26) homologous gene, we tested for transcripts in this area. Extensive RT-PCR analysis of polyA+RNA isolated from patient and control lymphoblastoid cells and from human nucleus caudatus by using primers from sequences surrounding DSC3 identified a transcribed fragment of 782 bp that is encoded by two exons separated by an intron of 987 bp. DSC3 is located in one of these exons (exon 4 of Fig. 1c). This transcript is encoded by the opposite strand of the CIS4 homologue. Location of the XDP-specific sequence alteration within this novel transcript prompted us to name the gene DYT3 (designation derived from XDP locus DYT3) in accordance with the nomenclature committee.
Structure of DYT3. To characterize the gene that includes the DSC3-containing exon, we screened several cDNA libraries by using a 316-bp fragment from the proximal part of exon 4 as a probe. Hybridization screening of four cDNA libraries with this probe did not yield a positive signal, indicating low frequency of the transcript. Therefore we applied a PCR-based enrichment method (see Materials and Methods) and isolated two phage clones (Fig. 1d) containing cDNA inserts of 1,313 bp (phage 7/02) and 1,683 bp (phage 10/02). Screening of an additional library subpool with a probe located outside the DSC3 region resulted in the isolation of a third phage with an insert of 1,252 bp (phage 3/03, Fig. 1d). Sequence analysis of the three phage clones and additional RT-PCR experiments demonstrated that the DYT3 gene represents a multiple transcript system. At least three independent transcription start sites define four distinct transcripts (transcripts 1-4).
Transcripts 1 (phage 7/02, Fig. 1d) and 2 (phage 3/03, Fig. 1d) use transcription start sites within the TAF1 (TATA-box-binding protein-associated factor) gene. Both transcripts include the distal exons 36 and 37 but not the last exon 38 of TAF1 (17). Because these transcripts are incomplete at their 5′ ends, we performed RT-PCR by using primers derived from exons 3 and 4 (see Materials and Methods) and various TAF1 exons. We obtained five different splice variants, none of which included exon 1 of transcript 1. Therefore, these PCR products are extensions of phage insert 3/03 and are designated as splice variants 2a-e (Fig. 1). The longest splice variants detected by RT-PCR include exons 30-37 of TAF1 but may be even longer at their 5′ ends. Splice variants 2a and 2b incorporate a small exon not known to be used by TAF1 (exon 35′ of Fig. 1c). Interestingly, a sequence homologous to exon 35′ appears to be used by the autosomal TAF1-like transcript (27). Variants 2a and b differ in the size of exon 35 used (35a, 35b). Whereas this exon is composed of 126 bp in variant 2a, the rare exon 35b of variant 2b does not contain the nine 3′ terminal base pairs (Table 2). Splice variant 2c uses TAF1 exons 30-37 not different from the published TAF1 sequence (17). Splice variant 2d includes an additional small exon of 6 bp, which is designated as 34′ (Fig. 1c, Table 2). Finally, variant 2e incorporates the exon 32′, which is 22 bp in size and overlaps with a long interspersed nuclear element sequence on the opposite strand.
Table 2. Exon-intron boundaries of DYT3.
| Exon | Exon size, bp | 3′Intron/5′exon-3′exon/5′intron | Intron size, bp | Features |
|---|---|---|---|---|
| 30* | (123) 82† | ...CTTGCT ----- CCAGATgtaagc | 734 | |
| 31* | 93 | tactagTCTTGG ----- CGTAAGgtgagt | 87 | |
| 32* | 85 | tcacagAACATC ----- ATAATGgtgggt | 17,457 | pPol II LINE in antisense |
| 32′* | 22 | ttctagGGTACA ----- GTGCAGgtttgt | 12,768 | CpG island |
| 33* | 68 | ttatagGACCTG ----- ACTGAGgtaggt | 503 | |
| 34* | 117 | tctcagTATGAT ----- CCTCAGgtgagt | 149 | |
| 34′* | 6 | tcaaagGCTAAGgtgagt | 3,228 | |
| 35a* | 126 | ccacagCCTCCT ----- ACACGgtagga | 783 | |
| 35b* | 117 | ccacagCCTCCT ----- AAGCAGgtaaca | 792 | |
| 35′* | 102 | actcagATGCGC ----- GCCCCAgtgagt | 300 | |
| 36* | 157 | attcagGAAGGT ----- TCTCTGgtaggc | 917 | pPol II |
| 37* | 178 | ttacagCTATCC ----- CATCAGgtaatc | 24,080 | |
| 1 | 110 | ttcaagGTGATG ----- CACAAGgtactt | <7,318 | |
| 2 | >686 | TTGGCC ----- TCCACTgcaagt | Rare splice donor | |
| 3 | 184 | ttacagGTACCA ----- TTCGCGgtgagt | 34,710 | |
| 3, var. 4 | >724 | TCAATA ----- TTCGCGgtgagt | <34,170 | |
| 4 | 692 | atgcagCTTCAG ----- TTCACGgtgagt | 987 | |
| 5 | >110 | gctcagTGGAAT ----- TTATGG... | 1,860 |
Note that intron 2 utilizes a rare splice donor site (gc).
