Abstract
E-cadherin loss in cancer is associated with de-differentiation, invasion, and metastasis. Drosophila DE-cadherin is regulated by Wnt/β-catenin signaling, although this has not been demonstrated in mammalian cells. We previously reported that expression of WNT7a, encoded on 3p25, was frequently downregulated in lung cancer, and that loss of E-cadherin or β-catenin was a poor prognostic feature. Here we show that WNT7a both activates E-cadherin expression via a β-catenin specific mechanism in lung cancer cells and is involved in a positive feedback loop. Li+, a GSK3β inhibitor, led to E-cadherin induction in an inositol-independent manner. Similarly, exposure to mWNT7a specifically induced free β-catenin and E-cadherin. Among known transcriptional suppressors of E-cadherin, ZEB1 was uniquely correlated with E-cadherin loss in lung cancer cell lines, and its inhibition by RNA interference resulted in E-cadherin induction. Pharmacologic reversal of E-cadherin and WNT7a losses was achieved with Li+, histone deacetylase inhibition, or in some cases only with combined inhibitors. Our findings provide support that E-cadherin induction by WNT/β-catenin signaling is an evolutionarily conserved pathway operative in lung cancer cells, and that loss of WNT7a expression may be important in lung cancer development or progression by its effects on E-cadherin.
Loss of chromosome 3p is one of the earliest and most frequent genetic events in lung cancer (1). Although one predominant 3p tumor suppressor gene has not been identified by mutation analysis, at least five distinct homozygous deletion regions have been described that include several genes with demonstrable relevance to lung cancer development and progression (2-6). Epigenetic silencing appears to be the major mechanism by which the expression of these genes is lost (3, 7).
We previously reported that the expression of WNT7a, encoded at 3p25, was absent or markedly reduced in most lung cancer cell lines and direct tumors (8). We also identified a homozygous deletion of β-catenin, encoded at 3p21, in the mesothelioma cell line, NCI-H28 (8). This deletion was independently confirmed and shown to be confined to the β-catenin gene (9). Signaling through the canonical WNT pathway inhibits phosphorylation of β-catenin by GSK3β thereby preventing its proteasome-mediated destruction (10). In turn, β-catenin binds and activates TCF/LEF transcription factors (11). More recently, β-catenin has been shown to activate transcription factors other than TCF/LEF, including the retinoic acid and vitamin D receptors, which promote differentiation (12, 13), and the androgen receptor (14, 15). Each appears capable of competing with TCF/LEF for β-catenin binding. Thus, WNT/β-catenin signaling may have very different consequences depending on the cellular context.
β-catenin has a second role linking E-cadherin to the actin cytoskeleton (10). Loss of E-cadherin induces an epithelialmesenchymal transition with increased tumorigenicity (16). We used a lung tumor microarray to report that loss of E-cadherin or β-catenin had a severe effect on patient survival (17). In a multivariate analysis, E-cadherin loss remained significant. Other than methylation, mechanisms regulating E-cadherin expression in lung cancer have not been thoroughly investigated. Although in Drosophila the E-cadherin homologue is known to be a Wnt target gene (18), this has not been observed in colon cancer cells where many of the known WNT targets have been identified. E-cadherin can be downregulated in epithelial cells by transcription factors that bind E-box elements in its promoter. Four such factors have been identified: Snail, Slug, ZEB, and E12/E47 (19-23). At least for Snail and ZEB, repression involves CtBP binding, which in turn recruits histone deacetylases (HDACs) leading to chromatin inaccessibility (24). Which factors predominate in lung cancer is unknown.
We were intrigued by the loss in lung tumors of two 3p-encoded WNT pathway genes and wondered whether WNT signaling could affect E-cadherin expression. Although Wnt7a up-regulates β-catenin in some contexts (25-27), during limb development Wnt7a signals through a non-β-catenin pathway (28). Our results demonstrate that apparent physiologic levels of WNT7a positively regulate E-cadherin expression in lung cancer cells via β-catenin. Moreover, downregulation of E-cadherin was uniquely correlated with ZEB1 expression and could be reversed by RNA interference against ZEB1. Pharmacologic reversal of E-cadherin and WNT7a losses could be achieved with the use of inhibitors of GSK3β, HDACs, or in some cases only with combined treatment. These findings have potential implications for lung cancer treatment and provide support for the hypothesis that E-cadherin regulation by WNT/β-catenin signaling is evolutionarily conserved.
