Abstract
This work describes a vertebrate homeobox gene, designated Homez (homeodomain leucine zipper-encoding gene), that encodes a protein with an unusual structural organization. There are several regions within Homez, including three atypical homeodomains, two leucine zipper-like motifs, and an acidic domain. The gene is ubiquitously expressed in human and murine tissues, although the expression pattern is more restricted during mouse development. Genomic analysis revealed that human and mouse genes are located at 14q11.2 and 14C, respectively, and are composed of two exons. The zebrafish and pufferfish homologs share high similarity to mammalian sequences, particularly within the homeodomain sequences. Based on homology of homeodomains and on the similarity in overall protein structure, we deliniate Homez and members of ZHX family of zinc finger homeodomain factors as a subset within the superfamily of homeobox-containing proteins. The type and composition of homeodomains in the Homez subfamily are vertebrate-specific. Phylogenetic analysis indicates that Homez lineage was separated from related genes >400 million years ago before separation of ray- and lobe-finned fishes. We apply a duplication-degeneration-complementation model to explain how this family of genes has evolved.
The homeobox-containing proteins play an important role in eukaryotic development (1). These factors possess a helix-turn-helix DNA-binding motif known as a homeodomain (HD), which is conserved in all eukaryotes (1). We have reported (2) the isolation of several proteins that bind the Hoxc8 early enhancer region. In this report, we analyzed the clone OH10, which encodes a previously unreported HD protein.
Materials and Methods
Mouse 129/SvJ genomic high-density gridded bacterial artificial chromosome (BAC) filters (FBAC-4422 library, Genome Systems, St. Louis) were screened with a 2.6-kb OH10 cDNA probe according to the standard protocol. The procedures for DNA isolation, pulse-inversion gel electrophoresis, and fluorescence in situ hybridization analysis were published elsewhere (3). DNA and protein sequences were examined against available public databases by using the various BLAST programs available through the network server at the National Center of Biotechnology Information (www.ncbi.nlm.nih.gov). The search for the ORFs in the genomic clone was done with the GENSCAN program (http://genes.mit.edu/GENSCAN.html). The analysis of the predicted proteins for conserved motifs was carried out with the help of the PFAM HMM database at http://pfam.wustl.edu./hmmsearch.shtml. Multiple alignments were performed by using the CLUSTALW program and PAM-350 scoring matrix. A clocklike maximum-likelihood rooted phylogenetic tree has been calculated with the use of PUZZLE software (4). Repeat elements were characterized by using REPEAT-MASKER2 (http://ftp.genome.washington.edu/cgi-bin/Repeat-Masker). Comparative percentage identity plot sequence alignment was done with PIPMAKER (http://bio.cse.psu.edu/pipmaker). Yeast one-hybrid and electromobility shift assays were perfomed essentially as described (2). The mouse Homez probe for whole-mount in situ hybridization studies was generated by PCR. The following oligonucleotides were used: sense 5′-CTCTTCTCTCTTTCACACAGCA-3′ and antisense with the T7 polymerase-binding site 5′-CCAAGCTTCTAATACGACTCACTATAGGGAGATCCTGACAGTCTGATT-3′. Whole-mount in situ hybridization, RNA dot blot, and Northern blot studies were performed according to standard procedures. The 32P-labeled probes for RNA dot and Northern blots were derived from the mouse 2.6-kb OH10 cDNA clone and the 1.8-kb cDNA of human EST AI332699, respectively.
The His-tagged Homez expression construct pTriExOH10 was made by subcloning a BamHI-NotI DNA fragment, derived from the mouse EST clone (accession no. BF454433), into the pTriEx-4 vector (Novagen). COS-1 cells were grown on coverslips and transfected with the pTriExOH10 expression vector. Cells were incubated with the anti-His monoclonal mouse Ab (Novagen) (1:2,500 dilution) in conjunction with Texas red 488 goat anti-mouse IgG (Molecular Probes) (1:250 dilution).
Results and Discussion
Isolation and Characterization of Homez cDNAs and Encoded Proteins. We used the yeast one-hybrid screen to identify potentially important regulatory proteins involved in Hoxc8 early expression (2). Several clones were identified as strong interactors of the proximal region of the Hoxc8 early enhancer. One of them, OH10, encodes a previously unreported homeobox protein. We termed this protein Homez, which stands for the homeodomain leucine zipper-containing factor. Homez has an interesting structural organization, including the presence of three HDs and two leucine zipper-like motifs.
