Abstract
Deletion of the SHOX region on the human sex chromosomes has been shown to result in idiopathic short stature and proposed to play a role in the short stature associated with Turner syndrome. We have identified a human paired-related homeobox gene, SHOT, by virtue of its homology to the human SHOX and mouse OG-12 genes. Two different isoforms were isolated, SHOTa and SHOTb, which have identical homeodomains and share a C-terminal 14-amino acid residue motif characteristic for craniofacially expressed homeodomain proteins. Differences between SHOTa and b reside within the N termini and an alternatively spliced exon in the C termini. In situ hybridization of the mouse equivalent, OG-12, on sections from staged mouse embryos detected highly restricted transcripts in the developing sinus venosus (aorta), female genitalia, diencephalon, mes- and myelencephalon, nasal capsula, palate, eyelid, and in the limbs. SHOT was mapped to human chromosome 3q25-q26 and OG-12 within a syntenic region on chromosome 3. Based on the localization and expression pattern of its mouse homologue during embryonic development, SHOT represents a candidate for the Cornelia de Lange syndrome.
Keywords: embryonic development, transcription factor, Cornelia de Lange syndrome
Homeobox genes encode proteins containing a 60-amino acid residue motif that represents a DNA binding domain. They have been characterized extensively as transcriptional regulators involved in pattern formation in both invertebrate and vertebrate species (for reviews see 1–4). Embryonic development is mediated through an interactive network created by the overlapping expression patterns of a number of transcription factors including the homeobox gene superfamily (5). This dynamic scaffold provides positional information for cell determination and differentiation underlying developmental processes from the simple determination of body axis to the formation of complex body structures (6, 7). This vital function of homeodomain proteins during embryonic development has initially been elucidated in lower organisms. More recently, several human genetic disorders including the Waardenburg syndrome (PAX3) (8), Aniridia (PAX6) (9), Synpolydactyly (HOXD13) (10), Schizencephaly (EMX2) (11), and the Rieger syndrome (RIEG) (12) have been shown to be caused by aberrations in human homeobox genes.
We have previously described the identification of a pseudoautosomal homeobox gene, SHOX, that is proposed to be responsible for idiopathic short stature and implicated to play a role in the short stature phenotype of Turner syndrome patients (13). This gene shows the highest homology to the murine OG-12 gene (14), which was consequently proposed as putative mouse homologue of SHOX. The homeodomains of SHOX (also called PHOG) (15) and OG-12 were shown to be identical suggesting that the two proteins bind to equivalent DNA elements and therefore trigger similar physiological pathways. Here we report the identification of a SHOX-related human gene showing a much higher degree of homology to OG-12 than SHOX. This gene, SHOT (for SHOX HOmologous gene on chromosome Three), is localized on chromosome 3q25-q26 and represents a candidate for the Cornelia de Lange syndrome, which was previously linked to this region (16).
MATERIALS AND METHODS
Oligonucleotides.
E1-for, 5′-GTG CCC CGG AGC TGA AAG ATC GC-3′; E1-rev, 5′-CCT CTG CTT GAT TTT GGT CTG GCCT TCG-3′; E2-rev, 5′-CTT TAT GGA GTT GAT TTT CTG G-3′; E3-for, 5′-GTG TTC TCA TAG GGG CCG CCA GC-3′; E4-for, 5′-ACG CCA ACG CCA GCA CCA ATG T-3′; E4-rev, 5′-CGC GGT TCC ACA CTC CCC TGT-3′; oligo(dT)-adaptor primer, 5′-GGC CAC GCG TCG ACT AGT AC [dT]20-3′.
cDNA Isolation.
Sequence information for the initial SHOT cDNA cloning procedure was taken from two human expressed sequence tags (AA425240 and AA479928) from the EMBL database and the Institute for Genomic Research gene index, respectively. Primers E1-for and E4-rev were used to amplify cDNA from a bone marrow fibroblast cell line. Sequence information of all subsequently established primers was derived from this initial PCR product. The 5′ and 3′ ends of SHOT were generated by rapid amplification of cDNA ends from a bone marrow fibroblast cDNA library that was constructed according to Rao et al. (13). For 5′ rapid amplification of cDNA ends experiments the primer E2-rev was used in combination with an AP1 adaptor primer (CLONTECH). A second round of amplification was performed using 1/100th of the PCR product and the primer E1-rev and the adaptor primer AP2. To clone the 3′ end of the SHOT transcript the primers E3-for and AP1 were used for the first round of amplification and E4-for, together with AP2, for the nested PCR.
