Abstract
B7 family proteins provide costimulatory signals that regulate T cell responses. Here we report the third set of B7 family-related T cell inhibitory molecules with the identification of a homolog of the B7 family, B7x. It is expressed in immune cells, nonlymphoid tissues, and some tumor cell lines. B7x inhibits cell-cycle progression, proliferation, and cytokine production of both CD4+ and CD8+ T cells. B7x binds a receptor that is expressed on activated, but not resting T cells that is distinct from known CD28 family members. Its receptor may be a recently identified inhibitory molecule, B and T lymphocyte attenuator. These studies identify a costimulatory pathway that may have a unique function in downregulation of tissue-specific autoimmunity and antitumor responses.
Keywords: costimulation, autoimmunity, tumor
It is now clear that T cell responses that follow T cell activation via antigen receptor engagement are regulated by costimulatory signals. Because of their fundamental biological importance and therapeutic potential, there has been considerable interest in the identification of molecules with costimulatory activity. The classic costimulatory molecules B7-1 and B7-2 provide critical positive costimulatory signals on interaction with CD28 on resting T cells, whereas interaction of B7-1 and B7-2 with cytotoxic T lymphocyte antigen-4 (CTLA-4) on recently activated cells can restrict T cell responses. Thus, the major role of the classical B7-1,B7-2/CD28,CTLA-4 system seems to be regulation of T cell responses at a fairly early stage in lymphoid tissues (1, 2). In the past few years, several new members of the B7 family have been discovered, including B7h, PD-L1, PD-L2, and B7-H3 (3-6). Although their precise roles in T cell regulation are just now being elucidated, they seem to have distinct, although partially overlapping functions. B7h (7) [also called B7RP-1 (8), GL50 (9), B7H2 (10), and LICOS (11)] binds to inducible costimulator (ICOS) on activated T cells and provides costimulation for activated T cells. The B7h/ICOS pathway plays a critical role in regulating Th2 cytokines, is essential for germinal center formation and T-B interactions, and may provide costimulation for activated T cells at sites of inflammation in nonlymphoid tissues. PD-L1 (12) [also termed B7-H1 in humans (13)] and PD-L2 (14) [also called B7-DC (15)] binds to programmed death 1 (PD-1) receptor on T and B cells, but at present the function of these molecules is controversial. Some reports have demonstrated that PD-L1 and PD-L2 have inhibitory effects on T cell responses (12, 14), whereas others have reported that both ligands (B7-H1 and B7-DC) positively regulate T cell proliferation and specifically enhance IL-10 or IFN-γ production (13, 15). Finally, B7-H3, another newly identified B7 homolog, binds an as yet currently unknown counterreceptor on activated T cells and seems to enhance proliferation of CD4+ and CD8+ T cells and selectively increases IFN-γ expression (16, 17). Collectively, these studies have revealed that members of the B7 family may play important roles at diverse stages and sites during the elaboration of cell-mediated immune responses.
In this article, we identify and characterize a B7 family member, B7x, which is expressed not only by cells of hematopoietic origin but also on some nonhematopoietic tissues and some tumor cells. B7x can inhibit proliferation and cytokine production by both CD4+ and CD8+ T cells. It binds a counterreceptor on activated, but not resting, T cells that is distinct from CD28, CTLA-4, ICOS, and PD-1. A B7x-Ig fusion protein binds to Th1 cells derived from WT mice, but not mice with induced null mutation for the newly identified CD28 homolog BTLA (B and T lymphocyte attenuator) (18). Together, these results suggest that the B7x pathway may represent a novel system for downregulation of T cell responses in nonlymphoid tissues and tumors.
Materials and Methods
Bioinformatic Analysis. Public databases were explored by using blast searches with protein and nucleotide sequences. All of the sequence alignment and homology comparison were done with
MACVECTOR 7.0. The phylogenetic tree was generated by paup* version 4.0b10 (19) by using sequence alignment by removal significant inserts and trimming C- and N-terminal extensions. All branches of the tree were supported by bootstrap confidence values of >50% after 100 replicates.
