Abstract
Comprehensive gene expression patterns generated from cDNA microarrays were correlated with detailed clinico-pathological characteristics and clinical outcome in an unselected group of 99 node-negative and node-positive breast cancer patients. Gene expression patterns were found to be strongly associated with estrogen receptor (ER) status and moderately associated with grade, but not associated with menopausal status, nodal status, or tumor size. Hierarchical cluster analysis segregated the tumors into two main groups based on their ER status, which correlated well with basal and luminal characteristics. Cox proportional hazards regression analysis identified 16 genes that were significantly associated with relapse-free survival at a stringent significance level of 0.001 to account for multiple comparisons. Of 231 genes previously reported by others [van't Veer, L. J., et al. (2002) Nature 415, 530-536] as being associated with survival, 93 probe elements overlapped with the set of 7,650 probe elements represented on the arrays used in this study. Hierarchical cluster analysis based on the set of 93 probe elements segregated our population into two distinct subgroups with different relapse-free survival (P < 0.03). The number of these 93 probe elements showing significant univariate association with relapse-free survival (P < 0.05) in the present study was 14, representing 11 unique genes. Genes involved in cell cycle, DNA replication, and chromosomal stability were consistently elevated in the various poor prognostic groups. In addition, glutathione S-transferase M3 emerged as an important survival marker in both studies. When taken together with other array studies, our results highlight the consistent biological and clinical associations with gene expression profiles.
Breast cancer patients with the same diagnostic and clinical prognostic profile can have markedly different clinical outcomes. This difference is possibly caused by the limitation of our current taxonomy of breast cancers, which groups molecularly distinct diseases into clinical classes based mainly on morphology. Microarray technology with its ability to simultaneously interrogate 10,000-40,000 genes has changed our thinking of molecular classification of human cancers (1). Two major reports have described the use of microarrays to assess the molecular classification of human breast cancer and have defined new subgroups based on expression that are relevant to patient management (2, 3). Sorlie et al. (2) investigated 51 carcinomas from a single patient cohort with locally advanced T3/T4 breast cancer with node involvement treated with primary chemotherapy. van't Veer et al. (3) studied 78 cases of patients with sporadic cancer all under the age of 55 with no lymph node involvement and not treated with adjuvant chemotherapy.
The tumors in both studies could be partitioned into two majors subgroups based on their estrogen receptor (ER) status as suggested by others (4, 5). Additionally, these expression cassettes could provide a refined estimate of prognosis, perhaps beyond those clinical indicators currently available to us. Sorlie et al. (3) identified a luminal subgroup (subgroup A) of ER-positive tumors associated with the best outcome. van't Veer et al. (3) addressed this problem, by investigating a narrow subset of node-negative breast cancer patients. They found 231 genes significantly associated with disease outcome as defined by the presence of distant metastasis at the 5-year mark. Van de Vijver et al. (6) provided a validation of the van't Veer predictor applied to 234 new patients from the same institution and using the same array platform.
In the present work, we have undertaken a population-based study from a regional cancer center where there are 350 new patients a year referred in from a population of 1.5 million. Over a 2-year period, 700 new cancer cases were seen, and of these 700 cases we analyzed 99 cases representative of the population. The overall survival of this group of 99 cases, adjusted for standard prognostic factors of tumor size and nodal status, is comparable to that of the 700 patients selected from the cohort seen in the years 1993-1995. Because patients were treated within existing standards of practice in those years, there was variation in patient management, reflecting the heterogeneity in clinical presentation of breast cancer.
Materials and Methods
Clinicopathological Characteristics of Breast Cancers. Tumor samples from 103 patients with primary local breast carcinoma were accessed from the John Radcliffe Hospital from January 1993 to December 1994. Four of the 103 samples were excluded from the analyses because of technical difficulties, leaving a total of 99 breast tumors. A detailed list of all samples and clinical and histopathological data for the patients is in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). All of the tumor samples were invasive ductal carcinomas; 46 individuals were node negative and 53 were node positive. Almost all of the patients received adjuvant treatment after surgery, consisting of radiotherapy (80 patients), chemotherapy (32 patients), and endocrine therapy (78 patients) according to accepted practice guidelines at that time. The chemotherapy regimen for the majority of the patients consisted of six cycles of cyclophosphamide, methotrexate, and 5-fluorouracil, and the endocrine therapy consisted of tamoxifen for at least 5 years after surgery. All tumor samples had been flash-frozen and stored at -80°C. All of the tumor samples contained >50% of tumor cells based on frozen sections adjacent to the selected samples.
