Abstract
Toll-like receptor 4 (TLR4) is a cell-surface receptor that activates innate and adaptive immune responses. Because it recognizes a broad class of pathogen-associated molecular patterns presented by lipopolysaccharides and lipoteichoic acid, TLR4 is a candidate gene for resistance to a large number of diseases. In particular, mouse models suggest TLR4 as a candidate gene for resistance to major agents in bovine respiratory disease and Johne's disease. The coding sequence of bovine TLR4 is divided into three exons, with intron/exon boundaries and intron sizes similar to those of human TLR4 transcript variant 1. We amplified each exon in 40 individuals from 11 breeds and screened the sequence for single-nucleotide polymorphisms (SNPs). We identified 32 SNPs, 28 of which are in the coding sequence, for an average of one SNP per 90 bp of coding sequence. Eight SNPs were nonsynonymous and potentially alter specificity of pathogen recognition or efficiency of signaling. To evaluate the functional importance of these SNPs, we used codon-substitution models to detect diversifying selection in an extracellular region that may physically interact with ligands. One nonsynonymous SNP is located within this region, and other substitutions are in adjacent regions that may interact with coreceptor molecules. The 32 SNPs were found in 20 haplotypes that can be assigned to geographic ranges of origin. Haplotype-tagging SNP analysis indicated that 12 SNPs need to be genotyped to distinguish these 20 haplotypes. These data provide a basic understanding of bovine TLR4 sequence variation and supply haplotype markers for disease association studies.
Toll-like receptors are a family of proteins that perform two functions: recognition of pathogen ligands and signaling to initiate innate and adaptive immune responses. Whereas the signaling domains of the 10 Toll-like receptors known in mammals are highly conserved, the leucine-rich repeat ligand-recognition domains are more diverse to accommodate recognition of different pathogen-associated molecular patterns. In conjunction with the coreceptor MD-2, Toll-like receptor 4 (TLR4) recognizes lipopolysaccharide and the structurally similar lipoteichoic acid, components of Gram-negative and Gram-positive bacterial cell walls, respectively (1-3).
Lack of TLR4 can cripple immune responses to pathogens that produce these ligands. Salmonella typhimurium is a Gram-negative pathogen to which mice have some natural resistance. A comparison of closely related strains of mice showed a reduction of LD50 from 2,000 or more organisms for the homozygous normal TLR4 strain, to <2 organisms for mice homozygous null at TLR4 (4).
Other phenotypes in model mammalian systems point to TLR4 as a candidate gene for resistance to two of the most devastating bovine disorders in the U.S., bovine respiratory disease and Johne's disease. Bovine respiratory disease complex (”shipping fever”) is a common disorder involving many component pathogens. However, one of the most important is Mannheimia haemolytica (formerly Pasteurella haemolytica), which normally resides in the upper respiratory tract of cattle, but on stress can invade the lower respiratory tract and cause disease. A related organism, Pasteurella pneumotropica, has similar features in murine infections. TLR4-null mutant mice were more likely to develop infection with experimental P. pneumotropica pneumonia than their wild-type counterparts (5). This finding suggests TLR4 may play a role in preventing the establishment of such related disorders as bovine shipping fever. TLR4 also has been shown to play a role in resistance to Streptococcus pneumoniae-induced pneumonia (6), further demonstrating the versatility of this important receptor for respiratory immune protection.
TLR4 is also a candidate gene for resistance to both bovine tuberculosis and Johne's disease. These disorders are caused by Mycobacterium bovis and Mycobacterium avium subsp. paratuberculosis, respectively (7, 8). It has recently been shown that TLR4 is required to control the infection of a related pathogen, Mycobacterium tuberculosis, in experimental mice (9). Whereas the modes of infection differ among these pathogens, the basic structures recognized by TLR4 suggest a possible role in resistance to both. Further, because of the broad range of pathogens and potential pathogens that produce its ligands, TLR4 may be considered a candidate gene for resistance to several other infectious diseases.
A cDNA sequence for bovine TLR4 has been reported, with 72% and 65% amino acid similarities to human and mouse TLR4, respectively (10), and TLR4 is known to reside on the distal tip of bovine chromosome 8 (11). However, several issues need to be resolved to further investigate TLR4 as a candidate disease-resistance gene in cattle. First, the genomic structure needs to be established and sufficient flanking intronic sequences gathered to enable simple PCR amplification of the coding portions of the gene. Then, a basic understanding of naturally occurring variation in bovine TLR4 needs to be obtained before alleles can be tested for disease association. Therefore, the present study assesses segregating variation, establishes haplotypic markers for association studies, and provides an insight into the evolution of bovine TLR4. Despite the relative abundance of data on TLR4 in mammalian species, the ligand-binding region has not been identified at a resolution finer than the extracellular domain. We also attempt to delineate a functional region of this domain to better evaluate the importance of the sequence variants we identified.