Exons 30-37 are designated according to the TAF1 nomenclature.
Size of exon 30 is based on amplification by using primer 82800F. The actual exon 30 of TAF1 is 123 bp.
Transcript 3 is defined by the cDNA insert from phage clone 10/02 and is composed of exons 2, 3, and 4 (Fig. 1c). Because this transcript could not be extended by RT-PCR, we assume it is initiated at a CpG island immediately upstream of exon 2. Exon 2 and the CpG island overlap with gene ING2 (inhibitor of growth family, member 2) that is transcribed from the opposite strand and might code for a tumor suppressor (18) belonging to the ING1p33 gene family. Variant 3 ends with a 3′ UTR and is probably terminated at the putative polyadenylation site AATAAA located 838 bp downstream of exon 4 (Fig. 1c).
Transcript 4 is a RT-PCR extension to 1,415 bp of the original 782-bp fragment. Given that the 5′ extended exon 3 was not detected in any phage clone, a third transcription initiation site is likely to exist in close proximity to the extended exon 3. Exons 3 and 4 of DYT3 cover part of a CIS4 homologue on the opposite strand.
Although most transcripts use different combinations of DYT3 exons, there are three exons (exons 1 and 2, and 5′ extended exon 3) used by only one transcript each. Taken together, the data suggest that DYT3 is composed of at least 16 exons (Fig. 1c, Table 2) spanning >109 kb of genomic DNA (Fig. 1c, Table 2).
There is no common long ORF represented by all four transcripts. ORFs of transcripts 1, and 2 a, b, c, and d are given by the TAF1 exons included in these transcripts. These exons (at least exons 35-37 but not the terminal exon 38) code for the C-terminal protein kinase of TATA box-binding protein associated factor (TAF)II250 (28). Exon 30 to the proximal region of exon 34 code for the second bromodomain of TAFII250. Transcript 1 starts with an ORF that codes for at least 72 aa before termination at a TGA codon in exon 1. A putative polyadenylation signal ATTAAA is located 39 bp 3′ of exon 5. The proximal ORFs of splice variants 2a, b, c, and d code for 338, 335, 304, and 306 aa, respectively. These four ORFs are terminated in exon 3. It needs to be stressed that a putative ATG start codon is not included in these exons. Possibly splice variants 2a, b, c, and d start at an even more 5′ exon of TAF1 where several CpG islands and putative Polymerase II promoters are located within introns. The insertion of exon 32′ in splice variant 2e interrupts the ORF defined by the TAF1 exons. The longest ORF in splice variant 2e starts with an ATG codon at the distal end of exon 32, terminates in exon 3, and codes for 228 aa.
Transcript 3 starts with an ORF in exon 2 of DYT3 (Fig. 1c), encodes 106 aa, and terminates at a TGA codon within this exon. In silico analysis suggests a 5′ extension of the ORF to code for 149 aa. Finally, transcript 4 would start in exon 3, encode 51 aa, and terminate in exon 4, including DSC3 within the transcript. This small ORF could also be used by the other three alternative transcripts. The DSC3 C→T transition results in the exchange of an Arg for a Cys in this ORF. It needs to be emphasized, however, that the putative start codon of transcript 4 is not within a classical “Kozak context” (29).