Relapse-free survival (RFS) was defined as the interval elapsed between the date of breast surgery and the date of diagnosed further episode of breast cancer, whether the breast cancer was classified as a recurrence or second primary, and whatever the histology. Breast cancer survival (BCS) was defined as the interval elapsed between the date of breast surgery and the date of breast cancer-related death (documented from hospital records). ER status was determined by using ligand-binding assays and immunohistochemistry. Grade was determined by using the Elston-Ellis grade system (7).
RNA Extraction and Probe Preparation. Isolation of RNA was performed by using the TRIzol method (Invitrogen) according to the manufacturer's instructions. RNA quality from each tumor biopsy was assessed by visualization of the 28S/18S ribosomal RNA ratio on 1% agarose gel. Total RNA was linearly amplified by using a modification of the Eberwine method (8, 9). Total RNA from the Universal Human Reference (Stratagene) was amplified and used as reference for cDNA microarray analysis. The cDNA microarray chips consisted of 7,650 total features and were manufactured at the National Cancer Institute microarray facility.
A detailed protocol for RNA amplification and cDNA probe labeling and hybridization is available at http://nciarray.nci.nih.gov/reference/index.shtml. genepix software (Axon Instruments, Union City, CA) was used to analyze the raw data, which were then uploaded to a relational database maintained by the Center for Information Technology at the National Institutes of Health.
Data Analysis. Images of all of the scanned slides were meticulously inspected for artifacts, and aberrant spots and slide regions were flagged for exclusion from analyses. Log (base 2) ratios for each spot were calculated as follows. In each channel, signal was calculated as foreground median minus background median. If the signal was <100 in any single channel, the signal value in that channel was set to 100. If the signal was <100 in both channels, the spot was flagged as unreliable and not used in any further analyses. Also, if >50% of the pixels in the foreground in either channel reached the saturation threshold, the spot was flagged and not used in analyses. For all remaining (nonflagged) spots, a log ratio was calculated as log2[(red signal)/(green signal)]. The log ratios were then normalized within each array by subtracting from each the median log ratio value across the spots on the array. The channel-specific intensity data and normalized log ratios of all 99 experiments are available in Tables 3-5, which are published as supporting information on the PNAS web site.
The first phase of the analysis was to compare expression profiles between specimens segregated according to values of standard prognostic variables. In particular, we considered the following comparisons: tumor grade 1 or 2 vs. 3; tumor size ≤2 cm vs. >2 cm; age <50 years vs. age ≥50 years (menopausal status); node negative vs. node positive; and ER-vs. ER+. These comparisons were made by parametric t tests using the statistical software splus (SPLUS 6.0 Professional, Insightful, Seattle). To control for multiple comparisons, we reported as significant genes only those that reached significance at level P = 0.001. Testing 7,650 probes at this significance level, we expect that the average number of spuriously significant (false positive) results will be eight or less.
Cluster analyses were conducted to search for natural groupings in the profiles. Before clustering, a screening procedure was applied to eliminate genes showing minimal variation across the set of 99 specimens. Specifically, for each gene, the 5th and 95th percentiles of the ratios were calculated. If the ratio of the 95th to 5th percentile was <3 that gene was not included in the cluster analysis. This process left 706 probe elements for the cluster analyses. Hierarchical agglomerative clustering using the statistical package brb-arraytools software (available at http://linus.nci.nih.gov/BRB-ArrayTools.html) was applied to these normalized log ratios by using both compact linkage and average linkage and both Euclidean and one minus Pearson correlation distance metrics. Normalized log ratios were median-centered within each gene for all of the cluster analyses. The clustering results obtained by using compact linkage with one minus Pearson correlation distance applied to the 706 probe elements appeared by visual inspection to yield the most distinctive clusters (remaining blinded to any clinical or outcome variables), and hence this was the clustering algorithm used for the unsupervised cluster analyses based on these probe elements (Fig. 1). The presence of significant clustering was assessed by applying the global test of clustering proposed by McShane et al. (10). The same techniques were applied for the clustering analyses using gene subsets sets derived from the van't Veer and Sorlie studies (Figs. 3, 4, 5, 6 which are published as supporting information on the PNAS web site).