Materials and Methods
Forty cattle were selected to represent a cross section of the diversity of domestic cattle populations. Thirteen Bos taurus indicus individuals were chosen, including 5 Brahman, 3 Nellore, 2 Gyr, 2 Ankole-Watusi, and 1 Boran. Twenty-seven Bos taurus taurus individuals were chosen, including 6 Angus, 6 Holstein, 5 Texas Longhorn, 4 Limousin, 3 Jersey, and 3 N′Dama. In each breed, individuals were selected to be as unrelated as possible.
To determine the intron/exon boundaries and flanking intronic sequences of bovine TLR4, primers were designed from a consensus sequence composed of coding sequence (GenBank AF310952) and a 3′ EST (GenBank BF889715), and amplicons were sequenced. Intron lengths were obtained by gel electrophoresis of amplicons that spanned the introns. To obtain 5′ UTR sequence, a bovine bacterial artificial chromosome (BAC) library (12) was screened, BACs containing bovine TLR4 were subcloned, and the subclones were sequenced. Primers were then designed to amplify each of the exons with small amounts of flanking sequence.
To screen the diversity panel, each exon was amplified twice for every individual. The separate replicates of each PCR were used for sequencing in the forward and reverse directions, to reduce the risk of reporting PCR artifacts as polymorphisms. The first two exons were amplified by using AmpliTaq Gold (Perkin-Elmer) with a 10-min step at 94°C, followed by 35 cycles of 94°C, the annealing temperature, and 72°C for 30 sec each, and a final 5-min extension at 72°C. The long third exon was amplified by using an Expand Long Template PCR System (Roche Applied Science) with a 2-min step at 95°C, followed by 35 cycles alternating 30 sec at 95°C and 3 min at 68°C, and a final 5-min extension at 68°C. Sequencing was performed on an ABI Prism 3100 with BigDye chemistry (Applied Biosystems). Primers used for PCR and sequencing are shown in Table 5, which is published as supporting information on the PNAS web site, www.pnas.org.
All single-nucleotide polymorphisms (SNPs) occurring in fewer than three individuals were subcloned to verify correct genotype scoring, and both alleles were identified in subclones for such heterozygotes. All polymorphisms were named based on coding nucleotide positions relative to the reference allele, GenBank accession no. AY297040. HAPLOTYPER (13) was used to predict haplotypes for each individual from the genotype data. For this analysis, individuals were pooled by subspecies. Two Ankole-Watusi individuals were considered to belong to the B. taurus indicus subspecies, given their pattern of observed SNPs in TLR4 and breed history of subspecies admixture. Four haplotypes were predicted with probabilities <95%, and were subject to further verification. One haplotype was confirmed by pooling African breed samples from both subspecies for analysis by Clark's method (14). For the remaining three individuals with low probability haplotype predictions, all informative exons were amplified, subcloned, and sequenced. In each case, at least five subclones were sequenced, including at least one copy of each allele.
To test apparent SNPs in adjacent coding positions 1947-1948, PCR-restriction fragment length polymorphism (PCR-RFLP) assays were designed that would allow haplotype testing for these two SNPs. PCR products were generated by using the PCRRFLP primers (see Table 5) to amplify all five heterozygous individuals and six homozygous controls. Eco0109I cut GG haplotypes only, and TfiI cut AA haplotypes only.
SNPTAGGER (15) was used to assess the numbers and positions of SNP markers necessary to distinguish observed haplotypes. MEGA 2.1 (16) was used to perform Z tests for purifying selection. These tests used the Nei-Gobojori method for computation of potential synonymous and nonsynonymous substitutions (17) and estimated variances of average substitution rates by bootstrapping with 1,000 replications. Positively selected codon sites were identified by using the PAML 3.12 software package (18). This analysis incorporated a neighbor-joining tree computed with MEGA 2.1, but the method is remarkably robust to deviations in tree morphology (19). TLR4 sequences used were from human, pygmy chimpanzee, gorilla, orangutan, mouse, hamster, rat, cat, horse, and cow. The PAML analysis used the discrete M3 model (19) with three classes of omega values and equilibrium codon frequencies calculated from average nucleotide frequencies at the three codon positions. A likelihood ratio test for positive selection was conducted by comparing likelihoods from the M0 and M3 models.