Methods
Immunohistochemistry of Lung Tumor Specimens. A 193 non-small-cell lung cancer (NSCLC) cohort has been described (17). A second independent cohort of 96 tumors included 33 squamous, 32 adeno, 11 large-cell, and 9 small-cell carcinomas, plus 11 others. Specimens were processed, stained, and scored by using standard methods (17).
Cell Culture. Ten NSCLC and three mesothelioma cell lines were used: squamous (NCI-H157 and H226), large-cell (H1334, H661, and H460), adeno (A549, H2122, H1648, and H1264), bronchioalveolar (H322), and mesothelioma (H513, H290, and H28). All lines were cultured in RPMI medium 1640 under standard conditions. Treatment with inhibitors [trichostatin A (TSA) and lithium chloride (LiCl)] was performed 24 h after cells were plated, with cultures 60-80% confluent. For Wnt1- and Wnt7a-conditioned media, stably transfected HEK293 cells were cultured for 3 days in RPMI medium 1640 containing 10% FBS. When nearly confluent, conditioned media were removed, centrifuged, and used immediately or stored at -80°C.
Cell Lysis and Western Blots. Cells were disrupted in lysis buffer (20 mM Tris·HCl, pH 8.0/100 mM NaCl/0.5% IGEPAL/0.5 mM PMSF/10 μg/ml leupeptin/5 μg/ml pepstatin A/2.1 μg/ml aprotinin) on ice. After sonication, the Bradford assay was used for protein quantification. Protein lysates (10 or 20 μg) were resolved by SDS/PAGE and analyzed by Western blot using PVDF membranes (Millipore) according to the manufacturer's instructions. Anti-β-catenin (C19220) and E-cadherin (C20820) antibodies (BD Biosciences Pharmingen/Transduction Laboratories), and anti-tubulin (Ab-4, NeoMarkers/LabVision) were used at 1:2,000, 1:2,500, and 1:2,000 dilutions, respectively, in PBS and 1% Tween (PBST) containing 1.0% nonfat dry milk. Detection used horseradish peroxidase-conjugated secondary antibodies and chemiluminescence (Western Lightning, Perkin-Elmer). Signals were quantified by densitometry using a Chemi-Imager from Alpha Inotech (San Leandro, CA). Extraction of free cytoplasmic β-catenin was performed as described by Sharma et al. (29). Cells treated with mWnt1- or mWnt7a-conditioned media were rinsed with PBS and scraped into digitonin lysis buffer (1% digitonin/150 mM Tris·HCl, pH 7.5/10 mM MgCl2). Lysates were centrifuged in an Eppendorf 5415C microfuge at 13,000 rpm for 10 min, and supernatants containing soluble cytoplasmic proteins were frozen at -80°C. Pellets containing cytoskeletal and nuclear components were solubilized in RIPA buffer and stored at -80°C.