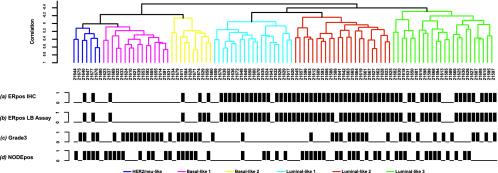
Fig. 1.
Dendrogram of 99 breast cancer specimens analyzed by hierarchical clustering analysis using 706 probe elements selected for the high variability across all tumors (see Materials and Methods). The tumors were separated into two main groups mainly associated with ER status as determined by the ligand-binding (LB) assay and confirmed by immunohistochemistry (IHC). The dendrogram further branched into smaller subgroups within the ER+ and ER-classes based on their basal and luminal characteristics: Her-2/neu subgroup, dark blue; basal-like 1 subgroup, pink; basal-like 2 subgroup, yellow; luminal-like 1 subgroup, light blue; luminal-like 2 subgroup, red; and luminal-like 3 subgroup, green. Black bars represent ER+ tumors assessed by IHC (a), ER+ tumors assessed by LB assay (b), grade 3 (c), and node-positive tumors (d).
Survival comparisons among clusters resulting from unsupervised cluster analysis were made by using Kaplan-Meier estimation and Cox proportional hazards regression. To assess univariate associations of individual genes (log ratios) with survival, Cox proportional hazards regression methods using the splus software were used.
Results
Classification of Tumors Samples Based on Clinical/Pathologic Characteristics. Clinical parameters such as ER status, nodal status, tumor size, tumor grade, and menopausal status of the patient affect the behavior of breast cancers. We asked whether these clinical/pathologic characteristics were associated with differential gene expression. Parametric t tests identified 606 probe elements of the 7,650 elements represented in our array that could segregate ER+ and ER-breast tumors (Table 1, P < 0.001). The detailed gene list is in Table 6, which is published as supporting information on the PNAS web site. Within this list were genes previously known to be estrogen responsive or associated with ER status such as LIV-1, TFF3, GATA 3, c-myb, and BTG2 (11-14). A total of 137 probe elements distinguished high-grade and intermediate/low-grade breast tumors (Table 1, P < 0.001), including genes involved in cell cycle progression such as topoisomerase II α, MCM2, BUB1, and proliferating cell nuclear antigen (PCNA) (15-17). The detailed gene list is included in Table 7, which is published as supporting information on the PNAS web site. Few genes, however, discriminated tumor size (3 probes, P < 0.001), nodal (11 probes, P < 0.001), and menopausal status (13 probes, P < 0.001). This finding suggests that ER status has a strong association with gene expression, and tumor grade has a moderate association. However, there is no strong evidence that nodal and menopausal status of the patient or tumor size is associated with the expression profiles of the tumors.
Table 1. No. of genes discriminating known clinico-pathological phenotypes in breast cancer.
| Clinico-pathological parameters | No. of significant expressed genes, P < 0.001* |
|---|---|
| ER status | |
| ER+ versus ER− | 606 |
| Grade status | |
| Grade 1/2 versus grade 3 | 137 |
| Node status | |
| Node positive versus negative | 11 |
| Tumor size | |
| ≤2 cm versus >2 cm | 3 |
| Menopausal status | |
| Premenopausal versus postmenopausal | 13 |
For 7,650 comparisons, the expected number of spuriously significant (false positive) findings at level P < 0.001 is ≈8 or less.