Results
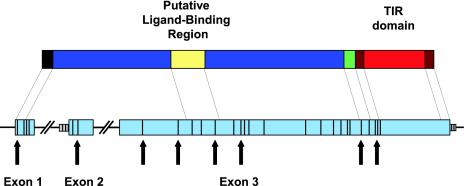
Bovine TLR4 has three exons, with splice sites similar to those of human TLR4 transcript variant 1 (20). Exon 1 includes coding base pairs 1-95, exon 2 consists of base pairs 96-260, and exon 3 comprises base pairs 261-2526. Each exon has been submitted to GenBank along with flanking sequences (accession nos. AY297041-AY297043). The whole genomic length is estimated to be ≈11 kb, of which the first intron comprises about 5 kb and the second, 3 kb.
Thirty-two SNPs were found among the 40 individuals in our panel, and 28 of these SNPs were in coding regions (cSNP). All SNPs have been submitted to dbSNP (accession nos. 9805774-9805805), and are listed in Table 1. Each cSNP is described relative to the reference allele found in GenBank accession AY297040, which is the allele found at the highest frequency in taurine cattle. SNPs outside coding regions are listed with reference to the nearest coding region. For example, E2-60 refers to a SNP 60 bp 5′ to exon 2. Summaries of nonsynonymous SNPs are shown in Table 2. Relative positions of all SNPs are shown in Fig. 1.
Table 1. List of observed haplotypes.
|
Haplotype
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | B1 | A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | C1 | D1 | E1 | F1 | G1 | H1 | H2 | I1 |
| 10 (C/T) | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | T |
| 64 (A/C) | A | A | A | A | A | A | A | A | A | C | C | C | C | A | A | A | A | C | C | C |
| 69 (C/A) | C | C | C | C | C | C | C | A | A | C | C | C | C | C | C | C | C | C | C | C |
| 75 (T/C) | T | T | T | T | T | T | T | T | T | C | C | C | C | T | T | T | C | C | C | C |
| E2-60 (T/G) | T | T | T | T | T | T | T | G | G | G | T | T | G | T | T | T | T | G | G | T |
| E2-26 (G/A) | G | G | A | G | G | G | G | A | G | A | A | G | G | G | G | G | G | G | G | G |
| 117 (G/A) | G | G | G | G | G | G | G | A | A | A | G | G | A | G | G | G | A | G | A | A |
| 148 (G/A) | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | A | A | G | G | G |
| 452 (A/C) | A | A | A | A | A | A | A | A | A | A | A | A | A | C | A | A | A | C | C | C |
| 714 (C/G) | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | G |
| 828 (C/T) | C | C | C | C | C | C | C | C | C | T | C | C | C | C | C | C | C | C | C | C |
| 897 (C/T) | C | C | C | C | C | C | C | C | C | T | C | C | C | C | C | C | C | C | C | C |
| 1040 (C/A) | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | A | A | A |
| 1119 (A/G) | A | A | A | A | A | A | G | G | G | G | G | A | G | A | G | A | G | G | G | G |
| 1142 (A/G) | A | A | A | A | A | A | A | A | A | A | A | A | A | A | G | A | G | A | A | A |
| 1153 (T/C) | T | T | T | T | T | T | T | T | T | C | T | T | T | T | C | T | C | T | T | T |
| 1167 (T/G) | T | T | T | T | T | T | G | G | G | G | G | T | G | T | G | T | G | T | T | T |
| 1279 (C/T) | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | T | C | C | C | C |
| 1521 (A/G) | A | A | A | A | A | A | G | G | G | G | G | A | G | A | G | A | G | G | G | G |
| 1656 (T/C) | C | C | C | C | C | T | T | T | T | T | T | C | C | T | T | T | T | T | T | T |
| 1767 (T/C) | T | T | T | T | T | T | C | C | C | T | T | T | T | T | T | T | T | T | T | T |
| 1827 (T/C) | T | T | T | T | T | T | T | T | T | T | C | T | T | T | T | T | T | C | C | C |
| 1866 (T/C) | T | T | T | T | T | T | T | T | T | T | T | T | C | T | T | T | T | T | T | T |
| 1875 (C/T) | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | T | T | T |
| 1947 (G/A) | G | G | G | G | G | G | G | G | G | G | G | G | A | G | G | G | G | A | A | A |
| 1948 (G/A) | G | G | G | G | G | G | G | G | G | G | G | G | A | G | G | G | G | A | A | A |
| 1992 (C/A) | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | A | A | A |
| 2013 (C/T) | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | T | C | C | C | C |
| 2021 (C/T) | T | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| 2028 (T/C) | T | T | T | T | T | T | C | C | C | C | T | T | T | T | T | T | T | T | T | T |
| E3 + 15 (T/C) | T | T | T | C | C | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| E3 + 18 (T/G) | T | T | T | T | G | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| Frequency | 10 | 2 | 18 | 2 | 2 | 28 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 |
Table 2. Nonsynonymous SNPs.