Discussion
We have described a multiple transcript system, DYT3, within the XDP critical region. There is evidence that the base change observed within exon 4 (DSC3) of this transcript system (Fig. 1) contributes to the pathogenesis of XDP. (i) DSC3 lies within the core haplotype of XDP. (ii) The mutation is not found in any of 629 X chromosomes from various populations, including 208 X chromosomes from Filipino controls. (iii) DSC3 is the only molecular alteration detected in a mature transcript within the XDP core haplotype. (iv) The exon harboring DSC3 is included in all alternative transcripts of the gene, thus arguing for an important function that is disturbed by the mutation. (v) Consistent with a function, the transcripts are present in the brain, including nucleus caudatus, a region affected in XDP (3, 4). Although there is presently no experimental evidence, one or several of the other XDP-specific sequence changes could contribute to the disease, e.g., by influencing splicing of transcripts.
The discovery of XDP-specific sequence changes now facilitates molecular diagnosis of the disorder.
DYT3 is a multiple transcript system. Such systems are being increasingly recognized and further blur the definition of a gene (30). One example is the RAY1/ST7 tumor suppressor locus that is composed of a multigene system. It includes many overlapping coding and noncoding transcripts, with the latter apparently being regulatory RNAs (31).
The function of DYT3 is not yet known. Two of the four main transcripts include portions of TAF1 coding for transcription factor TAFII250. DYT3, however, is not just an extension of TAF1 for several reasons. There are two transcripts (transcripts 3 and 4) that do not use exons of TAF1 at all. Transcripts 1 and 2 that include portions of TAF1 do not use the large distal exon 38 of this gene and incorporate novel exons not found in TAF1 transcripts (17); and one of the novel exons, i.e., exon 32′ (transcript 2e), interrupts the ORF of TAF1.
Transcripts 1 and 2 code for proteins that share structural homology with the distal part of TAFII250. This portion of a hypothetical DYT3 protein includes the C-terminal protein kinase and one bromodomain of TAFII250. The two bromodomains located in the C-terminal portion of TAFII250 have an intrinsic histone-acetyltransferase activity (32) and promote protein-protein interactions, especially with the acetylated H4 histones. This is important for the function of TAFII250, which together with TATA-binding protein and 10-12 additional TAFIIs is part of transcription factor IID (32, 33). Possibly the putative proteins derived from transcripts 1 and 2 of DYT3 that use several exons of TAF 1 represent novel TAFIIs that might function within transcription complexes.
The presence of ING2 and of the CIS4 homologue on the opposite strand of DYT3 might indicate regulation of these two genes by at least some transcripts of DYT3. Transcript 3 covers portions of ING2 and therefore could be involved in its regulation by antisense RNA, and all four transcripts partially overlap with the CIS4 homologue. Given that all four transcripts include DSC3 containing exon 4, potentially all transcripts could cause XDP by interfering with the function of the CIS4 homologue, provided it is not a pseudogene. An example of potential antisense regulation by a disease gene is SCA8, the ill-defined gene implicated in spinocerebellar ataxia, type 8 (34). Here portions of the SCA8 gene might be a natural antisense RNA, because the SCA8 gene partially overlaps with the Kelch-like 1 (KLHL1) gene that is encoded by the opposite strand (35).
Another possibility of the molecular pathological mechanism of DSC3 action is a missense mutation in variant 4. This transcript potentially encodes a small polypeptide of 51 aa, and the base change would result in an exchange of an arginine for a cysteine. It remains to be clarified, however, whether such small polypeptide does indeed exist. Database searches neither indicate a known polypeptide motif nor point to a potential function.
XDP is a homogeneous disorder introduced by founder effect in the Filipino population (2). Therefore, all XDP cases are caused by the same common sequence change(s) within DYT3. Additional mutations within DYT3 that might contribute to a better understanding of the pathological mechanism of this disease do not exist. Thus, to clarify the molecular pathology of XDP, all possible disease mechanisms need to be investigated experimentally. It will be especially fascinating to learn how DYT3 relates to other genes that have been implicated in various forms of Parkinson's disease and dystonia (36).