Unsupervised Clustering Identifies Natural Groups Corresponding to Breast Lineage Markers. Using an unsupervised hierarchical clustering approach, we sought to define natural subclasses of breast tumors as determined by gene expression profiles. We performed the unsupervised clustering on the 706 probe elements selected as exhibiting high variability across all tumors (see Materials and Methods). Included in this list are genes corresponding to the standard prognostic parameters, such as ER and HER-2/neu status. Application of the global test of clustering and reproducibility measures of McShane et al. (10) showed borderline statistically significant evidence for clustering (P = 0.057), and the robustness indices suggested (robustness >75%) that the most reproducible clustering structure was evident when the dendrogram was cut at two clusters. Our results show that the tumor samples could be confidently separated into two main groups primarily associated with ER status as determined by the ligand-binding assay and confirmed by immunohistochemistry (Fig. 1). The entire cluster diagram including gene expressions is included in Fig. 7, which is published as supporting information on the PNAS web site. Eighty-two percent of the tumors in the left main branch are ER-, and 88% of the tumors in the right main branch are ER+ by immunohistochemistry. This finding corroborated the earlier analysis (Table 1) that the major factor discriminating the expression phenotype is ER status. As expected, the ER-cluster had a higher percentage of high-grade tumors than the ER+ cluster (Fig. 1).
The dendrogram further branched into smaller subgroups within the ER+ and ER-classes. Although our sample size was not large enough to establish high reproducibility of smaller subgroups, we were able to locate in our dendrogram several subgroupings previously identified in other studies. We describe here what we observed in our data relative to those clusters that have been reported by others. Within the ER-cluster are tumors with “basal”-like expression characteristics as defined by higher gene expression of keratin 5, keratin 6, metallothionein 1X, and fatty acid binding protein 7 as reported (Table 8, which is published as supporting information on the PNAS site) (2, 18). Furthermore, they exhibited higher expression of the secreted frizzled-related protein 1 (SFRP1) and the oncogene c-kit and lower expression of fibronectin 1 and mucin 1. Basal 1 subgroup was differentiated by higher expression of matrix metalloproteinase 7 and cell growth-related genes such as topoisomerase II α, mitotic feedback control protein Madp2 homolog (MAD2L1), cell division control protein 2 homolog (CDC2), and PCNA, suggesting a signature for a high proliferation rate (Table 8). In contrast, the basal 2 subgroup was distinguished by higher expression of many components of the transcriptional factor AP-1 such as c-fos, c-jun, and fos B, as well as by overexpression of activating transcription factor 3, caveolin 1 and 2, hepatocyte growth factor, and transforming growth factor β receptor II. A subgroup distinct from the basal-like groups in the ER-subset is defined by a high rate of HER-2/neu overexpression (HER-2/neu: 5/7 tumors). This HER-2/neu subgroup was further distinguished from the basal-like subgroups by the higher expression of MDR1, S100 calcium-binding protein P, fatty acid synthase, RAL-B, RAB6A, fibronectin 1, and syndecan 1 and lower expression of c-kit and c-myc.
The ER+ subgroup showed differential expression of genes associated with ER activation such as LIV-1, trefoil factor 3, neuropeptide Y receptor Y1, keratin 8, GATA3, and X-box binding protein 1. These are also genes that define the ER+ cluster as having “luminal” characteristics as defined (2, 18). Moreover, the ER+ cluster could be further segregated into three smaller subclasses: luminals 1, 2, and 3 (Fig. 1).