All PCRs were carried out in a final volume of 50 μl with 100 pg–200 ng template DNA, 20 pmol of each primer, 200 μM dNTPs (Pharmacia), 1.5 mM MgCl2, 75 mM Tris⋅HCl (pH 9.0), 20 mM (NH4)2SO4, 0.01% Tween 20, and 2 units of Goldstar DNA Polymerase (Eurogentec, Brussels). Cycling was carried out in a GeneE Thermocycler (Techne Laboratories, Princeton, NJ) under the following conditions: 94°C for 2 min, 94°C for 30 sec, 68°C for 30 sec, 72°C for 1 min for 10 cycles; 94°C for 30 sec, 64°C for 30 sec, 72°C for 1 min for 15 cycles; 94°C for 30 sec, 62°C for 30 sec, 72°C for 1 min for 15 cycles, and a final extension for 5 min at 72°C. All PCR-generated fragments were cloned into the pCR2.1-TOPO vector (Invitrogen) or pBluescript (Stratagene) for further analysis.
Screening of Genomic Libraries and Southern Analysis.
Genomic clones corresponding to the OG-12 and SHOT genes were isolated from a mouse cosmid (A. Frischauf, unpublished data) and segment one of the Roswell Park Cancer Institute human P1-derived artificial chromosome library (17). The cosmid library was screened with a 607-bp probe (corresponding to positions 926 to 1533) of the OG-12a cDNA (accession no. U66918), yielding the cosmid B212cos. The P1-derived artificial chromosome library was screened using a 929-bp fragment (corresponding to positions 1 to 929), which contained the complete coding region of SHOTb under conditions recommended by the Roswell Park Cancer Institute (17). Hybridization was carried out in a buffer containing 0.5 M NaPi (pH 7.2), 7% SDS, 1 mM EDTA at 65°C and filters washed with 40 mM NaPi, 1% SDS at 65°C. Southern blots were hybridized and washed with the same buffers at 60°C for low stringency and 65°C for high stringency conditions.
Chromosomal Mapping.
Fluorescence in situ hybridization was carried out using a 17 kb EcoRI fragment from the mouse cosmid B212cos (OG-12) and the entire human PAC clones 262M18 and 83I04 (SHOT). The cosmid fragment and the P1-derived artificial chromosome DNAs were labeled by nick translation with biotin-11-dUTP and hybridized to metaphase chromosomes as described (18, 19). Cohybridization experiments of the murine and human genomic clones were carried out with digoxygenin-11-dUTP-labeled chromosome 3 specific painting probes. Hybridization signals were visualized via avidin-conjugated fluorescein isothiocyanate and Cy3-conjugated anti-digoxygenin antibodies, respectively. Human chromosomes were counterstained with 4′,6-diamidino-2-phenylindole; mouse chromosomes with propidium iodide. Images were taken with a cooled charge coupled device camera system (Photometrics, Tucson, AZ) and processed on a Macintosh computer using the nu200 2.0 software package (Photometrics) and the Adobe photoshop program.
Antisense in Situ Hybridization.
Mouse embryos and fetuses were obtained from matings between C57/Bl6 mice. Embryos were fixed in 4% paraformaldehyde (pH 7.2) overnight, dehydrated through an ethanol series, cleared in toluene and embedded in paraffin. Sections (5 μm) were cut for each stage. Sense and antisense probes were generated from two different OG-12 cDNA fragments corresponding to cDNA positions 171–578 (OG-12a specific) and 926–1,533 (identical between OG-12a and OG-12b), respectively. In situ prehybridizations and hybridizations as well as probe synthesis were carried out as described (20). Slides were dipped in Kodak NTB2 emulsion diluted 1:1 with water, exposed at 4°C for 5–10 days and developed with Kodak D19 developing solution and Kodakfix at 15°C for 4 min. Sections were stained with eosin and hematoxylin and visualized using a Zeiss Axiophot microscope. Photographs were taken in darkfield, color inverted using the Adobe photoshop program and superimposed on lightfield images of the same section.
Reverse Transcription–PCR.
Poly(A)+ RNA from human heart, pancreas, placenta, skeletal muscle, fetal kidney, and fetal liver was purchased from CLONTECH. Total RNA from a bone marrow fibroblast cell line was isolated according to Rao et al. (13). For the generation of first strand cDNA, 100 ng of poly(A)+ selected or 10 μg of total RNA were reverse transcribed using a Superscript first strand cDNA synthesis kit from GIBCO/BRL and the oligo(dT)-adaptor primer. Subsequent PCRs were performed with 5 μl of an 1/10 dilution of this reaction. SHOT transcripts were detected by PCR with the primers E1-for and E3-rev. SHOX specific cDNA fragments were generated with the primers described previously (13).