The partial cDNA sequence, derived from OH10, was used in a BLASTN search of the EST database (dbEST) and as a result several highly homologous human and mouse EST clones were identified. The human and mouse ESTs (GenBank accession nos. AI332699 and BF454433) were completely sequenced. The DNA sequence alignment revealed that the human clone is identical to the deposited entry KIAA1443 (accession no. AB037864) and corresponds to UniGene EST cluster Hs.156051. Based on amino acid sequence alignment results, we concluded that these clones are derived from the same gene. We have also isolated and mapped the mouse genomic BAC clone 416m21, which contains the complete Homez sequence. Based on the UniGene EST cluster Mm.73805 and the sequence derived from the one-hybrid clone OH10, the murine Homez orthologous sequence was deduced. The BLASTN search also identified a rat genomic clone CH230-272D8 (accession no. AC119293) that contains a complete Homez ortholog. The multiple alignment of deduced sequences is shown in Fig. 1. The ORF encodes a putative protein of 549 aa with calculated Mr of 61,111 kDa and pI of 4.89 in humans, and a protein of 518 aa with the Mr of 58,197 kDa and pI of 4.88 in mice, respectively. The rat gene encodes a 513-aa protein with Mr of 57,677 and pI of 4.98. Amino acid sequence alignment indicates almost 79% identity between human and rat homologs in a 525-residue overlap. The amino acid identity between human and mouse orthologs is 77% in a 528-aa overlap and between mouse and rat orthologs is >89% in a 519-aa overlap. The sequence around the first methionine in these genes is in agreement with the Kozak consensus sequence (5). We have observed that the human Homez has an additional 24-aa sequence at its N-terminal end.
Fig. 1.
Amino acid sequence and structural organization of Homez. (A) Amino acid sequence alignment of human, mouse, and rat proteins. Homeodomains are in yellow, leucine zipper motifs are in green, proline-rich motifs are in violet, the serine-rich sequence is in dark blue, and the acidic domain is in blue. Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus.
The putative human 549-aa sequence was analyzed by comparison against the PFAM database of protein domains and hidden Markov models to identify conserved motifs. This secondary structure prediction indicates the presence of three regions within Homez that could form α-helices with a helix-turn-helix motif, a characteristic signature of the homeodomain. The only basic region within the first helix was found in the HD2 of Homez. This region also has a putative nuclear localization signal RKTKRK (Fig. 1). The region within the first helix in the HD3 is rich in proline residues that would alter the structure of the amino-terminal arm. It is highly unlikely that the HD3 possesses a DNA-binding ability, although it could be involved in mediating protein-protein interactions. In addition to three atypical HDs, there are several protein-binding regions, including two leucine zippers, proline- and serine-rich motifs, and an acidic domain (Fig. 1). The presence of these structural motifs within the protein sequence and the fact that Homez binds to DNA in one-hybrid assays implicate it as a transcriptional regulator.
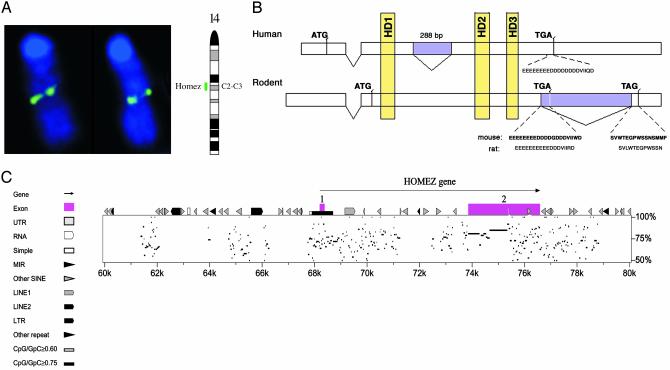
Isolation of Homez Genomic Clones and Analysis of Alternatively Spliced Products. A search of National Center for Biotechnology Information databases with the Homez cDNA sequence identified a human BAC genomic clone R-124D2 (accession no. AL049829) from chromosome 14. Based on sequence alignment, we deduced that the gene is localized at the 14q11.2 region (www.ensembl.org) and is encoded by two exons. The first exon (≈120 bp) contains a translational start codon, and the second exon (3,128 bp) contains most of the coding sequence as well as 1, 515 bp of 3′ UTR. The two exons are separated by an intronic region of 8,725 bp (Fig. 2B). Direct comparison of cDNA and genomic sequences revealed alternative splicing of human Homez mRNAs: some transcripts (BQ650619 and BQ064437 in dbEST) lack a segment between the first and second homeodomains. This segment has all of the characteristic features of an intron and is spliced in frame so that this optional intron does not change the downstream amino acid sequence of Homez. This alternatively spliced transcript encodes a 418-aa protein with Mr of 47,310 and pI of 4.67. Variation of nucleotide sequences make the generation of a similar mRNA isoform in mouse impossible, indicating that it is a human-specific variant.