Mice and Cells. C57/BL6 and BALB/c mice were purchased from The Jackson Laboratory and used at age 6-9 wk. Animals were housed in accordance with the Animal Care and Use Committee regulations at the University of California, Berkeley. T cells, cells, and CD11c+ dendritic cells were isolated from the spleen. T cells were purified by negative selection by using the Pan T cell isolation kit (Miltenyi Biotec, Auburn, CA). B cells were positively selected with B220 Dynabeads (Dynal, Oslo). CD11c dendritic cells were positively selected from the spleen after treatment with type II collagenase for 60 min by using CD11c (N418) microbeads (Miltenyi Biotec) to >75% purity. To purify macrophages, mice were injected i.p. with 2 ml of 4% thioglycolate for 48-96 h before washing out the peritoneal exudates cells. Peritoneal macrophages were adhered overnight, and nonadherent cells were removed by washing with PBS. Macrophages were typically >90% F4/80+. The D011.10 Th1 and DO11.10 Th1 BTLA-/- cells were from K.M. All cells were cultured in DMEM supplemented with 10% FCS, 2 μM L-glutamine, 100 units/ml penicillin and streptomycin (all from BioWhittaker), and 2 μM 2-mercaptoethanol (Sigma).
Production of Fusion Protein. B7x-Ig was prepared by fusing the coding region of the extracellular domain of B7x to a chimeric sequence containing the CH2-CH3 domain of mouse IgG1 and a Myc-His tag in pcDNA4 (a gift from William Sha, University of California, Berkeley). The construct was linearized with Bgl and transfected into 293T cells with FuGENE 6 transfection reagent (Roche Molecular Biochemicals). Stable transfectants were selected in 1 mg/ml of Zeocin (Invitrogen). To produce fusion protein, stable transfectants were cultured in serum-free DMEM for 72 h, the supernatant was collected, and B7x-Ig was purified by affinity column chromatography over His-Bind resin (Novagen). We also produced B7x-Ig in an inducible secreted serum-free Drosophila expression system. The coding region of the extracellular domain of B7x was fused to a human IgG1 Fc tag of plasmid pMT/BiP (a gift from Chen Dong, University of Washington School of Medicine, Seattle). The construct was cotransfected into Drosophila cell line S2 with a hygromycin resistance plasmid. The stable transfected cell line was induced with CuSO4 to secrete B7x-Ig in Drosophila serum-free medium (Invitrogen), and B7x-Ig was purified on a ImmunoPure Plus protein G column (Pierce). The purity of the fusion protein was confirmed by SDS/PAGE and immunoblotting with Abs against Myc, mouse IgG, or human IgG.
Real-Time PCR Analysis. cDNA was prepared from total RNA extracted from the isolated cells by using TRIzol reagent (Sigma) by reverse transcription with oligo(dT). Real-time PCR was performed in an ABI Gene Amp 5700 machine with the SYBR green mastermix kit (Applied Biosystems) for B7x and a reference gene (GAPDH). A standard curve of known quantities of PCR template was used for both B7x and GAPDH detection. CT values were set manually above a threshold that reflected linear amplification and converted into arbitrary units by using the known standard curve. B7x expression data were then normalized relative to GAPDH. Primers used were: B7x-For171, 5′-TGGCTTTGGCATTTCAGGC-3′; B7x-Rev287, 5′-CCGTTGAGT T TGATGTCAGGT TC-3′; GA PDH-For46, 5′-AATGGTGAAGGTCGGTGTGAAC-3′; GAPDH-Rev159, 5′-AGGTCAATGAAGGGGTCGTTG-3′; and GAPDHRev500, 5′-CCTTTTGAGGTGCGGATGTAAC-3′.
Retrovirus Constructs. B7x-GFP fusion protein constructs were generated by using PCR to amplify the coding sequence of B7x without the stop codon and then cloned into the pEGFPN3 vector (CLONTECH). After confirmation by DNA sequencing, the constructs of B7x-GFP fusion protein, B7-2/IRES/GFP or GFP alone, were cloned into a mouse stem cell virus retroviral expression vector (a generous gift from William Sha). Retrovirus was produced by transient transfection of the Pheonix-GP packaging cell line. For infection of Chinese hamster ovary (CHO) cells, retroviruses were pseudotyped with vesicular stomatitis virus G-glycoprotein. Stable clones were selected by flow cytometric single-cell sorting.
CHO Cell Stimulation of T Cells. Purified T cells (105 per well) were incubated with mitomycin C-treated CHO transfectants (105 per well) in 96-well plates precoated with anti-CD3 (500A2). Cultures were pulsed with 1 μCi (1 Ci = 37 GBq) of [3H]thymidine per well for the last 16 h of a 72-h incubation.