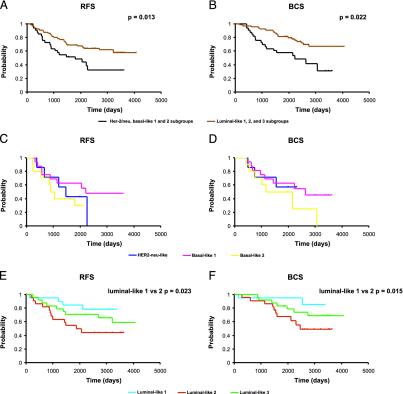
We then asked whether the array-derived tumor groups demonstrated any differences with respect to survival. When the basal/neu (predominantly ER-) and the luminal-like (predominantly ER+) cluster were compared by Kaplan-Meier and Cox regression analysis, the luminal-like subgroup had a significant advantage in both RFS and BCS (Fig. 2 A and B). The three subclasses within the basal/neu (ER-) cluster appeared to have similarly poor survival characteristics (Fig. 2 C and D). By contrast, the three subgroups within the luminal-like (predominantly ER+) cluster showed distinct differences in survival (Fig. 2 E and F). Luminal 1 had the best outcome with an 80% 10-year RFS. This luminal 1 subgroup was also correlated with lower-grade tumors and was further characterized by differential higher expression of c-kit, hepatocyte growth factor, insulin-like growth factor-binding protein-3, ATF-3, and components of the AP-1 transcriptional factor such as c-fos, c-jun, fosB, and jun-D, and as expected by lower expression of cell growth related genes such as topoisomerase II α, mitotic kinesin-like protein-1, PCNA, CDC2, BUB1, and MAD2L1 (Table 8). This was significantly different from the luminal 2 subgroup that had the worst outcome with a 10-year RFS of 40% (P = 0.022 luminal-like 1 vs. luminal-like 2). This subgroup showed higher expression of a protein tyrosine phosphatase type IVA member, tumor necrosis factor receptor-associated factor 3, RAD21, and BRCA1-associated protein 1 (BAP1) and lower expression of FGFR1, CXCR4, ATF-3, and vascular cell adhesion molecule 1. The luminal 3 subgroup had an intermediate survival outcome of 60% at 10 years. Intriguingly, although some of the ER+ tumors were HER-2 overexpressors (6 of 66), they were dispersed among the luminal subgroups.
Fig. 2.
RFS and BCS analysis of the 99 breast cancer patients based on the gene expression cluster analysis classification. RFS (A) and BCS (B) of the predominantly ER- and predominantly ER+ clusters (the Her-2/neu and basal-like 1 and 2 subgroups were considered in one group and the luminal-like subgroups 1, 2, and 3 were considered in the other group). RFS (C) and BCS (D) of the Her-2/neu and basal-like 1 and 2 subgroups. RFS (E) and BCS (F) of the luminal-like 1, 2, and 3 subgroups.
In the Sorlie et al. study (3), tumor classification was performed based on the 456 cDNA clones (427 unique genes) in their “intrinsic” gene list. To assess how these genes could perform in classifying our tumor set, we sought the overlap between either the intrinsic gene list and our cDNA array or the overlap between the intrinsic gene list and the 706 variably expressed probe elements in this study. We found 332 (285 unique genes) that overlapped with the full set of probe elements on our cDNA array and 105 (96 unique genes) probe elements that overlapped with our 706-gene list. Interestingly, in both situations hierarchical cluster analysis segregated our tumors into two distinct subgroups mainly based on their basal (predominantly ER-) and luminal (predominantly ER+) characteristics (Figs. 5 and 6).
The luminal-like tumors were further segregated into at least two (possibly three) smaller subgroups, which may correspond to luminal A, B, and C subtypes. Additional analysis revealed that 100% of the specimens in the luminal A subtype in the 332-gene cluster and 77% in the 105-gene cluster were assigned as luminal-like 1 tumors in our unsupervised cluster analysis (based on the 706-element cluster, Fig. 1). In contrast, the specimens in the luminal B and C subtypes were assigned either as luminal-like 2 or luminal-like 3 tumors.
Although the luminal A subtype showed a favorable clinical outcome when compared with other luminal subtypes, this difference was not statistically significant.
Identification of Gene Clusters Associated with Survival. Our database includes the survival data of all patients studied with a median follow-up of 6.1 years. To determine the genes associated with improved RFS, we performed Cox proportional hazards regressions on each of the 7,650 probe elements on our array. A group of 485 different probe elements were identified that could separate RFS in the 99 patients with a P value <0.05, but only 16 probe elements were significant at the more stringent P < 0.001 level used to control for multiple comparisons (Table 9, which is published as supporting information on the PNAS web site). We discuss results for the larger list of 485 so that our results may be compared with results of previous studies. Of the genes reported to be associated with prognosis in breast cancer, only PCNA and topoisomerase II α were found to be elevated in the poor prognostic group (HER2-neu/basal-like). Interestingly, this gene list did not include the ER, Her-2/neu, or p53. This is consistent with the findings of van't Veer et al. (3), who found no association between HER2 and ER expression levels and survival in the node-negative cases included in their study.