Fig. 2.
(A) Chromosomal mapping of the mouse Homez locus by fluorescence in situ hybridization. Location of the 416m21 BAC is identified by the fluorescent signal (pseudocolored green). (B) Genomic structure of the Homez genes. (C) The percentage identity plot sequence alignment analysis.
We have isolated the mouse BAC clone 416m21 and used it in fluorescence in situ hybridization analysis. Our mapping data show that Homez localized at the 14 C2-C3 region (Fig. 2A). Sequence analysis of public database identified the mouse genomic clone RP24-90 (accession no. 116591.4), which was mapped in silico to 14C1 cytoband (www.ensembl.org). This mouse cytoband is a region of known synteny to human 14q11.2 region (Oxford Grid; www.informatics.jax.org/searches/oxfordgrid_form.shtml) and, consequently, this finding confirmed the orthologous relationship of murine and human Homez.
Based on comparison of these sequences, it was deduced that the second exon of the mouse ortholog is interrupted by an additional intron at the end of the coding sequence. This results in different C termini of mouse Homez and formation of a protein of 513 aa (Fig. 2B). A longer mouse isoform of 518 aa, which contains an acidic tail similar to the human protein, was isolated by the one-hybrid screen (2). The high level of sequence homology suggests that two similar C-terminal isoforms also could exist in rat (Fig. 2B), but the absence of rat cDNAs in dbEST prevents a confident identification at the present time. In contrast, the human genome sequence is divergent from rodents, and nucleotide sequence analysis shows only one variant of human C terminus, specifically a long acidic domain (Fig. 2B). In mouse and rat, the first exon is noncoding and translation starts from the second exon. Sequences within the first exon in human and rodents retain significant homology, so that the sliding of the translation start is likely to result from the usual accumulation of neutral mutations during evolution as observed frequently for orthologs.
Mouse and human genomic sequences were used for percentage identity plot analysis to verify that the deduced genomic structure of the Homez genes is complete. Data in this analysis, shown in Fig. 2C, indicate that homology between two species is limited to already known exon sequences.
Orthologs from Different Species. The BLASTN search identified a Takifugu rubripes genomic sequence (clone S001210) in the Fugu Genomics Project database (Medical Research Council Human Genome Mapping Project Resource Centre; http://fugu.hgmp.mrc.ac.uk) and Danio rerio cDNA sequence sharing a high similarity to Homez (accession nos. AF531077 and CAD60854). The T. rubripes protein is 589-aa-long and has a calculated Mr of 65,973 and pI of 6.25. There is almost 28% identity in a 521-residue ovelap between human and pufferfish homologs (Fig. 3). The D. rerio ortholog encodes a 650-aa protein with Mr of 72,997 and pI of 8.74. The identity is >27% in a 577-aa overlap between human and zebrafish homologs (Fig. 3). Homology between fish and human is limited mainly to homeodomain sequences in which the similarity of HD is maintained at a high level: 77% and 71% for HD1, 76% and 78% for HD2, and 63% and 63% for HD3 between human and zebrafish and between human and pufferfish, respectively. The coding sequence of zebrafish Homez is located within an entire exon, whereas the pufferfish ortholog contains an intron between HD1 and HD2 (different from the human optional intron). Alternative splicing at the C terminus of Homez in both fish species is likely to be absent. In contrast to human Homez, fish homologs lack the C-terminal acidic domain.