RNA, Primers, and Quantitative Real-Time RT-PCR. Quantitative RTPCR assays were performed as described (8). Unique PCR products, verified by gel analysis and dissociation curves, were confirmed initially by DNA sequencing. The absence of DNA contamination was verified by “no template” and “no reverse-transcriptase” controls. Amplification data were analyzed by using GENEAMP 5700 SDS software and converted into cycle numbers at a set cycle threshold (Ct values). To normalize for the amount of input cDNA, Ct values for the housekeeping gene GAPDH or 18S RNA were subtracted from the Ct values for each specific gene generating a ΔCt value. All statistical correlations were performed by using ΔCt values and the Spearman correlation function. Primer sequences (5′ to 3′) were: β-catenin (forward) GAG CCT GCC ATC TGT GCT CT, (reverse) ACG CAA AGG TGC ATG ATT TG; E-cadherin (forward) CGG GAA TGC AGT TGA GGA TC, (reverse) AGG ATG GTG TAA GCG ATG GC; Snail (forward) CGC GCT CTT TCC TCG TCA G, (reverse) TCC CAG ATG AGC ATT GGC AG; Slug (forward) AAT ATG TGA GCC TGG GCG C, (reverse) CTC TGT TGC AGT GAG GGC AAG; ZEB1 (forward) AGC AGT GAA AGA GAA GGG AAT GC, (reverse) GGT CCT CTT CAG GTG CCT CAG; mWnt1 set 1 (forward) CCT CCA CGA ACC TGT TGA CG, (reverse) GTT CTG TCG GAT CAG TCG CC; mWnt1 set 2 (forward) TCC CTC CCC TCA CGA CCT C, (reverse) TGT TGC AAG CTC GTC CAG C; hWNT5a (forward) CGG TGT ACA ACC TGG CTG ATG, (reverse) CAC CTT GCG GAA GTC TGC C; mWnt7a set 1 (forward) CCG TTG GAA CTG CTC AGC G, (reverse) CCG CAG CGA TAA TCG CAT AG; mWnt7a set 2 (forward) GCT GGA CCA CAC TGC CAC AG, (reverse) CTT GTT TCG ACT GGC ACG C; 18S RNA (forward) TTT TCG GAA CTG AGG CCA TG, (reverse) CTT GGC AAA TGC TTT CGC TC. Primers for hWNT7A and GAPDH were as described (8).
Methylation-Specific PCR. E-cadherin promoter methylation was determined by the method of Herman et al. (30) using primers specific for methylated and unmethylated E-cadherin sequences of the CpG island as described (31). Specificity of products was verified by DNA sequencing. DNA from normal lymphocytes, treated or not treated with SssI methyltransferase (New England Biolabs), was used as positive controls for the methylated and unmethylated forms, respectively.
RNA Inhibition and Adenoviral Infection. RNA interference inhibition was performed as described (32). Ten microliters of 500 nM double stranded oligonucleotides prepared by using the Silencer kit from Ambion (Austin, TX) were transfected by using Oligofectamine (Invitrogen) into 1 × 105 H661 cells, which were plated 24 h before. Cells were harvested at 48 h. Effective sequences were (5′ to 3′) ZEB188 (antisense), AAT GAT CAG CCT CAA TCT GCA CCT GTC TC; ZEB188 (sense), AAT GCA GAT TGA GGC TGA TCA CCT GTC TC. Adenoviral infections with a S37A β-catenin-encoding adenovirus were performed on NCI-H661 cells by using a multiplicity of infection between 1 and 10 under standard conditions.
Results
Coordinate Expression and Loss of E-Cadherin/β-Catenin Proteins in Lung Cancers. To determine the relationship between E-cadherin and β-catenin protein levels in lung cancer, we analyzed two sets of resected tumors. In 96 tumors from Grenoble, France, E-cadherin and β-catenin protein levels were positively correlated (Pearson product moment correlation = 0.581, P < 0.001, data not shown). Similar correlations (Pearson product moment correlation = 0.658, P < 0.001) were observed in 193 microarrayed NSCLC samples. To identify lung cancer cell lines with reduced E-cadherin, we screened 10 NSCLC and 3 mesothelioma cell lines by Western blot. Five NSCLC lines (NCI-H226, H157, H661, H460, and H1264) and one mesothelioma (H28) had markedly reduced or absent E-cadherin protein. The resected patient samples showed lower frequencies (11-18%) of severe E-cadherin loss, although ∼30% had intermediate loss. The higher frequency of E-cadherin loss in the NSCLC cell lines may reflect their derivation from metastatic sites and advanced tumors (33), which is consistent with studies suggesting that severe E-cadherin loss is associated with aggressive, metastatic disease (17, 34, 35).