Cytokine ELISA. Aliquots of supernatants were collected at 48 h after initiation of cell cultures. IL-2 and IFN-γ were measured with mAbs and recombinant cytokine standards from PharMingen.
Flow Cytometry. After incubation with the anti-Fc receptor Ab 24G2 for Fc receptor blocking, cells were stained with B7x-Ig or mouse IgG1 or human IgG1 as a control for 45 min on ice and then stained with phycoerythrin (PE)-conjugate anti-mouse or human IgG (Caltag, South San Francisco, CA). In some experiments, cells were stained with PE- or FITC-conjugated anti-humaster IgG, PE-conjugated anti-ICOS (eBioscience, San Diego), anti-F480 (eBioscience), anti-CD4 and anti-CD8 (Caltag), or biotin-conjugate anti-B7.2 (PharMingen), anti-CD28 (PharMingen), anti-PD-1 (eBioscience), and then stained with PE-Streptavidin (Caltag). The cells were analyzed on an EPICS XL-MCI analytical cytometer (Coulter).
Carboxyfluorescein Diacetate Succinimidyl Ester (CFSE) Staining and Analyses. Purified T cells (107/ml) were washed with Hanks' balanced salt solution, labeled with 2.5 μM CFSE (Molecular Probes) for 10 min at 37°C, and then washed twice with cold completed DMEM. T cells were stimulated with plate-bound anti-CD3 500A2 and the indicated CHO transfectants. On day 4 of culture, cells were stained with PE-anti-CD4 or PE-anti-CD8 and analyzed by flow cytometry.
Results
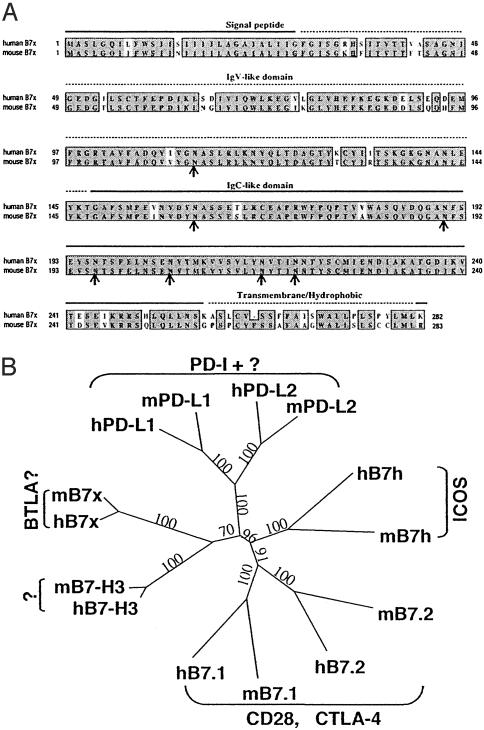
Identification, Cloning, and Characterization of Mouse and Human B7x. By searching the mouse EST database of the National Center for Biotechnology Information with all published mouse B7 family members, we identified three overlapping ESTs with significant homology (GenBank accession nos. BB551556, BB666051, and BI454643), suggesting the existence of a previously unrecognized B7 family member. Using primers derived from these sequences and mRNA from various mouse cell lines, including the dendritic cell line DC2.4, macrophage line J774, and the fibrosarcoma cell line SA1/N, we obtained by RT-PCR cDNAs containing the full-length ORF of the predicted gene. This gene, designated mouse B7x, encodes a putative 283-aa protein (Fig. 1A) and shares varying degrees of amino acid identity with mouse B7-1 (13%), B7-2 (13%), B7h (14%), PD-L1 (20%), PD-L2 (16%), and B7-H3 (24%).
Fig. 1.
B7x is a member of the B7 family. (A) Comparison of human B7x with mouse B7x. Predicted signal peptide, Ig V-like and C-like domains, and the hydrophobic/transmembrane region are indicated. Identical amino acids are highlighted in black, and similar residues are shaded in gray. The potential N-glycosylation sites are indicated by arrows. (B) Phylogenetic tree of mouse and human B7 family generated by paup* version 4.0b10. Numbers show the percentage of bootstrap support for each clade.