Fig. 3.
Sequence alignments of Homez homologs. Human, rat, zebrafish, and pufferfish proteins were aligned. Homeodomain-containing regions are shown on a yellow background. Dr, Danio rerio; Hs, H. sapiens; Rn, R. norvegicus; Tr, Takifugu rubripes.
We detected several ESTs from different vertebrate species that encode partial sequences with high similarity to the human gene. The human sequence shares >77% identity with the Sus scrofa EST 365727 (GenBank accession no. BI340452) in a 179-aa overlap, 85% identity with the Canis familiaris EST ha78g02.g1 (accession no. BM537152) in a 182-residue overlap, and >75% identity with the Bos taurus EST 463756 (accession no. BI682619) in a 138-aa overlap (data not shown).
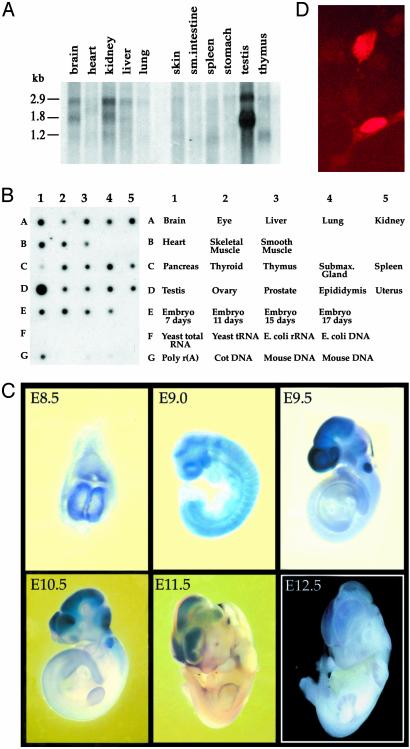
Expression and DNA-Binding Activity. The expression and abundance of Homez mRNA transcripts in human and mouse tissues was examined by Northern blot and RNA dot blot analysis. The RNA dot blot analysis demonstrated a ubiquitous distribution in all human tissues examined with strong expression in adult testis and kidney as well as fetal lung and kidney (data not shown). We have also checked the expression pattern of the murine ortholog. The mouse gene shows a strong expression of a 2.9-kb transcript in testis, kidney, brain, and liver, 1.8-kb transcript in kidney, brain, and liver, and a unique 1.6-kb transcript in testis (Fig. 4A). The RNA dot blot analysis shows expression in all murine tissues examined with a high level of expression in the testis (Fig. 4B). The mouse Homez starts to express early in embryogenesis: we noticed the RNA transcripts in embryonic day (E) 7. The embryonic expression pattern was examined by whole-mount in situ hybridization. At E8.5-E9.0, RNA transcripts were expressed in all developing organs. Later on, during embryogenesis Homez shows a more restricted expression pattern. At E9.5-E12.5, we detected relatively strong expression in the developing brain, the optic vesicle, and the otic placode (Fig. 4C). The subcellular localization of Homez was determined by transfection studies in COS-1 cells. When cells were transfected with epitope-tagged Homez, the fluorescence was preferentially detected in the nucleus (Fig. 4D).
Fig. 4.
Distribution of Homez mRNA transcripts and protein in various mouse tissues and cells. (A and B) Mouse RNA dot blot array (Clontech) and Northern blot (Origene, Rockville, MD) were hybridized according to the manufacturer's recommendations. (C) Expression of the Homez gene during embryogenesis. Whole-mount in situ hybridization of developing embryos from E8.5 to E12.5 with the Homez-specific RNA probe. (D) Subcellular localization of Homez in COS-1 cells.
Yeast one-hybrid and gel-shift experiments indicate a significant interaction between Homez and the Hoxc8 regulatory region, confirming that Homez possesses a sequence-specific DNA-binding activity (data not shown).