| SNP | Amino acid | Charge | Domain | Conservation | Reference allele | Substitute allele |
|---|---|---|---|---|---|---|
| 10 (C/T) | R4C | +/polar | Signal | Low | Divergent | Moderate |
| 148 (G/A) | D50N | −/polar | Extracellular | Low | Conserved | Moderate |
| 452 (A/C) | N151T | Polar/polar | Extracellular | Moderate | Conserved | Conserved |
| 714 (C/G) | N238K | Polar/+ | Extracellular | Moderate | Conserved | Divergent |
| 1040 (C/A) | A347E | Nonpolar/− | Extracellular | Low | Conserved | Divergent |
| 1142 (A/G) | K381R | +/+ | Extracellular | High | Divergent | Divergent |
| 1948 (G/A) | V6501 | Nonpolar/nonpolar | Transmembrane | Moderate | Conserved | Conserved |
| 2021 (C/T) | T6741 | Nonpolar/polar | Transmembrane/cytoplasmic | Moderate | Conserved | Conserved |
Fig. 1.
Positions of SNPs on genomic DNA, and position of putative ligand-binding region in protein. Positions of SNPs are shown with black lines on the blue coding regions and gray introns. Nonsynonymous SNPs are marked with arrows. Protein domains shown, with included amino acid residues (left to right): signal sequence (1-23), putative coreceptor-binding region 1 (24-273), putative ligand-binding region (274-368), putative coreceptor-binding receptor 2 (369-632), transmembrane region (633-653), proximal cytoplasmic region (654-672), Toll/IL-1 receptor (TIR) domain (673-819), and distal cytoplasmic region (820-841).
Twenty haplotypes were predicted to exist in the panel individuals. PCR-RFLP was used to confirm that all individuals heterozygous for the adjacent SNPs at base pairs 1947-1948 had AA/GG haplotypes, as predicted by HAPLOTYPER (data not shown). Four haplotypes were assigned probabilities of less than 95% by HAPLOTYPER. Of these, one was an African haplotype at low frequency in our sample, and it was confirmed by Clark's method (14) on pooled African breed samples from both subspecies. For the remaining three lower-confidence haplotypes, all informative exons were subcloned and sequenced. Final haplotypes revised to incorporate these data are included in Table 1. Haplotype assignments for all individuals is shown in Table 6, which is published as supporting information on the PNAS web site. Diversity statistics for both subspecies are found in Table 3.
Table 3. Diversity statistics, by subspecies.
|
No. of SNPs segregating
|
Avg. frequency of SNPs, %
|
Nucleotide level
|
Amino acid level
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subspecies | n | Total | Synonymous | Noncoding | Non-synonymous | Synonymous and noncoding | Non-synonymous | No. of alleles | % heterozygosity | No. of alleles | % heterozygosity |
| B. taurus | 26 | 5 | 1 | 3 | 1 | 23.6 | 19.2 | 6 | 76.9 | 2 | 30.8 |
| taurus | |||||||||||
| B. taurus | 13 | 29 | 21 | 3 | 5 | 20.4 | 12.3 | 15 | 100.0 | 6 | 46.2 |
| indicus | |||||||||||
One Longhorn individual was found to have one typical taurine haplotype and one haplotype more typical of indicine cattle (14 SNPs removed from the most similar taurine haplotype). This may have resulted from a “breeding up” strategy that incorporated some B. taurus indicus genetic material in the background of this otherwise taurine individual. To avoid misrepresentation of taurine diversity, this individual was omitted from diversity and evolutionary inference statistics, although its haplotypes and SNPs are reflected elsewhere in this work.
Single-tailed Z tests provide significant support for overall purifying selection in B. taurus indicus (P < 0.001), but not in Bos t. taurus (P = 0.209; see note below Table 3 concerning population composition for these tests). However, the likelihood ratio test strongly indicates the existence of positively selected sites (P << 0.001) in TLR4 across mammals. The discrete model M3 used three classes with estimated ω values of 0.001, 0.596, and 2.239 and probabilities of 0.379, 0.515, and 0.106, respectively. The overall average ω value was 0.545. Table 4 shows a list of amino acid sites with posterior probabilities for positive selection P(ω > 1) > 0.90. Twenty-one sites had probabilities > 0.90, twelve > 0.95, and two > 0.99.