DNA Sequence Analysis.
All PCR-generated clones were sequenced using the Autoread Thermo Sequenase kit from Amersham with Cy5-labeled vector primers M13, universal and reverse, and analyzed on an ALF express automated sequencer (Pharmacia). The following exon-specific primers were used: E1-for and E1-rev; E4-for and E4-rev. All sequences were evaluated using the gcg software package and the facilities of the EMBL database.
RESULTS
cDNA and Predicted Protein Sequence of the Human Homeobox Gene SHOT.
We have previously shown that the human SHOX and murine OG-12 genes are closely related and share identical homeodomains (13). To identify additional SHOX-related genes we carried out database homology searches using the complete SHOX and OG-12 cDNAs as query sequences. Two nonoverlapping expressed sequence tags (AA479928 and AA425240) with extensive homologies to the 5′ and 3′ portion of SHOX and OG-12 were identified. Using primers from both expressed sequence tags we were able to generate a 964-bp cDNA fragment from bone marrow fibroblast reverse transcribed RNA, showing that both expressed sequence tags belong to a single human gene that is homologous but not identical to SHOX. We will refer to this gene as SHOT (SHOX HOmologous gene on chromosome Three). To derive full-length cDNA clones, sequence information from this fragment was subsequently used to design primers for 5′ and 3′ rapid amplification of cDNA ends experiments. 5′ rapid amplification of cDNA ends experiments using the E2-rev/E1 and AP1/AP2 primer combinations revealed two different 5′ splice variants, whereas internal PCRs demonstrated the existence of an alternatively spliced exon within the C-terminal portion of the SHOT gene. In summary, sequence analysis of the generated fragments revealed the existence of at least two distinct cDNA isoforms, SHOTa and SHOTb. SHOTa (1,948 bp) and SHOTb (1,541 bp) encode proteins of 331 and 190 amino acids, respectively. The putative start codons of SHOTa and -b are in good agreement with Kozak’s criteria for translation initiation sites (21) and correspond to the translation start sites of OG-12a and OG-12b, respectively.
Comparison of SHOT, OG-12, and SHOX Gene and Protein Sequences.
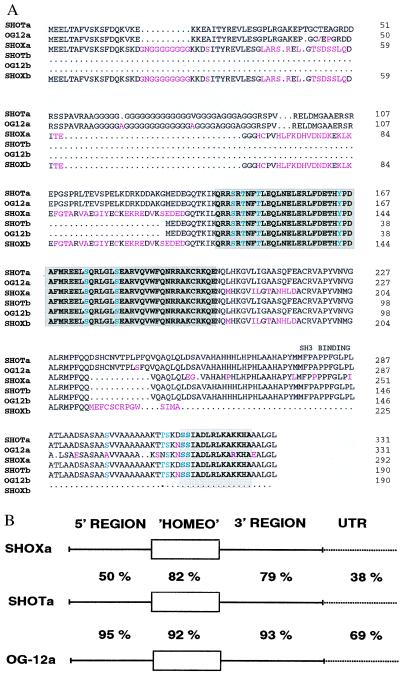
SHOTa and SHOTb are predicted to encode proteins with a homeodomain identical to murine OG-12 and human SHOX (Fig. 1A). This homeodomain shows the highest homology to the homeodomains of paired-related proteins, including Arx (22), Prx2/S8 (23), Phox2 (24), aristaless (25), Pax-3 (26), and Drg11 (27). In addition to the homeodomain itself, several potential phosphorylation sites, a putative SH3 binding domain and a 14-amino acid residue motif at the C-terminal end (“OAR-domain”) (28) are highly conserved among the OG-12, SHOX, and SHOT proteins (Fig. 1A). The predicted SHOT and OG-12 proteins are 99% identical over their entire length while the overall homology between SHOT and SHOX amounts to only 83% at the amino acid level, demonstrating that the human SHOT is related more closely to the murine OG-12 than to SHOX.
Figure 1.
(A) Alignment of the predicted amino acid sequences of mouse OG12a and -b and human SHOXa and -b with human SHOTa and -b. The homeodomain is shaded in dark gray. The 14-amino acid domain in the C terminus (OAR-domain) (28) is shaded in light gray. Letters in red indicate amino acid changes compared with SHOT. Letters in green indicate potential phosphorylation sites. (B) Nucleotide sequence comparison between SHOTa and SHOXa and SHOTa and OG-12a, respectively. “Homeo” indicates the homeobox region and untranslated region the 3′ untranslated regions.