In the van't Veer et al. data set, 231 genes were noted to be prognostic for RFS in the node-negative breast cancer patients. We asked first whether any of these potentially prognostic genes were included in our 7,650-element array. We observed that 93 probe elements representing 56 unique genes overlapped, and based on their expression levels hierarchical cluster analysis we could separate our patients into two distinct subgroups. When Kaplan-Meier analysis was performed, a statistically significant survival difference was seen between these two groups (P = 0.03, Fig. 3). This finding demonstrated that a subset of the genes identified by van't Veer et al. to be prognostic in untreated node-negative patients could be confirmed to have an association with clinical outcome in an independent cohort of treated individuals with mixed nodal status. To identify a minimal number of the most important prognostic genes, we sought the overlap between our optimal survival list of 485 probe elements and the 231 genes in the van't Veer et al. prognostic gene set. This overlap survival gene list consisted of only 11 unique genes represented by 14 probe elements. As expected (because these 14 elements were among those selected because of their observed significant univariate association with survival), these 14 elements separated our patients into two major groups, showing a significant difference in survival (as visualized in Fig. 4). Intriguingly, 5 of the 11 unique genes, RFC4, MCM6, MAD2L1, BUB1, and CKS2 appear to be involved in DNA replication and chromosomal stability, and all were up-regulated in the poor prognostic group. This finding suggests that differences in replicative potential distinguish the prognostic groups.
Discussion
Microarray analyses on breast cancers have identified gene expression profiles able to separate tumor classes associated with patient survival (1). Perou et al. (18) and Sorlie et al. (2) showed that the expression profiles primarily distinguished ER+ from ER-tumors and called them luminal and basal subtypes because of their respective luminal and basal characteristics. van't Veer et al. (3) had similar observation but extended this to gene expression (or genetic profile) associations with survival in an untreated, node-negative cohort.
Here, we present an analysis on 99 tumors from node-positive and node-negative patients, the majority receiving adjuvant treatment according to accepted practice guidelines at the time of the diagnosis. Our results were significant in their concordance with those of the earlier studies despite the differences in patient populations, treatments used, and technology platforms used. Thus, our results provide supporting evidence for the prognostic importance of genes identified in previous reports on a completely independent patient cohort with an independent microarray platform. We found that the ER status of the tumor was, indeed, the most important discriminator of expression subtypes and that tumor grade was a distant second. Other clinical features, namely lymph node positivity, menopausal status, and tumor size were not strongly reflected in the expression patterns obtained with the 7,650-feature microarray in this investigation. This finding confirms that ER biology plays a central role in breast carcinogenesis defining the configuration of the final tumor. Furthermore, investigation of gene expression in primary tumors may be unlikely to identify a set of genes whose expression reliably correlates with lymph node metastasis. This finding is consistent with data showing that only a small fraction of cells in a tumor mass have metastatic potential (19, 20). The genetic signature from this metastatic fraction would be “diluted” by the signals from nonmetastasizing cells.