Supplementary Material
Acknowledgments
We are grateful to Petra Happel for superb technical assistance and to Angelika Köhler for culturing lymphoblastoid cells. Thanks to P. Propping (Bonn, Germany) for donation of DNA samples from healthy Japanese, to M. Caballero (Havana, Cuba) for DNA samples from healthy Cubans, and to Filipino members of the Deutsch-Philippinischer Freundschaftskreis, Bad Homburg, for contributing blood samples. The technical assistance of M. K. Steen-Müller and U. Aulepp is highly appreciated. We gratefully acknowledge the Dystonia Medical Research Foundation for initial funding of this project (1989-1992) and the Deutsche Forschungsgemeinschaft for support since 1993 (Grants Mu668/3 and 6).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TAFII250, TATA box-binding protein associated factor; XDP, X-linked dystonia parkinsonism syndrome; SNP, single-nucleotide polymorphism; DSC, disease-specific single-nucleotide change; STRP, short tandem repeat polymorphism.
Database deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos.: human DYT3 transcripts, variant 4, AJ549245; variant 3, AJ549246; variant 1, AJ549247; variant 2a, AJ549248; variant 2b, AJ549249; variant 2c, AJ549250; variant 2d, AJ555148; and variant 2e, AJ555149).
References
- 1.Lee, L. V., Kupke, K. G., Caballar-Gonzaga, F., Hebron-Ortiz, M. & Müller, U. (1991) Medicine 70, 179-187. [DOI] [PubMed] [Google Scholar]
- 2.Graeber, M. B. & Müller, U. (1992) Brain Pathol. 2, 287-295. [DOI] [PubMed] [Google Scholar]
- 3.Waters, C. H., Faust, P. L., Powers, J., Vinters, H., Moskowitz, C., Nygaard, T., Hunt, A. L. & Fahn, S. (1993) Movement Disorders 8, 387-390. [DOI] [PubMed] [Google Scholar]
- 4.Altrocchi, P. H. & Forno, L. S. (1983) Neurology 33, 802-805. [DOI] [PubMed] [Google Scholar]
- 5.Kupke, K. G., Lee, L. V. & Müller, U. (1990) Neurology 40, 1438-1442. [DOI] [PubMed] [Google Scholar]
- 6.Kupke, K. G., Graeber, M. B. & Müller, U. (1992) Am. J. Hum. Genet. 50, 808-815. [PMC free article] [PubMed] [Google Scholar]
- 7.Wilhelmsen, K. C., Weeks, D. E., Nygaard, T. G., Moskowitz, C. B., Rosales, R. L., dela Paz, D. C., Sobrevega, E. E., Fahn, S. & Gilliam, T. C. (1991) Ann. Neurol. 29, 124-131. [DOI] [PubMed] [Google Scholar]
- 8.Graeber, M. B., Kupke, K. G. & Müller, U. (1992) Proc. Natl. Acad. Sci. USA 89, 8245-8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller, U., Haberhausen, G., Wagner, T., Fairweather, N., Chelly, J. & Monaco, A. P. (1994) Genomics 23, 114-117. [DOI] [PubMed] [Google Scholar]
- 10.Haberhausen, G., Schmitt, I., Köhler, A., Peters, U., Rider, S., Chelly, J., Terwilliger, I. D., Monaco, A. P. & Müller, U. (1995) Am. J. Hum. Genet. 57, 644-650. [PMC free article] [PubMed] [Google Scholar]
- 11.Németh A. H., Nolte, D., Dunne, E., Niemann, S., Kostrzewa, M., Peters, U., Fraser, E., Bochukova, E., Butler, R., Brown, J., et al. (1999) Genomics 60, 320-329. [DOI] [PubMed] [Google Scholar]
- 12.van der Maarel, S. M., Scholten, I. H., Huber, I., Philippe, C., Suijkerbuijk, R. F., Gilgenkrantz, S., Kere, J., Cremers, F. P. & Ropers, H. H. (1996) Hum. Mol. Genet. 5, 887-897. [DOI] [PubMed] [Google Scholar]