To obtain additional information about the evolutionary relationship between SHOT, SHOX, and OG-12 we compared the 3′ untranslated regions. The human SHOT and SHOX cDNAs exhibit a nucleotide sequence identity of 38% in their 3′ untranslated regions, whereas SHOT-encoded cDNAs show a 69% identity to the murine OG-12 cDNAs in that region. In contrast, SHOX and OG-12 are only 40% identical in their 3′ untranslated regions. Fig. 1B shows the comparison between SHOXa, SHOTa, and OG-12a at the nucleotide level in the coding and noncoding regions. These homologies indicate a common origin of SHOT and OG-12.
To determine if the murine genome contains any genes as closely related to SHOX as OG-12 is to SHOT, we performed a series of Southern blot hybridizations using SHOT and SHOX cDNAs as probes. As expected, SHOT and OG-12 are sufficiently homologous to exhibit cross-hybridization under high stringency conditions, whereas SHOX detects the OG-12 specific band when lowering the stringency. Consistent with that, OG-12 detects strong SHOT-specific and weak SHOX-specific bands on human DNA at high stringency hybridization conditions (data not shown).
Chromosomal Localization of SHOT.
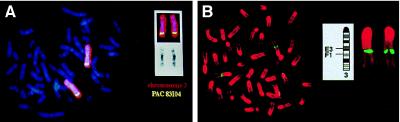
To determine whether SHOT was also sex-linked, fluorescence in situ hybridization was carried out. For this purpose, two overlapping PAC clones, 83I04 and 262M18, were isolated and hybridized to metaphase chromosomes. We assigned the chromosomal localization of SHOT to human chromosome 3. This genomic localization was confirmed by cohybridizations using a human chromosome 3 specific painting probe. By superimposing the specific hybridization signals on G-banded chromosomes, it was possible to assign SHOT to chromosome 3q25-q26 (Fig. 2A).
Figure 2.
Chromosomal localization of SHOT and OG-12. (A) Cohybridization of biotinylated human PAC 83I04 (yellow) and digoxygenin-labeled chromosome 3 specific painting probe. Chromosomes 3 are displayed separately as gray scale image demonstrating the 4′,6-diamidino-2-phenylindole bands and as full-color image showing the specific hybridization signals. (B) Hybridization of the biotinylated B212cos/17 kb EcoRI fragment, containing the entire OG-12 gene to mouse metaphase chromosomes. Metaphase spreads were prepared from methotrexate-synchronized and BrdUrd (BrdU)-treated mouse spleen cultures as previously described (19). The hybridized probe was detected using avidin-fluorescein isothiocyanate (Vector Laboratories) and is pseudocolored in yellow. The chromosomes are counterstained with propidium iodide and pseudocolored in red. The probe hybridized to chromosome 3, band E3-F1. Ideogram of mouse chromosome, representing schematically the G-banding pattern. The two homologues of mouse chromosome 3 are enlarged and displayed next to the ideogram.
Expression of the SHOT Gene.
The identification of cDNA clones from heart, liver, lung, and skeletal muscle cDNA libraries demonstrated conclusively that SHOT is expressed in the corresponding organs. Reverse transcription–PCR analysis of RNA derived from adult heart, skeletal muscle, bone marrow fibroblasts, pancreas, and placenta confirmed that SHOT is expressed in a wide range of tissues (data not shown).
Chromosomal Localization of OG-12.
The chromosomal localization of OG-12 was determined by fluorescence in situ hybridization, using a 17-kb EcoRI fragment of cosmid B212cos containing the entire OG-12 gene. OG-12 was assigned to chromosome 3, which was confirmed by cohybridization experiments using a chromosome 3 specific painting probe and by superimposing the hybridization signal on propidium iodide-banded chromosomes as inverted gray scale images. OG-12 was hereby mapped to mouse chromosome 3, band E3-F1 (Fig. 2B). OG-12 and SHOT therefore map to a region that was previously shown to be syntenic between human and mouse (29).
Expression Pattern of OG-12 During Mouse Embryonic Development.