By searching the National Center for Biotechnology Information database with mouse B7x, we identified two human epithelial cell cDNAs, both encoding a polypeptide (previously called hypothetical protein FLJ22418) with similarity to mouse B7x. We obtained two EST clones containing portions of these cDNAs and sequenced the inserts to obtain an accurate sequence of the entire ORF. This gene, designated human B7x (Fig. 1A), encodes a putative 282-aa protein with 87% amino acid identity with the mouse B7x. This level is much higher than the 40-46% identity between human and mouse B7-1 or B7-2. Human B7x shares varying levels of identity with human B7-1 (12%), B7-2 (13%), B7h (16%), PD-L1 (18%), PD-L2 (18%), and B7-H3 (24%). This B7x gene appears to be the same as the recently discovered B7-S1 (20) and B7-H4 (21).
The B7x molecule belongs to the Ig superfamily. It has a signal peptide in the N terminus, an extracellular domain with IgV- and IgC-like domains, seven sites for N-linked glycosylation, and a hydrophobic/transmembrane region (Fig. 1A). Like other members of the B7 family, B7x has four conserved cysteine residues that are involved in the formation of IgV- and IgC-like domains. The absence of a heptad structure and B30.2 domains distinguishes B7x from the butyrophilins and myelin oligodendrocyte glycoproteins (22-24). The mouse and human B7x genes have similar organization and size, each consisting of six exons occupying ≈70 kb. In both species the genes have chromosomal locations distinct from other known B7 family members: The mouse B7x is in the F3 region of chromosome 3 and the human B7x is in the p12/13.1 region of chromosome 1.
The identification of B7x brings to seven the known members of the B7 family. A phylogenetic comparison of the family performed with paup* version 4.0b10 (19) is shown in Fig. 1B. This analysis divides the B7 family into three groups: group I including B7-1, B7-2, and B7h; group II consisting of PD-L1 and PD-L2, and group III containing B7x and B7-H3 (Fig. 1B). Interestingly, the receptors for group I (CD28, CTLA-4, and ICOS) are close homologs. The receptor for group II, PD-1, is more distantly related from the receptors for the group I B7 members. The clustering in this tree suggests that the members of group III (B7x and B7-H3) may share the same or closely related receptors distinct from CD28, CTLA-4, ICOS, or PD-1.
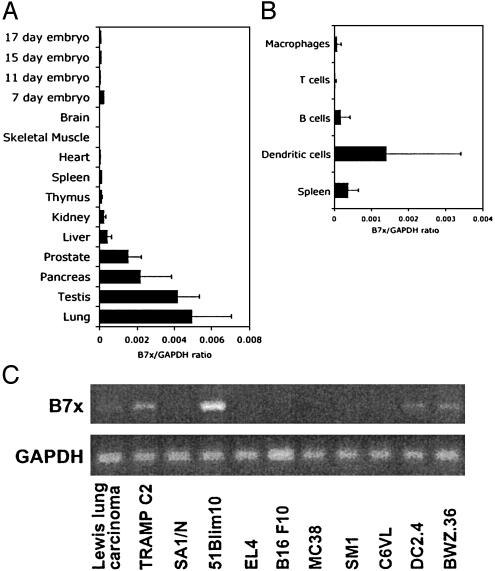
B7x Is Expressed in Lymphoid and Nonlymphoid Tissues and By Some Tumor Lines. The tissue distribution of mouse B7x expression was analyzed by real-time PCR. B7x was detected in most tissues tested, including the spleen and thymus. Interestingly, B7x is most highly expressed in nonlymphoid tissues such as the lung, testis, pancreas, and prostate instead of the spleen. When the expression of B7x was compared in immune cells, we found that B7x was expressed at relatively high levels on splenic CD11c+ dendritic cells and at a low level on splenic B cells, T cells, and thioglycolate-induced peritoneal macrophages (Fig. 2B). Interestingly, EST database searches revealed that five of eight mouse B7x ESTs were derived from mammary tumors, and three of six human B7x ESTs originated from ovarian and uterine tumors, suggesting that some tumor cells highly express B7x. RT-PCR analyses of a panel of murine tumor cell lines revealed that B7x is most highly expressed in 51Blim10 (colon carcinoma), followed by TRAMP C2 (prostate adenocarcinoma), DC2.4 (dendritic cell line), Lewis lung carcinoma, and SA1/N (fibrosarcoma).
Fig. 2.