- 13.Dong, B., Horowitz, D. S., Kobayashi, R. & Krainer, A. R. (1993) Nucleic Acids Res. 21, 4085-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallier, M., Tavitian, A. & Moreau-Gachelin, F. (1996) J. Biol. Chem. 271, 11177-11181. [DOI] [PubMed] [Google Scholar]
- 15.Brancaccio, M., Guazzone, S., Menini, N., Sibona, E., Hirsch, E., De Arndr, M., Rocchi, M., Alruda, F., Tarone, G. & Silengo, L. (1999) J. Biol. Chem. 274, 29282-29288. [DOI] [PubMed] [Google Scholar]
- 16.Ruppert, S., Wang, E. H. & Tjian, R. (1993) Nature 362, 175-179. [DOI] [PubMed] [Google Scholar]
- 17.Nakashima, T., Sekiguchi, T., Sunamoto, H., Yura, K., Tomoda, S., Go, M., Kere, J., Schlessinger, D. & Nishimoto, T. (1994) Gene 141, 193-200. [DOI] [PubMed] [Google Scholar]
- 18.Jaeger, D. Stockert, E., Scanlan, M. J., Gure, A. O., Jager, E., Knuth, A., Old, L. J. & Chen, Y. T. (1999) Cancer Res. 59, 6197-6204. [PubMed] [Google Scholar]
- 19.Lubas, W. A., Frank, D. W., Krause, M. & Hanover, J. A. (1997) J. Biol. Chem. 272, 9316-9324. [DOI] [PubMed] [Google Scholar]
- 20.Kreppel, L. K., Blomberg, M. A. & Hart, G. W. (1997) J. Biol. Chem. 272, 9308-9315. [DOI] [PubMed] [Google Scholar]
- 21.Nolte, D. & Müller, U. (2002) Mamm. Genome 13, 62-64. [DOI] [PubMed] [Google Scholar]
- 22.Nolte, D., Ramser, J., Niemann, S., Lehrach, H., Sudbrak, R. & Müller, U. (2001) Neurogenetics 3, 207-213. [DOI] [PubMed] [Google Scholar]
- 23.Marchese, A., Heiber, M., Nguyen, T., Heng, H. H., Saldivia, V. R., Cheng, R., Murphy, P. M., Tsui, L. C., Shi, X., Gregor, P., et al. (1995) Genomics 29, 335-344. [DOI] [PubMed] [Google Scholar]
- 24.Peters, U., Haberhausen, G., Kostrzewa, M., Nolte, D. & Müller, U. (1997) Hum. Genet. 100, 569-572. [DOI] [PubMed] [Google Scholar]
- 25.Thomas, M. G., Hesse, S. A., Foss, Y. J. & Farzaneh, F. (1997) Methods Mol. Biol. 67, 405-418. [DOI] [PubMed] [Google Scholar]
- 26.Masuhara, M., Sakamoto, H., Matsumoto, A., Suzuki, R., Yasukawa, H., Mitsui, K., Wakioka, T., Tanimura, S., Sasaki, A., Misawa, H., et al. (1997) Biochem. Biophys. Res. Commun. 239, 439-446. [DOI] [PubMed] [Google Scholar]
- 27.Wang, P. J. & Page, D. C. (2002) Hum. Mol. Genet. 11, 2341-2346. [DOI] [PubMed] [Google Scholar]
- 28.Dikstein, R., Ruppert, S. & Tjian, R. (1996) Cell 84, 781-790. [DOI] [PubMed] [Google Scholar]
- 29.Kozak, M. (1996) Mamm. Genome 7, 563-574. [DOI] [PubMed] [Google Scholar]
- 30.Snyder, M. & Gerstein, M. (2003) Science 300, 258-260. [DOI] [PubMed] [Google Scholar]
- 31.Vincent, J. B., Petek, E., Thevarkunnel, S., Kolozsvari, D., Cheung, J., Patel, M. & Scherer, S. W. (2002) Genomics 80, 283-294. [DOI] [PubMed] [Google Scholar]
- 32.Mizzen, C. A., Yang, X. J., Kokubo, T., Brownell, J. E., Bannister, A. J., Owen-Hughes, T., Workman, J., Wang, L., Berger, S. L., Kouzarides, T., et al. (1996) Cell 87, 1261-1270. [DOI] [PubMed] [Google Scholar]
- 33.Goodrich, J. A. & Tjian, R. (1994) Curr. Opin. Cell Biol. 6, 403-409. [DOI] [PubMed] [Google Scholar]
- 34.Koob, M. D., Moseley, M. L., Schut, L. J., Benzow, K. A., Bird, T. D., Day, J. W. & Ranum, L. P. W. (1999) Nat. Genet. 21, 379-384. [DOI] [PubMed] [Google Scholar]
- 35.Nemes, J. P., Benzow, K. A. & Koob, M. D. (2000) Hum. Mol. Genet. 9, 1543-1551. [DOI] [PubMed] [Google Scholar]
- 36.Pulst, S. M., ed. (2003) Genetics of Movement Disorders (Academic, New York).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.