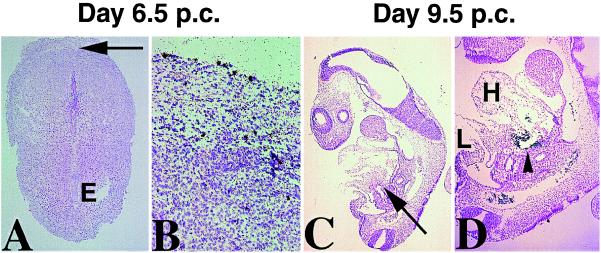
A clue to the biological function of SHOT may be provided by studying the expression pattern of its mouse equivalent, OG-12. The spatial and temporal patterns of OG-12 expression were examined by in situ hybridizations on serial sections of staged mouse embryos between day 5 postconception (p.c.) and 18.5 p.c. as well as on fetal and newborn animals. Although two different probes were used (see Materials and Methods), there were no differences detectable in the expression of the two isoforms, OG-12a and b. Transcription of OG-12 was not detected in the embryo before day 8 p.c. but was evident in a few cells of the mother’s mesometrium (Fig. 3 A and B). From day 8 onwards expression in the embryo could be detected in mesoderm derivatives that contribute to bone and cartilage formation in the limbs, ribs, palate, nasal capsula, prepuce, and spinal cord (Figs. 4 and 5). Craniofacial expression was observed in cells condensing around the developing eyelid (Fig. 5A), nasal cavity, and palate (Fig. 4A). As the palate fuses and the nasal capsula differentiates, this expression is down-regulated and only remains detectable in the tips of the fusing palate (Fig. 6B). Furthermore, heart (Figs. 3D and 5D) and metanephric mesoderms were also positive for OG-12 expression. Interestingly, in the heart OG-12 expression was restricted to the developing outflow track and the developing aorta (Figs. 3D and 5D) and could not be detected in the heart itself. In the ectoderm, expression of OG-12 was found in the developing brain, spinal, and otic ganglia (Fig. 5 B and C; and Fig. 6 A and B).
Figure 3.
Expression of OG-12 up to day 9.5 p.c. (A) Sagittal section through the decidua of a 6.5-day pregnant female. Expression is not detected in the embryo (E) but several cells in the mesometrium strongly express OG-12 (arrow in A indicates the region magnified in B. Positive signals are seen as black grains on stained sections in all figures. (C) Day 9.5 p.c. embryo, sagittal section. The arrow indicates the region magnified in (D) Strong expression is observed in the developing outflow track of the heart (arrowhead). Mesoderm cells in the limb (L) are also positive.
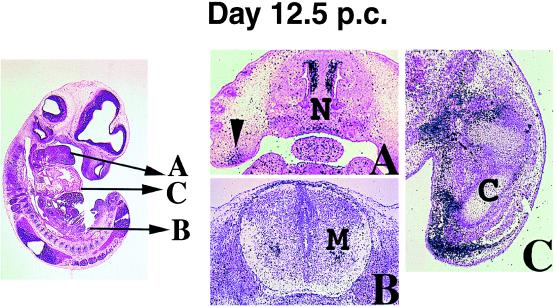
Figure 4.
Expression of OG-12 on day 12.5 p.c. The levels of transverse sections in A–C are indicated by arrows in the sagittal lightfield image. (A) A section through the nasal septum showing expression in condensing cartilaginous mesenchyme in the nose (N) and palate (arrowhead). (B) In the spinal cord, developing motor neurons are positive (M). (C) In the fore limb bud transcripts are restricted to undifferentiated mesenchyme condensing around the developing bone (C).
Figure 5.
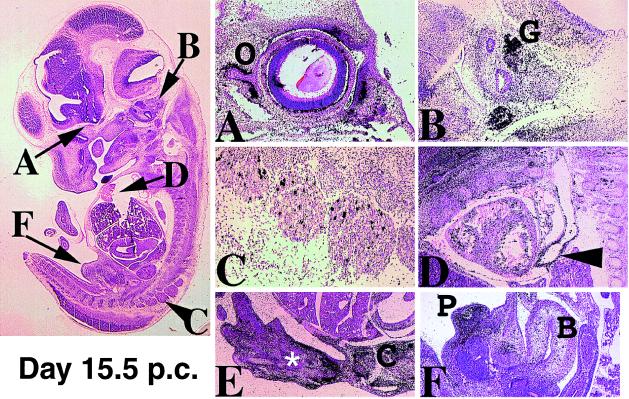
Expression in day 15 p.c. embryos. The figure on the left indicates the plane of the section magnified in A–F. (A) In the developing eye, transcripts are restricted to the developing eyelids and mesenchyme condensing around the optic nerve (O). (B) In the otic vesicle the vestibulocochlear ganglia (G) strongly express OG-12. (C) A distinct subpopulation of cells in the spinal ganglia is positive. (D) Sections through the developing heart indicate that transcripts are found in the developing aorta (arrowhead). Blood cells within the heart reflect strongly and therefore the signal observed is not real. (E) Mesenchymal cells in the hind limb surrounding the developing digits express OG-12. Expression in the developing carpels is weak (white asterisk), however in the joints (C) expression is also seen within bone cells. (F) Sections through the developing genitals (P) show strong expression in prepuce mesenchyme. At earlier stages of sex determination OG-12 transcripts are not detectable. Other urinary structures such as the bladder (B) do not express OG-12.