Table 4. Amino acid sites in bovine TLR4 with probabilities of positive selection P(ω > 1) > 0.90.
| Position | Residue | Position | Residue | Position | Residue | Position | Residue |
|---|---|---|---|---|---|---|---|
| 5 | A | 274* | R | 341* | D | 514* | T |
| 54* | I | 293* | L | 344 | K | 822* | Q |
| 68 | H | 295 | K | 349 | K | ||
| 119* | W | 312 | V | 351* | S | ||
| 319* | S | 360 | D | ||||
| 321 | G | 364** | I | ||||
| 322* | S | 368** | T | ||||
| 334 | H |
*, Amino acid sites with probabilities > 0.95; **, sites with probabilities > 0.99.
Discussion
Bovine TLR4 shares genomic structure with human and mouse TLR4. In each case the intron/exon boundaries are conserved, and the intron lengths are variable but reasonably similar. The overall length of bovine TLR4 is ≈11 kb, which compares to ≈10 kb for human and ≈14 kb for mouse. Most of the differences in length are found in lengths of the introns.
As in human and mouse, TLR4 is a highly polymorphic gene (20, 21). In our sample of 40 individuals from 11 breeds, we observed 32 SNPs, 28 of which are cSNPs. This gives an average of 1 SNP per 90 bp of coding for bovine TLR4, which is higher than other reports for bovine coding sequence (22). The bovine data also show more polymorphisms in equal or smaller sample size than human or mouse (20, 21), which is consistent with previous reports that indicate cattle are more polymorphic than humans (23, 24). This finding is not surprising, given that breeding populations of cattle are represented by two divergent subspecies. However, our data do confirm the overall trend of high interspecies sequence conservation but simultaneously high intraspecies diversity in innate immune receptors (25).
The cSNPs we observed are located in most of the predicted domains, except for the Toll/IL-1 receptor/resistance (TIR) signaling domain. The absence of variation in TIR domain is not surprising, given its high level of conservation across other mammalian species. The nonsynonymous cSNPs (Table 2) are distributed almost evenly in the remaining domains, and this distribution is consistent with the reduced interspecies conservation of these regions.
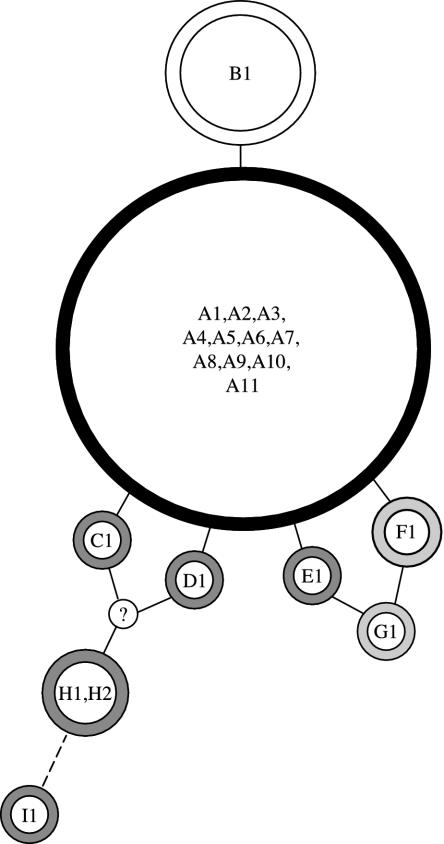
The B. taurus indicus subspecies was found to be more diverse than B. taurus taurus at TLR4, which is consistent with data from nuclear microsatellites (26). Indicine cattle have more alleles at both the nucleotide and amino acid levels, as well as higher heterozygosity at both levels (see Table 3). However, it should be noted that the five Brahman individuals we sampled were all homozygous for one allele, that with the A designations in Fig. 2, at the amino acid level. This actually represents lower diversity than we observed in taurine cattle, which show two alleles at the amino acid level. Therefore, to examine the effects of TLR4 diversity on disease-resistance phenotypes, populations will have to be used that have alleles from different Brahman individuals or from other indicine breeds such as Nellore and Gyr, which seem to contain more of the diversity present in the B. taurus indicus subspecies.
Fig. 2.
Haplotype relationships. Haplotypes at the amino acid level are shown as circles that include all equivalent haplotypes defined at the nucleotide level. Solid lines connect haplotypes related by a single amino acid substitution. A broken line indicates a distance of two amino acid substitutions. The frequency of each haplotype is depicted by circle size. Circle color denotes subspecies and historical continent of origin: white, taurine cattle only; black, common to all populations examined; light gray, found only in B. taurus indicus cattle of African descent; dark gray, found in indicine cattle. The haplotype denoted by “?” was not observed in our sample, but because of relationships with observed haplotypes it is predicted either to exist or to have existed in the B. taurus indicus subspecies outside our sample.