B7x is widely expressed in multiple tissues and tumor lines. (A) Real-time PCR was performed on cDNA from multiple mouse tissues. cDNA from the CLONTECH mouse multiple-tissue cDNA panel I was used as well as cDNA made from tissues dissected out of two C57/BL6 mice. The error bars represent the SD among the different mouse cDNA samples. (B) Real-time PCR was performed on CD11c+ dendritic cells, B cells, and T cells that were purified from the spleen and compared to the whole spleen. Thioglycolate-induced macrophages were purified by overnight adherence and removal of nonadherent cells. The results shown are the average and SD among 4-10 individual mouse samples. (C) RT-PCR analysis of B7x expression in mouse tumor cell lines.
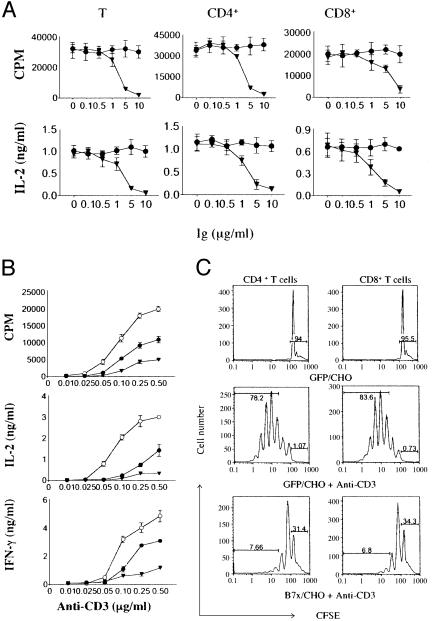
B7x Inhibits T Cell Antigen Receptor-Mediated Responses. We used two approaches to examine the effect of B7x on T cell activation. Initial experiments used purified T cells activated with plate-bound anti-CD3 in the presence of varying amounts of immobilized B7x-Ig. As shown in Fig. 3A, the presence of B7x-Ig decreased proliferation and IL-2 production in a dose-dependent fashion by unfractionated T cells, as well as purified CD4+ and CD8+ T cells. We next used a conventional costimulation assay in which purified T cells were activated with different amounts of plate-bound anti-CD3 in the presence of CHO transfectants expressing GFP, B7-2, or B7x. As expected, T cells stimulated in the presence of B7-2/CHO exhibited enhanced proliferation, IL-2, and IFN-γ production compared with control GFP/CHO (Fig. 3B). In contrast, B7x/CHO significantly reduced proliferation and cytokine production. Together, these results show that B7x can inhibit T cell antigen receptor-mediated T cell proliferation and cytokine production.
Fig. 3.
B7x inhibits T cell activation. (A) Purified T cells and CD4+/CD8+ subsets from BALB/c mice were stimulated with plate-bound anti-CD3 (0.25 μg/ml for CD4+ and total T cells, 2 μg/ml for CD8+ T cells) and plate-bound B7x-Ig (▾) or control Ig (•). Aliquots of supernatants were collected 48 h after initiation of cultures, cytokines were measured by ELISA, and proliferation was measured after 72 h by thymidine incorporation. Error bars indicate SD of triplicate cultures. These data are representative of three independent experiments. (B) Purified T cells were stimulated with plate-bound anti-CD3 and CHO transfectants expressing GFP (•), B7-2 (○), or B7x (▾). Cytokines and proliferation were measured as above. These data are representative of five independent experiments. (C) T cells were labeled with CFSE and stimulated with or without plate-bound anti-CD3 and CHO transfectants expressing GFP or B7x. On day 4, cells were harvested, stained with PE-anti-CD4 or PE-anti-CD8, and analyzed by flow cytometry. Percentages refer to fraction of cells in the nondividing peak or cells that have divided more than twice. These data are representative of three independent experiments.
We further investigated the mechanism of B7x action by labeling T cells with CFSE and stimulating them with various CHO transfectants in the presence or absence of plate-bound anti-CD3. Cells were harvested on day 4 and stained for CD4 and CD8 expression. B7x-mediated inhibition was determined by electronically gating on CD4+ or CD8+ T cells populations and measuring CFSE fluorescence intensity. T cells did not proliferate when incubated with GFP/CHO only (Fig. 3C). When stimulated with anti-CD3 and GFP/CHO, some T cells went through at least seven to eight divisions, with most CD4+ and CD8+ T cells dividing more than two times. However, when T cells were incubated with anti-CD3 and B7x/CHO, they were limited to three to four divisions (Fig. 3C). Further, only ≈1% of T cells did not divide when stimulated with anti-CD3, whereas 31.4% of CD4+ and 34.3% of CD8+ T cells could not divide in the presence of B7x. These differences in the number of divisions and the percentage of nondividing cells indicate that the interaction of B7x and its receptor on T cells leads to decreased proliferation by limiting the number of both CD4+ and CD8+ T cells that enter the cell cycle and reducing their division rate.