Figure 6.
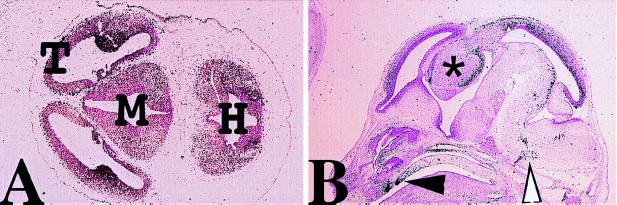
Expression in the developing brain. (A) Transverse section through the brain of a day 12.5 p.c. embryo showing intense expression in the ganglionic eminences of the telencephalon (T). Weaker expression is also detected in mesencephalon (M) and myelencephalon (H). (B) Sagittal section of a day 15.5 day p.c. head showing expression in the fusing palate (solid arrowhead), nose, and brain. The telencephalon and dorsal thalamus (asterisk) express OG-12 strongly. In the hind brain expression is seen in distinct cell populations of the medulla and the developing inferior and superior olivary nuclei (open arrowhead).
In the developing extremities transcription of OG-12 was observed on day 9.5 p.c. in mesodermal cells on the dorsal side of the limb bud (Fig. 3D). As the limb bud develops, this expression intensifies in the mesoderm of the progress zone and in undifferentiated mesoderm condensing around the ossification centers. On day 12.5 p.c. OG-12 transcripts were detected in the mesoderm of the anterior tip of the growing limb and in mesenchymal cells condensing around the cartilage primordium of fore and hind limb bones (Fig. 4C). On day 14.5 p.c. when digital differentiation becomes obvious, additional expression could be detected in mesodermal condensations at the periphery of the developing phalanges. At this stage transcripts are first detected in differentiating presumptive bone regions (Fig. 5E). Although expression of OG-12 decreases between day 14.5 and 16.5 p.c., it was still detectable in ossification centers of long bones as limb skeletal parts chondrify (not shown).
DISCUSSION
We have described the isolation and characterization of a human homeobox containing gene, SHOT. This gene is located on chromosome 3q25-q26 and is predicted to encode at least two proteins, SHOTa and SHOTb, that are 99% identical to the previously described mouse OG-12a and OG-12b (14), respectively. This high degree of sequence homology and the localization of the genes within a human/mouse syntenic region on chromosome 3 identify SHOT as the closest human equivalent of the murine OG-12 gene.
Evolutionary and Functional Relationship Between SHOT, SHOX, and OG-12.
The presence of two closely related human genes and only one mouse equivalent (39) has raised the question as to their origin and possible functions. The extremely high degree of conservation between the mouse OG-12 and human SHOT genes clearly argues for an indispensable role of these genes in both species. SHOX, on the other hand, seems to have considerably drifted and might therefore exhibit new or additional primate specific functional properties. The existence of a supplementary pseudoautosomal gene copy without a close murine homologue is not unprecedented and reflects the situation e.g., of the adenine nucleotide translocase gene family (30). It might be exactly the translocation of the duplicated gene onto the pseudoautosomal region on the human sex chromosomes that has allowed a considerable rate of divergence, possibly related to the high recombination frequencies in this region (31).
The homeodomains between all SHOT-, OG-12-, and SHOX-encoded proteins have been shown to be identical, suggesting that their DNA binding potential is also identical. Especially the amino acid residue at position 9 within the third helix motif of the homeodomain (recognition helix) appears to be crucial for the specificity of binding to the two bases following the TAAT core motif of the respective target sequences (32). The conservation of a glutamine-residue at this position in all SHOT-, OG-12-, and SHOX-encoded homeodomains may imply the binding of these proteins to equivalent target genes, thereby activating or repressing a similar subset of genes. Considering the putative role of SHOX in short stature phenotypes, it is tempting to speculate on the growth-related functions of SHOT-encoded proteins in human development. It will also be interesting to find out if these growth-related functions do not only affect height but, in a more general way, act as growth/proliferation regulators in certain tissues.
Relationship of SHOT to Other Homeodomain Protein Family Members.