From our data, it appears that purifying selection has been at work on TLR4 in the B. taurus indicus subspecies of cattle, as with TLR4 in humans (21). A codon-based, one-tailed Z test shows evidence for purifying selection in that subspecies (P < 0.001). Further, nonsynonymous SNPs have a lower average allele frequency than synonymous SNPs, which is consistent with purifying selection. Statistical tests of historical selection pressure are inconclusive for taurine cattle, but this may be because of the small number of cSNPs in that subspecies. These overall conclusions based on our segregating data are consistent with patterns evident from TLR4 evolution in many mammals. Our analysis relied on the widely used statistic ω, which is based on the ratio of nonsynonymous/synonymous polymorphisms. ω values < 1 indicate purifying selection, ω = 1 indicates neutral selection, and ω > 1 indicates purifying selection. As described for other genes (19), the PAML M3 discrete model best fit the TLR4 data, giving an average ω value of 0.545, which indicates the overall pattern of purifying negative selection in TLR4 among mammals.
However, this model also detected several sites under diversifying selection (see Table 4). Most of these codon sites are included in the region 274-368, which is located approximately in the middle of the extracellular domain (see Fig. 1). Given the continually evolving structures of its ligands and the evidence that TLR4 makes ligand contact (27), it seems reasonable that such a positively selected region could be a primary ligand-binding region, as is the case with MHC class I molecules (28). This hypothesis is supported by a multiple alignment including not only mammalian but also chicken TLR4. Conservation of amino acid identity drops dramatically in this region relative to both adjacent portions of the extracellular domain (8% vs. 33%). Further, there are two segregating polymorphisms in the extracellular domain of human TLR4 with noted effects on lipopolysaccharide responsiveness, D299G and T399I (21, 29). The D299G substitution is responsible for most of the blunted response to lipopolysaccharide (29), and it lies in this putative ligand-binding region. It should be noted that we have defined this region conservatively based on the locations of residues under strong positive selection, so it may be that the actual ligand-binding region is somewhat larger than we have assumed. Finally, the hypothesis that this region is a primary ligand-binding region suggests other roles for the remaining, more highly conserved portions of TLR4's extracellular domain. The functions of these other two extracellular regions may be explained as sites for coreceptor interaction and endogenous ligand interaction, based on experimental evidence for the importance of such coreceptors as MD-2 and CD14 (30, 31) and endogenous ligands such as hsp60, hyaluronan oligosaccharides, and β-defensin 2 (32-34).
Given this model of the TLR4 extracellular domain, one might expect to observe a higher number of nonsynonymous substitutions currently segregating in the putative ligand-binding region than in adjacent regions of TLR4. Our data do not show this for cattle, and neither do data for mouse and human TLR4 (20, 21), but several factors may be involved. First, in each case there is a small sample of nonsynonymous substitutions, and the results may be because of sampling effects. Second, selection pressure may be of variable intensity, which fits with the sporadic emergence of important pathogen variants. It could be that each population is currently between episodes of selection for TLR4 variants. A third possibility is that selective pressures may be of such low intensity that it is difficult to detect at specific time points in a population. Regardless, the overall trend in mammalian TLR4 indicates positive selection in this putative ligand-binding region.
It is interesting to note two bovine SNPs that lie in or close to this putative ligand-binding region and result in evolutionarily divergent amino acid substitutions. Position 347 is very divergent with the negatively charged A347E substitution, which may indicate that this is a deleterious allele. In the case of position 381, cattle are segregating two alleles, but both are positively charged amino acids, not the polar serine found in all other species studied to date, including chicken. Either position could result in unique aspects of bovine TLR4 biology. Given the proximity of both segregating polymorphisms to the putative ligand-binding domain, both merit further investigation.
Another potentially important observation is an apparently recent SNP that leads to the I674T substitution. It was found on only one haplotype, B1 (Fig. 2), which occurs only in taurine cattle, but at relatively high frequency. This substitution is predicted to be in either the transmembrane or the proximal cytoplasmic domain, close to the highly conserved TIR domain. It may confer some type of selective advantage and thus it may have been under positive selection. However, it is also possible that other population genetic forces, such as drift, could have elevated the allele frequency.