Similar to the findings of Sorlie et al. (2), unsupervised hierarchical clustering analysis segregated the tumors into two main clusters based on their basal (predominantly ER-) and luminal (predominantly ER+) characteristics. Furthermore, within each of these clusters we could identify smaller subgroups that were characterized by distinct gene expression signatures involving potential different oncogene-specific pathways. A HER-2/neu subgroup was characterized by higher expression of the oncogene her-2/neu and higher expression of genes involved in the ras pathway such a Ras-related GTPases, RALB, and RAB6A. Convergence of neu and ras pathways in breast cancer tumorigenesis has already been documented (21). In contrast, basal 1 and 2 subgroups were characterized by higher expression of the oncogenes c-kit, c-myc, and SFRP1. SFRP1 is a modulator of Wnt signaling. Recently, aberrations of Wnt signaling were reported to be involved in the pathology of various human neoplasms (22). Activation of the Wnt signaling pathway appears to lead to the cytosolic stabilization of a transcriptional cofactor, β-catenin, that can regulate the transcription from a number of target genes including the cellular oncogene c-myc. In breast carcinoma, SFRP1 expression has been associated with loss of ER and the presence of lymphoplasmocytic reaction around the tumor associated with a more aggressive disease (23, 24). Furthermore, basal 1 type exhibited higher expression of genes involved in cell cycle and growth such as PCNA, CDC2, and BUB1 whereas the basal 2 type showed higher expression of transcription factors such as c-fos and ATF3, expression signatures that both could be modulated by c-myc (21). These data raise the possibility that Myc, which is amplified in 15% of breast cancers, may have a more important role in determining the expression profile of a breast cancer than previously thought.
Moreover, as noted in earlier studies, our basal and luminal subgroups also showed the expected differences in survival with a better outcome in the luminal group. More interestingly, we found that the 231 genes described by van't Veer et al. (3) as separating survival groups in node-negative untreated patients may have distinct prognostic capabilities in a more heterogeneous population of node-positive/negative patients treated with adjuvant therapy. Using the 93 probe elements, 56 unique genes, from the van't Veer prognostic set represented in our microarrays, we could easily separate our 99 patients into two prognostic subsets. This finding appears to confirm the importance of some subset of these 56 genes as bona fide prognostic markers. In particular, the overlap between the van't Veer 231 genes and our 485 probes associated with survival (at P < 0.05 level) was 14 probe elements representing 11 unique genes. Five of the 11 genes in this set are involved in cell replication and chromosomal stability and were up-regulated in the poor prognostic setting, suggesting a molecular mechanism for this clinical outcome. An intriguing question is what might be a “minimal” set of genes necessary to establish a reproducible prognostic classification. In leukemia, with well-defined genetic changes, gene profiles segregate with particular translocations (25). In our breast cancer series the major groupings follow previously defined signaling pathways such as ER+, ER-, and c-erbB2/ras. Our study design did not permit us to relate BRCA1 and BRCA2 status to these expression patterns.
Finally, gene profiles that relate to prognosis may help define new therapeutic targets. In our study, cell cycle regulation is clearly important and suggests continued use of antiproliferatives is a rational approach. However, the melanoma tumor antigen PRAME was highlighted and should be further investigated in breast cancer as a potential tumor antigenic target. Also the glutathione S-tranferase pathway, well recognized to have a role in drug resistance, was associated with poor outcome and appeared to be strongly correlated with survival in both our study and that of van't Veer et al. (3).
Supplementary Material
Acknowledgments
This work was supported in part by Fonds National de la Recherche Scientifique Grant Ext. 260 V6/5/2-ILF 14773 (to C.S.), the National Cancer Institute, and the Genome Institute of Singapore.
Abbreviations: ER, estrogen receptor; RFS, relapse-free survival; BCS, breast cancer survival; PCNA, proliferating cell nuclear antigen.