Recently, a 14-amino acid C-terminal sequence motif (“OAR-domain”) was discovered by virtue of its conservation among a subclass of homeodomain proteins with high expression levels in craniofacial tissues including Ptx1/POTX (33, 34), Prx-1 (35), and Prx-2 (23), Cart-1 (36), Drg11 (27), OTP (37), and solurshin, involved in the Rieger syndrome (12). Although a direct function has not yet been assigned to this motif, analysis of OTP mutants lacking this region suggests an involvement of this domain in transactivation (37). SHOT shares 13 of 14 identical amino acids with SHOX and has 10 of 14 amino acids in common with solurshin, Ptx1, and Cart-1 within this motif. This clearly places SHOT, OG-12, and SHOX in the same category of homeobox genes such as solurshin, Ptx1, and Cart-1.
Expression Pattern of the Murine SHOT Homologue OG-12.
The expression analysis of OG-12 using two different probes has not revealed any differences in the spatial or temporal expression pattern of the two isoforms, OG-12a and OG-12b. Expression of OG-12 was detected during embryonic development in mesoderm derivatives that contribute to bone and cartilage formation and in ectodermal tissues including brain, spinal cord, and ganglia. The highest levels of expression were found in mesodermal tissues of the face involved in nose and palate formation, the developing eyelid and tissue surrounding the optic nerve, as well as in the developing heart mesoderm and in the mesoderm condensing around the chondrification centers of the limb. Interestingly, this pattern partly overlaps with the expression previously described for the homeobox genes RIEG, Prx-1, Prx-2, and Cart-1, predicted to be closely related to SHOT.
Potential Role of SHOT in Human Developmental Disorders.
SHOT is located on chromosome 3q25-q26, a chromosomal region involved in the development of the Cornelia de Lange syndrome. This syndrome was first described in 1933 and is characterized by growth and mental retardation (microcephaly), distinctive facial deformities including cleft palate, abnormally situated eyelids, and nose and ear deformities, as well as heart defects and reductive limb development (38). Interestingly, the expression of the mouse SHOT homologue, OG-12, is in perfect agreement with the features seen in Cornelia de Lange syndrome. It shows high expression levels in craniofacial tissues including the palate, nasal capsula, eyelid, and ear, as well as in heart (aorta), brain, and developing limbs. Thus, the chromosomal localization and the expression pattern of its murine homologue make SHOT an excellent candidate for the Cornelia de Lange syndrome gene.
Acknowledgments
We thank Katrin Schiebel and Stefan Kirsch for critical comments and Prof. Bartram for his support. The authors are grateful to Dagmar Bock for expert technical assistance. This work was funded by the Deutsche Forschungsgesellschaft Ra 380/9–1 to G.R. and the Deutsche Forschungsgemeinschaft and the European Community to A.P.M.
Footnotes
References
- 1.Gehring W J, Affolter M, Bürglin T. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 2.Manak, J. R. & Scott, M. P. (1994) Development (Cambridge, U.K.), Suppl. 61–77. [PubMed]
- 3.Krumlauf R. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 4.Boncinelli E. Curr Opin Genet Dev. 1997;7:331–337. doi: 10.1016/s0959-437x(97)80146-3. [DOI] [PubMed] [Google Scholar]
- 5.Mc Ginnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 6.Johnston D S, Nüsslein-Volhard C. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 7.Ingham P W, Arias A M. Cell. 1992;68:221–235. doi: 10.1016/0092-8674(92)90467-q. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin C T, Hoth C F, Amos J A, da-Silva E O, Milunsky A. Nature (London) 1992;355:637–638. doi: 10.1038/355637a0. [DOI] [PubMed] [Google Scholar]
- 9.Glaser T, Walton D S, Maas R L. Nat Genet. 1992;2:232–238. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 10.Muragaki Y, Mundlos S, Upton J, Olson B J. Science. 1996;272:548–551. doi: 10.1126/science.272.5261.548. [DOI] [PubMed] [Google Scholar]
- 11.Brunelli S, Faiella A, Capra V, Nigro V, Simeone A, Cama A, Boncinelli E. Nat Genet. 1996;12:94–96. doi: 10.1038/ng0196-94. [DOI] [PubMed] [Google Scholar]