Induction of E-Cadherin by GSK3β or HDAC Inhibition and WNT7a/β-Catenin Signaling. Because canonical WNT signaling leads to inhibition of GSK3β, we began our studies by using Li+, a known inhibitor of GSK3β (36). Also, because some transcriptional repressors of E-cadherin use HDACs (24), we wished to compare the Li+ effects against an HDAC inhibitor, TSA. In the large-cell carcinoma, H661, LiCl (20 mM), or a single dose of TSA (0.33 μM) led to a similar induction of E-cadherin protein (Fig. 1A) but with different kinetics, consistent with Li+ and TSA acting through distinct mechanisms. Thus, we reasoned that lower doses of each agent might result in an additive or synergistic effect when combined. The combination of 10 mM Li+ and 41 nM TSA resulted in substantially higher levels of E-cadherin protein at each time point (Fig. 1B).
Fig. 1.
Induction of E-cadherin by Li+ and TSA in lung tumor cell lines. (A) Cells were exposed to Li+ (20 mM) or TSA (0.33 μM) and equal amounts (10 μg) of protein lysates analyzed by Western blot with anti-E-cadherin antibodies. Equal loading was confirmed by parallel gels stained with Coomassie blue (not shown). (B) Additive effects of combined Li+ (10 mM) and TSA (41 nM) in H661 cells. (C) Myo-inositol does not block E-cadherin induction by Li+. H661 cells were treated with 20 mM Li+ for 24 h with or without 2 mM myo-inositol and analyzed by Western blot. Subsequent tubulin detection confirmed equal loading. (D) Effect of 24 h treatment with Li+ (20 mM), TSA (0.33 μM), or the combination on E-cadherin protein induction in additional lung cancer cell lines as described in the text.
Li+ is known to affect inositol signaling through inhibition of inositol monophosphatase (IMPase), an effect that can be rescued by the addition of myo-inositol (37). We treated H661 cells with LiCl in the presence or absence of 2 mM myo-inositol; the addition of myo-inositol had no effect on E-cadherin induction (Fig. 1C). This was confirmed by real-time RT-PCR where 2 mM inositol for 24 h produced a 2-fold or less increase in E-cadherin mRNA compared with a 12.9-fold increase by 20 mM Li+ (not shown). Moreover, 50-250 μM of the specific IMPase inhibitor, L690,330 (38), failed to induce E-cadherin mRNA or protein. These results are consistent with Li+ inducing E-cadherin by effects on GSK3β.
Although responses to Li+ and TSA were robust in H661 cells, it was important to determine the generality of these observations in other lung tumor cells. Li+ (20 mM) as a single agent induced E-cadherin in H290 cells, whereas only TSA (0.33 μM) showed activity in H460 (Fig. 1D). The absence of demonstrable E-cadherin in H290 treated with TSA was caused by apoptosis, as evidenced by activation of caspase-3 (not shown). In H157, only the combination of Li+ plus TSA led to detectable E-cadherin. In contrast, no effects were observed in H226, and only minimal induction by TSA occurred in H1264 (data not shown). H28, which lacks β-catenin because of a homozygous deletion, was unresponsive to Li+, as would be expected if the effect was caused by GSK3β inhibition. Thus, Li+ alone induced E-cadherin in two of six lung tumors having low/absent initial levels, and Li+ had an additive or synergistic effect with TSA in an additional line.
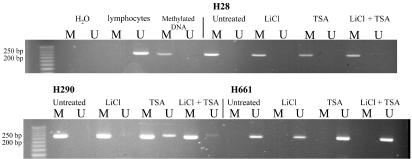
Induction of E-cadherin was observed only in cell lines with intrinsically low E-cadherin expression. Interestingly, WNT7a mRNA was concurrently up-regulated in responding cells as assessed by real-time RT-PCR (not shown). These findings suggest that WNT7a is involved in a positive autoregulatory loop, as has been noted for wingless in Drosophila (39), and that WNT7a might affect E-cadherin induction. In contrast, other genes reported to be WNT targets in colorectal tumors (e.g., c-MYC, cyclin D1, fibronectin, and PPARδ; refs. 40-43) were unaffected by Li+. Lastly, we examined the DNA methylation status of the E-cadherin promoter. Each line with absent/low E-cadherin was predominantly methylated with the exception of H661 (Fig. 2). This may explain the robust inducibility of E-cadherin in this line.