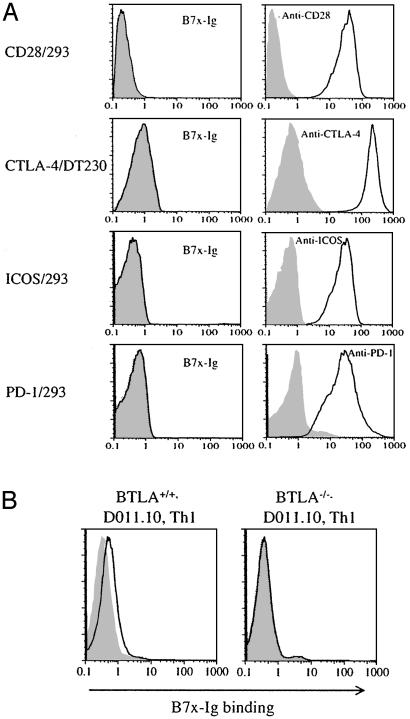
B7x Interacts with a Receptor on Activated T Cells Distinct from CD28, CTLA-4, ICOS, and PD-1. To determine whether T cells express a counterreceptor for B7x, we performed flow cytometric analysis by using a B7x-Ig fusion protein. B7x-Ig did not bind to resting T cells. However, stimulation of T cells with phorbol 12-myristate 13-acetate and ionomycin or anti-CD3/CD28 resulted in acquisition of B7x-Ig binding (data not shown). The relationship of the B7x receptor to known members of the CD28 family was assessed by using B7x-Ig and transfected cell lines that expressed high level of CD28, ICOS, or PD-1 or a tailless form of CTLA-4. As shown in Fig. 4A, expression of these molecules on the transfectants was readily detectable with the appropriate mAbs, but there was no detectable binding of B7x-Ig to any of the cell lines. Also, although CTLA-4-Ig stained B7-2-transfected CHO cells, it did not show detectable binding to B7x-transfected cells (data not shown). These results suggest that B7x has a counterreceptor on activated T cells that is distinct from CD28, CTLA-4, ICOS, and PD-1.
Fig. 4.
B7x has a putative counterreceptor on activated T cells distinct from CD28, CTLA-4, ICOS, and PD-1. (A) The 293 transfectants expressing CD28, ICOS, and PD-l or the DT230 transfectant expressing cell surface CTLA-4 were stained with B7x-Ig fusion protein (open histograms) or mouse IgG1 (shaded histograms) as a control, and were then stained with a PE-conjugate anti-mouse-IgG. All transfectants were also stained with specific mAbs (open histograms) or control Abs (shaded histograms). (B) DO11.10 T cell antigen receptor transgenic Th1 cells from WT or BTLA-/- mice were incubated with irradiated BALB/c spleen cells in the presence of 5 μg/ml OVA323-339 peptide for 4 days. Cells were then incubated with B7x-Ig (open histograms) or human IgG1 (shaded histograms) as a control and stained with a PE-conjugated anti-human-IgG.
BTLA is a member of the CD28 family that is expressed on activated T cells and resting B cells. Interestingly, BTLA has two immunoreceptor tyrosine-based inhibitory motif (ITIM) motifs in its cytoplasmic tail and seems to inhibit T cell responses (18). To determine whether B7x might be a ligand for BTLA, we compared the ability of B7x-Ig to bind to cells from Th1 lines established from WT or BTLA-/- mice. As shown in Fig. 4B, B7x-Ig clearly bound to the WT cells, but there was no detectable binding to BTLA-/- cells. This differential binding strongly suggests that BTLA and B7x-Ig are counterreceptors.
Discussion
Here we have described B7x, a member of the extended B7 family that can inhibit cytokine production and proliferation of both CD4+ and CD8+ T cells. Our preliminary data suggests that a receptor for B7x that is expressed on activated T cells may be the recently identified BTLA (18). Along with B7-1(B7-2)/CTLA-4 and PD-L1(PD-L2)/PD-1, B7x/BTLA is the third set of the B7 family system that mediates downregulation of T cell responses.