The haplotypes we observed can all be assigned to subspecies and historical continents of origin. Fig. 2 shows alleles that differ by at least one amino acid substitution. In this figure, reticulations probably indicate historical recombination events. As shown, there are many haplotypes that are found in only one subspecies and/or historical continent of origin. However, there are several alleles in both subspecies that differ at the nucleotide level, but are equivalent to allele A in Fig. 2.
These haplotype data could be used to produce several haplotype marker sets for different purposes. Analysis with SNPTAGGER (15) indicates that only 12 SNPs (of 32) need to be genotyped to distinguish the 20 complete haplotypes we found. If one considers only amino acid substitutions, just six SNPs need to be genotyped to distinguish the nine haplotypes we observed. Only one SNP must be genotyped to distinguish the two most common amino acid haplotypes, which total 87% of observed haplotypes in our sample, 100% of observed in taurine cattle, and more than 60% of those observed in indicine cattle, by frequency. However, the broad diversity of indicine haplotypes suggests that this single SNP analysis might miss meaningful information if the bovine population sampled has a large percentage of indicine genetic background. Examples of each of these sets of haplotype-tagging SNPs (htSNPs) are shown in Table 7, which is published as supporting information on the PNAS web site.
In summary, these data show bovine TLR4 to be highly polymorphic. We have defined a spectrum of common variation in this gene, against which future variants can be meaningfully compared, and we suggest a putative ligand-binding region with adjacent coreceptor-binding regions in the extracellular domain of TLR4. Additionally this study developed a set of haplotype markers for use in disease association studies with the many pathogens that produce ligands of TLR4, including important pathogens involved in bovine shipping fever, tuberculosis, and Johne's disease.
Supplementary Material
Acknowledgments
We thank Dr. Loren Skow for helpful discussions and suggestions in the review process; Janice Elliott, Elaine Owens, and Natalie Halbert for assistance and support; Christopher Seabury and Dr. Jim Derr for providing some of the DNA samples used in this project; and Avni Santani, Dr. Bhanu Chowdhary, and Dr. Terje Raudsepp for helpful discussions and studies underpinning our work. This work was supported by a Programs of Excellence grant from the Life Sciences Task Force of Texas A&M University, U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service National Research Initiative Grant 99-35205-8534, and Grant 517-0186-2001 from the State of Texas Advanced Technology Program.
Abbreviations: TLR4, Toll-like receptor 4; SNP, single-nucleotide polymorphism; cSNP, SNP in a coding region; TIR domain, Toll/IL-1 receptor/resistance domain; PCR-RFLP, PCR-restriction fragment length polymorphism.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY297040-AY297043). The polymorphism data in this paper have been deposited in the dbSNP database (accession nos. 9805774-9805805).
References
- 1.Chow, J. C., Young, D. W., Golenbock, D. T., Christ, W. J. & Gusovsky, F. (1999) J. Biol. Chem. 274, 10689-10692. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi, O., Hoshino, K., Kawai, T., Sanjo, H., Takada, H., Ogawa, T., Takeda, K. & Akira, S. (1999) Immunity 11, 443-451. [DOI] [PubMed] [Google Scholar]
- 3.Lien, E., Means, T. K., Heine, H., Yoshimura, A., Kusumoto, S., Fukase, K., Fenton, M. J., Oikawa, M., Qureshi, N., Monks, B., et al. (2000) J. Clin. Invest. 105, 497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Brien, A. D., Rosenstreich, D. L., Scher, I., Campbell, G. H., MacDermott, R. P. & Formal, S. B. (1980) J. Immunol. 124, 20-24. [PubMed] [Google Scholar]
- 5.Chapes, S. K., Mosier, D. A., Wright, A. D. & Hart, M. L. (2001) J. Leukocyte Biol. 69, 381-386. [PubMed] [Google Scholar]