Analysis of Homez-Related Homeobox Genes in the Genome. The three HDs of Homez possess a very weak similarity to each other. All of them contain residues that are highly conserved throughout the HD superfamily (1). Residues on helix 3 of a helix-turn-helix motif provide energetically significant contacts in the major groove of DNA. The Trp and Phe residues at positions 48 and 49 of the HD are conserved in almost all homeobox proteins. Residue 50 within the recognition helix is important for DNA-binding specificity because it is responsible for the base preference and is a signature residue for different types of HDs. This position is occupied by Met in HD1, by Gly in HD2, and by Asp in HD3. These amino acid positions are unique for Homez. Most HDs have an Asn at position 51, whereas in Homez HD1 has an Ala, HD2 has an Asp, and HD3 has a Ser. Asn makes contacts with a conserved alanine in the binding site of all HDs. The positions 53 and 55, occupied by Arg and Lys residues in other homeodomains, are responsible for direct electrostatic interactions with the DNA. The Arg residue at 53 is conserved in HD1 and -2, although this position is occupied by the leucine in HD3. The Lys at 55 is not conserved in Homez HDs: in HD1 it is Arg, and in HD2 and -3 it is replaced by Ala and Gln, respectively. The Tyr at 25 and Ile at 47 are involved in hydrophobic interactions with DNA. Tyr is conserved in HD1 only, whereas Trp and Glu occupy this position in HD2 and -3. Ile is conserved in HD2, and this position is occupied by Thr and Asp in HD1 and -3, respectively. The basic region proceeding helix 1 provides an energetically significant interaction in the minor groove. The HD1 and -3 of Homez lack this basic region; in fact, HD3 has a proline-rich stretch, which alters the configuration of the N-terminal arm. Based on the pattern of conservation of the critical amino acid residues within Homez HDs, we conclude that HD3 is an atypical HD. It is unlikely that HD3 makes a contact with DNA, although it may have a function(s) in protein-protein interactions. The HD2 has preserved many features of a canonical HD and is likely to be responsible for DNA interactions. However, we cannot exclude the possibility that HD1 has also retained nucleic acid-binding ability. Recent studies showed that the basic N-terminal arm of the HD is not critical in DNA binding (6).
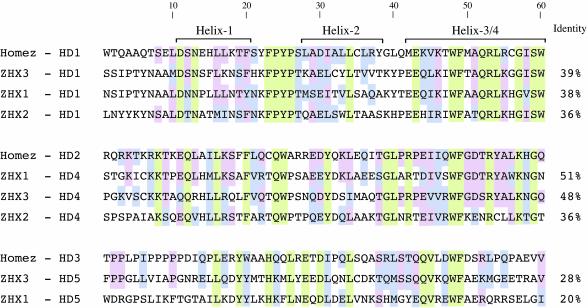
Alignment of Homez proteins from mammals and fish revealed that the homeobox sequences are more conserved than the inter-domain regions (Fig. 3). Because of this, individual HDs of human Homez were used as template in GenBank homology search. The results of this search are summarized in Fig. 5. The HD1 is most closely related to the first HD of zinc finger homeodomain (ZHX)3/KIAA0395 (amino acid 304-364, accession no. CAA18538) (39% identity, 65% similarity), a human zinc finger homeobox protein, followed by the HD1 of ZHX1 (amino acid 284-344, accession no. NP_009153) (38% identity, 71% similarity), and HD1 of ZHX2/KIAA0854 (amino acid 262-322, accession no. NP_055758) (36% identity, 65% similarity). The HD2 shares similarity with the HD4 (amino acid 660-720) of ZHX1 (51% identity, 74% similarity), the HD4 (amino acid 764-824) of ZHX3 (48% identity, 75% similarity), and the HD4 (amino acid 628-688) of ZHX2 (36% identity, 64% similarity). The HD3 is related to the last homeodomain (HD5) of ZHX1 and ZHX3 (28% and 20% identity and 52% and 33% similarity, respectively).
Fig. 5.
Genomic search for Homez-related genes. Alignment of the Homez homeodomains to the known homeodomain sequences in database. Residues identical in all HD sequences are shown on a green background, similar amino acids are on a violet background, and amino acids identical to Homez are on a pink background. Percentage identities to the Homez homeodomains are shown on the right.