- 12.Semina E V, Reiter R, Leysens N J, Alward W L M, Small K W, Datson N A, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel B U, et al. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 13.Rao E, Weiss B, Fukami M, Rump A, Niesler B, Mertz A, Muroya K, Binder G, Kirsch S, Winkelmann M, et al. Nat Genet. 1997;16:54–63. doi: 10.1038/ng0597-54. [DOI] [PubMed] [Google Scholar]
- 14.Rovescalli A C, Sadamitsu A, Nirenberg M. Proc Natl Acad Sci USA. 1996;93:10691–10969. doi: 10.1073/pnas.93.20.10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison J W, Wardak Z, Young M F, Robey P G, Laig-Webster M, Chiong W. Hum Mol Genet. 1997;6:1341–1347. doi: 10.1093/hmg/6.8.1341. [DOI] [PubMed] [Google Scholar]
- 16.Ireland M, English C, Cross I, Houlsby W T, Burn J. J Med Genet. 1991;28:639–640. doi: 10.1136/jmg.28.9.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ioannou P A, Amemiya C T, Garnes J, Kroisel P M, Shizuya H, Chen C, Batzer M A, de Jong P J. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. http://bacpac.med.buffalo.edu . (See also: http://bacpac.med.buffalo.edu). ). [DOI] [PubMed] [Google Scholar]
- 18.Lichter P, Cremer T. Human Cytogenetics: A Practical Approach. Oxford: IRL/Oxford Univ. Press; 1992. [Google Scholar]
- 19.Schröck E, Badger P, Larson D, Erdos M, Wynshaw-Boris A, Ried T, Brody L. Hum Genet. 1996;97:256–259. doi: 10.1007/BF02265277. [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson D G, editor. In Situ Hybridization: A Practical Approach. New York: IRL; 1992. [Google Scholar]
- 21.Kozak M. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 22.Miura H, Yanazawa M, Kato K, Kitamura K. Mech Dev. 1997;65:99–109. doi: 10.1016/s0925-4773(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 23.Leussink B, Brouwer A, el Khattabi M, Poelmann R E, Gittenberger-de Groot A C, Meijlink F. Mech Dev. 1995;52:51–64. doi: 10.1016/0925-4773(95)00389-i. [DOI] [PubMed] [Google Scholar]
- 24.Valarche I, Tissier-Seta J P, Hirsch M R, Martinez S, Goridis C, Brunet J F. Development (Cambridge, UK) 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 25.Schneitz K, Spielmann P, Noll M. Genes Dev. 1993;7:114–129. doi: 10.1101/gad.7.1.114. [DOI] [PubMed] [Google Scholar]
- 26.Goulding M D, Chalepakis G, Deutsch U, Erelius J R, Gruss P. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito T, Greenwood A, Sun Q, Anderson D J. Mol Cell Neurosci. 1995;6:280–292. doi: 10.1006/mcne.1995.1022. [DOI] [PubMed] [Google Scholar]
- 28.Furukawa T, Kozak C A, Cepko C L. Proc Natl Acad Sci USA. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Bry R W, Seldin M F. Genomics. 1996;33:337–35. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- 30.Ellison J W, Li X, Francke U, Shapiro L J. Mamm Genome. 1996;7:25–30. doi: 10.1007/s003359900007. [DOI] [PubMed] [Google Scholar]
- 31.Rappold G. Hum Genet. 1993;92:315–324. doi: 10.1007/BF01247327. [DOI] [PubMed] [Google Scholar]
- 32.Hanes S D, Riddihough G, Ish-Horowicz D, Brent R. Mol Cell Biol. 1994;14:3364–3375. doi: 10.1128/mcb.14.5.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamonerie T, Tremblay J J, Lanctot C, Therrien M, Gauthier Y, Drouin J. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 34.Szeto D P, Ryan A K, O’Connell S M, Rosenfeld M G. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nohno T, Koyama E, Myokai F, Taniguchi S, Ohuchi H, Saito T, Noji S. Dev Biol. 1993;158:254–264. doi: 10.1006/dbio.1993.1184. [DOI] [PubMed] [Google Scholar]
- 36.Zhao G Q, Eberspaecher H, Seldin M F, Crombrugghe B. Mech Dev. 1994;48:245–254. doi: 10.1016/0925-4773(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 37.Simeone A, D’Apice M R, Nigro V, Casanova J, Graziani F, Acampora D, Avantaggiato V. Neuron. 1994;13:83–101. doi: 10.1016/0896-6273(94)90461-8. [DOI] [PubMed] [Google Scholar]
- 38.Berg J M, McCreay B D, Ridler M A C, Smith G F. The de Lange Syndrome. New York: Pergamon; 1970. [Google Scholar]
- 39.van Schaick H S A, Smidt M P, Rovescalli A C, Luijten M, van der Kleij A A M, Asoh S, Kozak C A, Nirenberg M, Burbach J P H. Proc Natl Acad Sci USA. 1997;94:12993–12998. doi: 10.1073/pnas.94.24.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]