- 6.Malley, R., Henneke, P., Morse, S. C., Cieslewicz, M. J., Lipsitch, M., Thompson, C. M., Kurt-Jones, E., Paton, J. C., Wessels, M. R. & Golenbock, D. T. (2003) Proc. Natl. Acad. Sci. USA 100, 1966-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thoen, C. O. & Himes, E. M. (1986) Prog. Vet. Microbiol. Immunol. 2, 198-214. [PubMed] [Google Scholar]
- 8.Bendixen, P. H. (1978) Nord. Vet. Med. 30, 163-168. [PubMed] [Google Scholar]
- 9.Abel, B., Thieblemont, N., Quesniaux, V. J., Brown, N., Mpagi, J., Miyake, K., Bihl, F. & Ryffel, B. (2002) J. Immunol. 169, 3155-3162. [DOI] [PubMed] [Google Scholar]
- 10.Werling, D. & Jungi, T. W. (2003) Vet. Immunol. Immunopathol. 91, 1-12. [DOI] [PubMed] [Google Scholar]
- 11.White, S. N., Kata, S. R. & Womack, J. E. (2003) Mamm. Genome 14, 149-155. [DOI] [PubMed] [Google Scholar]
- 12.Cai, L., Taylor, J. F., Wing, R. A., Gallagher, D. S., Woo, S. S. & Davis, S. K. (1995) Genomics 29, 413-425. [DOI] [PubMed] [Google Scholar]
- 13.Niu, T., Qin, Z. S., Xu, X. & Liu, J. S. (2002) Am. J. Hum. Genet. 70, 157-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark, A. G. (1990) Mol. Biol. Evol. 7, 111-122. [DOI] [PubMed] [Google Scholar]
- 15.Ke, X. & Cardon, L. R. (2003) Bioinformatics 19, 287-288. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 17.Nei, M. & Gojobori, T. (1986) Mol. Biol. Evol. 3, 418-426. [DOI] [PubMed] [Google Scholar]
- 18.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555-556. [DOI] [PubMed] [Google Scholar]
- 19.Yang, Z., Nielsen, R., Goldman, N. & Pedersen, A. M. (2000) Genetics 155, 431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smirnova, I., Poltorak, A., Chan, E. K., McBride, C. & Beutler, B. (2000) Genome Biol. 1, 1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smirnova, I., Hamblin, M. T., McBride, C., Beutler, B. & Di Rienzo, A. (2001) Genetics 158, 1657-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heaton, M. P., Grosse, W. M., Kappes, S. M., Keele, J. W., Chitko-McKown, C. G., Cundiff, L. V., Braun, A., Little, D. P. & Laegreid, W. W. (2001) Mamm. Genome 12, 32-37. [DOI] [PubMed] [Google Scholar]
- 23.Konfortov, B. A., Licence, V. E. & Miller, J. R. (1999) Mamm. Genome 10, 1142-1145. [DOI] [PubMed] [Google Scholar]
- 24.Kruglyak, L. & Nickerson, D. A. (2001) Nat. Genet. 27, 234-236. [DOI] [PubMed] [Google Scholar]
- 25.Lazarus, R., Vercelli, D., Palmer, L. J., Klimecki, W. J., Silverman, E. K., Richter, B., Riva, A., Ramoni, M., Martinez, F. D., Weiss, S. T. & Kwiatkowski, D. J. (2002) Immunol. Rev. 190, 9-25. [DOI] [PubMed] [Google Scholar]
- 26.MacHugh, D. E., Shriver, M. D., Loftus, R. T., Cunningham, P. & Bradley, D. G. (1997) Genetics 146, 1071-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poltorak, A., Ricciardi-Castagnoli, P., Citterio, S. & Beutler, B. (2000) Proc. Natl. Acad. Sci. USA 97, 2163-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes, A. L., Ota, T. & Nei, M. (1990) Mol. Biol. Evol. 7, 515-524. [DOI] [PubMed] [Google Scholar]
- 29.Arbour, N. C., Lorenz, E., Schutte, B. C., Zabner, J., Kline, J. N., Jones, M., Frees, K., Watt, J. L. & Schwartz, D. A. (2000) Nat. Genet. 25, 187-191. [DOI] [PubMed] [Google Scholar]
- 30.Shimazu, R., Akashi, S., Ogata, H., Nagai, Y., Fukudome, K., Miyake, K. & Kimoto, M. (1999) J. Exp. Med. 189, 1777-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akashi, S., Ogata, H., Kirikae, F., Kirikae, T., Kawasaki, K., Nishijima, M., Shimazu, R., Nagai, Y., Fukudome, K., Kimoto, M. & Miyake, K. (2000) Biochem. Biophys. Res. Commun. 268, 172-177. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi, K., Burkart, V., Flohe, S. & Kolb, H. (2000) J. Immunol. 164, 558-561. [DOI] [PubMed] [Google Scholar]
- 33.Termeer, C., Benedix, F., Sleeman, J., Fieber, C., Voith, U., Ahrens, T., Miyake, K., Freudenberg, M., Galanos, C. & Simon, J. C. (2002) J. Exp. Med. 195, 99-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biragyn, A., Ruffini, P. A., Leifer, C. A., Klyushnenkova, E., Shakhov, A., Chertov, O., Shirakawa, A. K., Farber, J. M., Segal, D. M., Oppenheim, J. J. & Kwak, L. W. (2002) Science 298, 1025-1029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.