Although members of the ZHX family are bigger than Homez and contain two extra HDs in the middle, their overall architechture is similar to that of Homez (7-10). The homeobox sequences are conserved in accordance with their position; i.e., the first HDs are more structurally related to each other than to the other homeoboxes within the same protein. The same can be said for the HD2 of Homez and the HD4 of the ZHX family. The HD3 of Homez lacking the N-terminal α-helix also has a similarly truncated homeodomain (HD5) at the C terminus of ZHX1 and ZHX3 (the C terminus of ZHX2/KIAA0854 is divergent). In addition to homology in the amino acid sequence, these genes along with Homez have a similar genomic organization when the whole or almost the whole coding sequence appears as a long continuous exon (data not shown). A high level of sequence homology between similarly positioned homeoboxes, as well as the similarity of exon-intron structure, allows the delineation of Homez and members of ZHX family as a discrete subset within the superfamily of homeodomain-containing proteins.
This conclusion is also supported by findings in fish. Although orthologous members were absent in the protein database, translated BLAST searches in dbESTs in combination with searches in genome databases allowed us to identify two previously unreported zebrafish proteins with clear homology to ZHX2 and ZHX3 (data not shown). In contrast, a protein, which contains segments with similarity to all homeoboxes of Homez, appears to be absent in Drosophila and in Caenorhabditis elegans and furthermore the level of similarity to HDs at the top list of the BLAST search is much lower. This indicates that both the type of homeoboxes in the Homez/ZHX subfamily and their composition are vertebrate-specific.
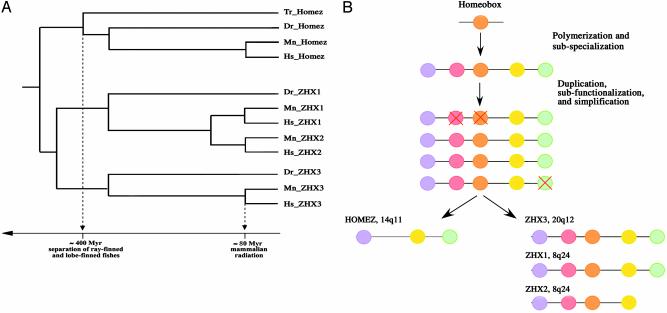
Evolution of Homez-Related Genes. Despite an obvious relation of Homez to the ZHX family, the reconstruction of an evolutionary tree from alignment and comparison of the available sequences shows that the Homez lineage was separated from related genes >400 million years ago, before the separation of ray-finned and -finned fishes (Fig. 6A). The prototype of the Homez/ZHX subfamily was likely to contain at least five homeoboxes. We propose that a variant of the duplication-degeneration-complementation model (11-13) can be applied to describe how this family of genes has evolved (Fig. 6B). First, polymerization of an ancestral homeobox has potentially lead to subspecialization of the descendant homeodomains. Second, duplication of these genes could occur both as a result of global genome duplication (ZHX3 and ZHX1 are located at 20q12 and 8q24, regions where some tetralogs grouped) and as a result of regional duplication (ZHX1 and ZHX2 both are located at 8q24.13). Third, subfunctionalization as a result of independent and complementary mutations in cis-regulatory elements decreases redundancy and allows duplicated genes to be retained in the genome. Selection can therefore act independently on each duplicate, increasing its functional specificity. Fourth, homeoboxes that lost their functions as a result of subfunctionalization and/or neo-functionalization of a whole gene, could be eliminated from such a gene. Homez protein, lacking two central HDs, perhaps is an example of this kind of simplification. Human Homez also shows how this could occur: its gene contains the optional coding segment (”optional intron”) between the first two HDs. The evolutionary fixation of “-” splice variant could be a mechanism that leads to further protein simplification (Fig. 6B).
Fig. 6.
(A) Phylogenetic tree of Homez-related genes. A clocklike maximum-likelihood rooted phylogenetic tree has been calculated with the use of puzzle software (4). The evolutionary sequences between sequences are not drawn to scale. (B) The evolution of Homez-related genes is explained by a duplication-degeneration-complementation model.