The wide distribution of B7x suggests that its function is likely to be quite distinct from the other B7 molecules with inhibitory activity. The expression of B7-1/B7-2 is largely restricted to “professional” antigen-presenting cells, and CTLA-4 mediated attenuation or termination of T cell expansion is thought to occur primarily in secondary lymphoid tissues. The function of the PD-1 system is less clear. Although PD-L1 can be expressed on endothelial cells after exposure to IFN-γ (25), the expression of both PD-L1 and PD-L2 is largely restricted to antigen-presenting cells (15, 26), suggesting that this system is also largely involved in downregulating T cells upon encounter with antigens on antigen-presenting cells, although this interaction could occur in peripheral tissues during an inflammatory response. We find that B7x, on the other hand, is widely expressed in many tissue types in the absence of inflammation or cytokine exposure. This high level of constitutive expression in the lungs, testis, pancreas, and prostate raises the intriguing possibility that B7x might have a role in downregulating responses to tissue-specific antigens in the tissues themselves. In this regard, it is of interest that experimental autoimmune encephalomyelitis is exacerbated in BTLA-/- mice (18).
Also interesting is the observation that B7x is constitutively expressed in some tumors, especially in nonlymphoid tumors. The fact that tumor cells rarely, if ever, express B7-1/B7-2 is thought to be a way by which they avoid direct CD28-mediated costimulation of naive T cells and hence minimize their capacity to initiate immune responses (27). It has been shown that some tumor cells exposed to IFN-γ express PD-L1 and that this exposure can downregulate responses in some instances by promoting T cell apoptosis (26). The constitutive expression of B7x by tumors suggests a mechanism of downregulating antitumor T cell responses at the level of the effector cell. This hypothesis has implications for the development of new strategies for tumor immunotherapy. We have shown that blockade of the inhibitory effects of CTLA-4 on T cell expansion can potentiate antitumor immunity in several animal models and some tumor patients (1, 28). Blockade of B7x offers the potential of enhancing antitumor responses in a way that should prove complementary to CTLA-4 blockade.
Acknowledgments
We thank Kathleen Noonan for invaluable assistance and Jackson Egen and Sujan Shresta for critical reading of the manuscript. This work was supported by the Howard Hughes Medical Institute and National Institutes of Health Grant CA40041 (to J.P.A.). X.Z. is a fellow of the Cancer Research Institute. P.L. is a recipient of the Wellcome International Research Fellowship. K.M. and J.P.A. are Investigators of the Howard Hughes Medical Institute.
Abbreviations: CTLA-4, cytotoxic T lymphocyte antigen 4; BTLA, B and T lymphocyte attenuator; ICOS, inducible costimulator; CFSE, carboxyfluorescein diacetate succinimidyl ester; PE, phycoerythrin; PD-1, programmed death 1; CHO, Chinese hamster ovary.
References
- 1.Egen, J. G., Kuhns, M. S. & Allison, J. P. (2002) Nat. Immunol. 3, 611-618. [DOI] [PubMed] [Google Scholar]
- 2.Sharpe, A. H. & Freeman, G. J. (2002) Nat. Rev. Immunol. 2, 116-126. [DOI] [PubMed] [Google Scholar]
- 3.Abbas, A. K. & Sharpe, A. H. (1999) Nat. Med. 5, 1345-1346. [DOI] [PubMed] [Google Scholar]
- 4.Coyle, A. J. & Gutierrez-Ramos, J. C. (2001) Nat. Immunol. 2, 203-209. [DOI] [PubMed] [Google Scholar]
- 5.Carreno, B. M. & Collins, M. (2002) Annu. Rev. Immunol. 20, 29-53. [DOI] [PubMed] [Google Scholar]