References
- 1.Liu, E. T. & Sotiriou, C. (2002) Breast Cancer Res. 4, 141-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorlie, T., Perou, C. M., Tibshirani, R., Aas, T., Geisler, S., Johnsen, H., Hastie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van't Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A., Mao, M., Peterse, H. L., van der Kooy, K., Marton, M. J., Witteveen, A. T., et al. (2002) Nature 415, 530-536. [DOI] [PubMed] [Google Scholar]
- 4.Gruvberger, S., Ringner, M., Chen, Y., Panavally, S., Saal, L. H., Borg, A., Ferno, M., Peterson, C. & Meltzer, P. S. (2001) Cancer Res. 61, 5979-5984. [PubMed] [Google Scholar]
- 5.West, M., Blanchette, C., Dressman, H., Huang, E., Ishida, S., Spang, R., Zuzan, H., Olson, J. A. J., Marks, J. R. & Nevins, J. R. (2001) Proc. Natl. Acad. Sci. USA 98, 11462-11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van de Vijver, M. J., He, Y. D., van't Veer, L. J., Dai, H., Hart, A. A., Voskuil, D. W., Schreiber, G. J., Peterse, J. L., Roberts, C., Marton, M. J., et al. (2002) N. Engl. J. Med. 347, 1999-2009. [DOI] [PubMed] [Google Scholar]
- 7.Pinder, S. E., Murray, S., Ellis, I. O., Trihia, H., Elston, C. W., Gelber, R. D., Goldhirsch, A., Lindtner, J., Cortes-Funes, H., Simoncini, E., et al. (1998) Cancer 83, 1529-1539. [PubMed] [Google Scholar]
- 8.Sotiriou, C., Khanna, C., Jazaeri, A. A., Petersen, D. & Liu, E. T. (2002) J. Mol. Diagn. 4, 30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sotiriou, C., Powles, T. J., Dowsett, M., Jazaeri, A. A., Feldman, A. L., Assersohn, L., Gadisetti, C., Libutti, S. K. & Liu, E. T. (2002) Breast Cancer Res. 4, 141-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McShane, L. M., Radmacher, M. D., Freidlin, B., Yu, R., Li, M. C. & Simon, R. (2002) Bioinformatics. 18, 1462-1469. [DOI] [PubMed] [Google Scholar]
- 11.El Tanani, M. K. & Green, C. D. (1997) J. Steroid Biochem. Mol. Biol. 60, 269-276. [DOI] [PubMed] [Google Scholar]
- 12.Prevot, D., Morel, A. P., Voeltzel, T., Rostan, M. C., Rimokh, R., Magaud, J. P. & Corbo, L. (2001) J. Biol. Chem. 276, 9640-9648. [DOI] [PubMed] [Google Scholar]
- 13.Hoch, R. V., Thompson, D. A., Baker, R. J. & Weigel, R. J. (1999) Int. J. Cancer 84, 122-128. [DOI] [PubMed] [Google Scholar]
- 14.Gudas, J. M., Klein, R. C., Oka, M. & Cowan, K. H. (1995) Clin. Cancer Res. 1, 235-243. [PubMed] [Google Scholar]
- 15.Wang, J. C. (2002) Nat. Rev. Mol. Cell. Biol. 3, 430-440. [DOI] [PubMed] [Google Scholar]
- 16.Tye, B. K. (1999) Annu. Rev. Biochem. 68, 649-686. [DOI] [PubMed] [Google Scholar]
- 17.Jallepalli, P. V. & Lengauer, C. (2001) Nat. Rev. Cancer 1, 109-117. [DOI] [PubMed] [Google Scholar]
- 18.Perou, C. M., Sorlie, T., Eisen, M. B., Van, D. R., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature 406, 747-752. [DOI] [PubMed] [Google Scholar]
- 19.Fidler, I. J. (1986) Cancer Metastasis Rev. 5, 29-49. [DOI] [PubMed] [Google Scholar]
- 20.Kerbel, R. S., Cornil, I. & Theodorescu, D. (1991) Cancer Metastasis Rev. 10, 201-215. [DOI] [PubMed] [Google Scholar]
- 21.Desai, K. V., Xiao, N., Wang, W., Gangi, L., Greene, J., Powell, J. I., Dickson, R., Furth, P., Hunter, K., Kucherlapati, R., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 6967-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polakis, P. (2000) Genes Dev. 14, 1837-1851. [PubMed] [Google Scholar]
- 23.Speirs, V. (2002) Breast Cancer Res. 4, 169-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ugolini, F., Charafe-Jauffret, E., Bardou, V. J., Geneix, J., Adelaide, J., Labat-Moleur, F., Penault-Llorca, F., Longy, M., Jacquemier, J., Birnbaum, D., et al. (2001) Oncogene 20, 5810-5817. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann, W. K., de Vos, S., Elashoff, D., Gschaidmeier, H., Hoelzer, D., Koeffler, H. P. & Ottmann, O. G. (2002) Lancet 359, 481-486. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.