Conclusion. In this report, we have described a homeobox factor with an interesting protein organization. The combination of a HD and a leucine zipper is not well known in vertebrates, however, it is a quite common protein structure in plants and fungi (14-16). The presence of leucine zippers suggests that Homez can function as a homo- or/heterodimer in the nucleus. The similarity of exon-intron organization allowed us to deliniate Homez and related members of ZHX family as a distinct subset within the superfamily of homeobox genes. The type of HDs in the Homez/ZHX subfamily and their composition is specific to vertebrate lineage. ZHX1 and ZHX3 members of the ZHX family of zinc finger HD proteins were characterized as transcription factors. The HD1 of ZHX1 and ZHX3 is involved in dimerization with the activation domain of the subunit A of nuclear factor-Y (NF-YA). The repressor domain was mapped to a region that includes the HD1 in ZHX3 and to the acidic domain in ZHX1 (8-10). The HD4 of ZHX1 is responsible for the DNA-binding activity. Based on the structural similarity between the HD1 and HD2 of Homez and the HD1 and HD4 of ZHX1 and ZHX3, respectively, we predict that the HD1 and the first leucine zipper of Homez are responsible for dimerization and that the HD2 possesses specific DNA-binding activity. Homez may also work as a transcriptional repressor because of the presence of a C-terminal acidic region. The fact that Homez has a specific DNA-binding activity and the presence of different structural domains, including serine and proline-rich motifs suggests that this protein is involved in a complex regulatory network.
Acknowledgments
We are grateful to Dr. Chi-hua Chiu and Hubert Lam for critical reading of our manuscript. This work was supported by National Institutes of Health Grant NS43525.
Abbreviations: BAC, bacterial artificial chromosome; En, embryonic day n; HD, homeodomain; ZHX1, zinc finger homeodomain 1.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF463423, AY258064, AY126016, AF531077, AY311507, and AY311506).
References
- 1.Gehring, W. J., Affolter, M. & Burglin, T. (1994) Annu. Rev. Biochem. 63, 487-526. [DOI] [PubMed] [Google Scholar]
- 2.Bayarsaihan, D. & Ruddle, F. H. (2000) Proc. Natl. Acad. Sci. USA 97, 7342-7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayarsaihan, D., Dunai, J., Greally, J. M., Kawasaki, K., Sumiyama, K., Enkhmandakh, B., Shimizu, N. & Ruddle, F. H. (2002) Genomics 79, 137-143. [DOI] [PubMed] [Google Scholar]
- 4.Strimmer, K. & von Haeseler, A. (1997) Proc. Natl. Acad. Sci. USA 94, 6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak, M. (1999) Gene 234, 187-208. [DOI] [PubMed] [Google Scholar]
- 6.Montclare, J. K. & Schepartz, A. (2003) J. Am. Chem. Soc. 125, 3416-3417. [DOI] [PubMed] [Google Scholar]
- 7.Yamada, K., Printz, R. L., Osawa, H. & Granner, D. K. (1999) Biochem. Biophys. Res. Commun. 261, 614-621. [DOI] [PubMed] [Google Scholar]
- 8.Kawata, H., Yamada, K., Shou, Z., Mizutani, T., Yazawa, T., Yoshino, M., Sekiguchi, T., Kajitani, T. & Miyamoto, K. (2003) Biochem. J. 373, 747-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada, K., Kawata, H., Shou, Z., Hirano, S., Mizutani, T., Yazawa, T., Sekiguchi, T., Yoshino, M., Kajitani, T. & Miyamoto, K. (2003) Biochem. J. 373, 167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada, K., Kawata, H., Matsuura, K., Shou, Z., Hirano, S., Mizutani, T., Yazawa, T., Yoshino, M., Sekiguchi, T., Kajitani, T. & Miyamoto, K. (2002) Biochem. Biophys. Res. Commun. 297, 368-374. [DOI] [PubMed] [Google Scholar]
- 11.Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L. & Postlethwait, J. (1999) Genetics 151, 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch, M. & Force, A. (2000) Genetics 154, 459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prince, V. E. & Pickett, F. B. (2002) Nat. Rev. Genet. 3, 827-837. [DOI] [PubMed] [Google Scholar]
- 14.Schena, M. & Davis, R. W. (1992) Proc. Natl. Acad. Sci. USA 89, 3894-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schena, M. & Davis, R. W. (1994) Proc. Natl. Acad. Sci. USA 91, 8393-8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakakibara, K., Nishiyama, T., Kato, M. & Hasebe, M. (2001) Mol. Biol. Evol. 18, 491-502. [DOI] [PubMed] [Google Scholar]