- 6.Liang, L. & Sha, W. C. (2002) Curr. Opin. Immunol. 14, 384-390. [DOI] [PubMed] [Google Scholar]
- 7.Swallow, M. M., Wallin, J. J. & Sha, W. C. (1999) Immunity 11, 423-432. [DOI] [PubMed] [Google Scholar]
- 8.Yoshinaga, S. K., Whoriskey, J. S., Khare, S. D., Sarmiento, U., Guo, J., Horan, T., Shih, G., Zhang, M., Coccia, M. A., Kohno, T., et al. (1999) Nature 402, 827-832. [DOI] [PubMed] [Google Scholar]
- 9.Ling, V., Wu, P. W., Finnerty, H. F., Bean, K. M., Spaulding, V., Fouser, L. A., Leonard, J. P., Hunter, S. E., Zollner, R., Thomas, J. L., et al. (2000) J. Immunol. 164, 1653-1657. [DOI] [PubMed] [Google Scholar]
- 10.Wang, S., Zhu, G., Chapoval, A. I., Dong, H., Tamada, K., Ni, J. & Chen, L. (2000) Blood 96, 2808-2813. [PubMed] [Google Scholar]
- 11.Brodie, D., Collins, A. V., Iaboni, A., Fennelly, J. A., Sparks, L. M., Xu, X. N., van der Merwe, P. A. & Davis, S. J. (2000) Curr. Biol. 10, 333-336. [DOI] [PubMed] [Google Scholar]
- 12.Freeman, G. J., Long, A. J., Iwai, Y., Bourque, K., Chernova, T., Nishimura, H., Fitz, L. J., Malenkovich, N., Okazaki, T., Byrne, M. C., et al. (2000) J. Exp. Med. 192, 1027-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, H., Zhu, G., Tamada, K. & Chen, L. (1999) Nat. Med. 5, 1365-1369. [DOI] [PubMed] [Google Scholar]
- 14.Latchman, Y., Wood, C. R., Chernova, T., Chaudhary, D., Borde, M., Chernova, I., Iwai, Y., Long, A. J., Brown, J. A., Nunes, R., et al. (2001) Nat. Immunol. 2, 261-268. [DOI] [PubMed] [Google Scholar]
- 15.Tseng, S. Y., Otsuji, M., Gorski, K., Huang, X., Slansky, J. E., Pai, S. I., Shalabi, A., Shin, T., Pardoll, D. M. & Tsuchiya, H. (2001) J. Exp. Med. 193, 839-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapoval, A. I., Ni, J., Lau, J. S., Wilcox, R. A., Flies, D. B., Liu, D., Dong, H., Sica, G. L., Zhu, G., Tamada, K. & Chen, L. (2001) Nat. Immunol. 2, 269-274. [DOI] [PubMed] [Google Scholar]
- 17.Sun, M., Richards, S., Prasad, D. V., Mai, X. M., Rudensky, A. & Dong, C. (2002) J. Immunol. 168, 6294-6297. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe, N., Gavrieli, M., Sedy, J. R., Yang, J., Fallarino, F., Loftin, S. K., Hurchla, M. A., Zimmerman, N., Sim, J., Zang, X., et al. (2003) Nat. Immunol. 4, 670-679. [DOI] [PubMed] [Google Scholar]
- 19.Swofford, D. L. (2000) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b10.
- 20.Prasad, D. V., Richards, S., Mai, X. M. & Dong, C. (2003) Immunity 18, 863-873. [DOI] [PubMed] [Google Scholar]
- 21.Sica, G. L., Choi, I. H., Zhu, G., Tamada, K., Wang, S. D., Tamura, H., Chapoval, A. I., Flies, D. B., Bajorath, J. & Chen, L. (2003) Immunity 18, 849-861. [DOI] [PubMed] [Google Scholar]
- 22.Linsley, P. S., Peach, R., Gladstone, P. & Bajorath, J. (1994) Protein Sci. 3, 1341-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry, J., Miller, M. M. & Pontarotti, P. (1999) Immunol. Today 20, 285-288. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes, D. A., Stammers, M., Malcherek, G., Beck, S. & Trowsdale, J. (2001) Genomics 71, 351-362. [DOI] [PubMed] [Google Scholar]
- 25.Eppihimer, M. J., Gunn, J., Freeman, G. J., Greenfield, E. A., Chernova, T., Erickson, J. & Leonard, J. P. (2002) Microcirculation 9, 133-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong, H., Strome, S. E., Salomao, D. R., Tamura, H., Hirano, F., Flies, D. B., Roche, P. C., Lu, J., Zhu, G., Tamada, K., et al. (2002) Nat. Med. 8, 793-800. [DOI] [PubMed] [Google Scholar]
- 27.Townsend, S. E. & Allison, J. P. (1993) Science 259, 368-370. [DOI] [PubMed] [Google Scholar]
- 28.Leach, D. R., Krummel, M. F. & Allison, J. P. (1996) Science 271, 1734-1736. [DOI] [PubMed] [Google Scholar]