Fig. 2.
E-cadherin promoter methylation analysis. E-cadherin-expressing and -negative cell lines were examined for promoter methylation. Only selected examples are shown. Lymphocyte DNA was used as a source of unmethylated DNA, and in vitro methylated DNA was used as a positive control. Cells were untreated, or treated for 24 h with Li+ (20 mM), TSA (0.33 μM), or both. After DNA extraction and sodium bisulfite treatment, PCR was performed for 35 cycles with primers specific for methylated (M) or unmethylated (U) templates, and the products were analyzed on agarose gels.
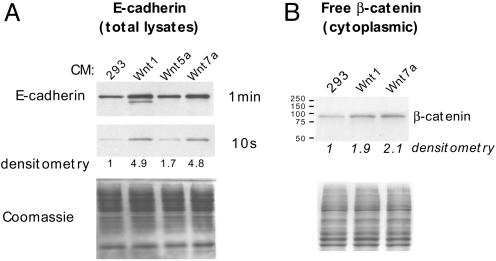
To directly test whether WNT7a could induce E-cadherin and up-regulate free β-catenin, we used cultured media from human HEK293 cells stably transfected with mouse Wnt1 (mWnt1), mWnt5a, or mWnt7a (44). Wnt1 is a classic inducer of canonical Wnt/β-catenin signaling (45), whereas Wnt5a usually signals by a non-β-catenin pathway (46). H661 cells exposed to either mWnt1 or mWnt7a up-regulated E-cadherin protein by ∼5-fold (Fig. 3A, see 10 s exposure). E-cadherin mRNA was similarly induced (7.7-fold, not shown). In contrast, mWnt5a had little effect. Because the mouse Wnts were not epitope-tagged (44), it was necessary to confirm their expression. This was done by using two independent primer sets for mWnt1 and mWnt7a (not shown), although we were unable to confirm the presence of mWnt5a. However, by real-time RT-PCR, human WNT5a was strongly expressed in HEK293 cells and thus could not be responsible for the observed differential effects. Also, by RTPCR neither human WNT1 nor WNT7a was expressed endogenously in 293 cells (not shown).
Fig. 3.
Wnt7a induces E-cadherin by canonical WNT/β-catenin signaling. (A) H661 cells were exposed to cultured supernatants of control 293 cells, or 293 cells stably transfected with mWnt1, mWnt5a, or mWnt7a. After 44 h, cells were harvested and 10 μg of protein extracts per lane were analyzed by Western blot for E-cadherin. Equal loading was confirmed by Coomassie blue staining. Densitometry values are relative to the 293 control supernatant set at 1.0. (B) Increased cytoplasmic β-catenin in H661 cells is induced by cultured supernatants from transfected 293 cells expressing mWnt1 and mWnt7a. H661 cells were exposed to cultured supernatants for 6 h. Twenty micrograms of the cytoplasmic fraction was analyzed for β-catenin. A parallel gel was stained with Coomassie blue for loading.
Because both mWnt1 and mWnt7a supernatants induced E-cadherin, it seemed likely that mWnt7a signaling would lead to increased levels of free β-catenin. To test this, we examined levels of cytoplasmic β-catenin. As shown in Fig. 3B, both mWnt1- and mWnt7a-cultured media induced a 2-fold increase in cytoplasmic β-catenin. Similarly, endogenous WNT7a mRNA was induced 5.6-fold by mWnt7a (not shown), consistent with a positive feedback loop involving β-catenin. The quantitative RT-PCR analysis demonstrated that mWnt7a expression in transfected 293 cells was only ∼2-fold higher than endogenous human WNT7a previously determined in short-term bronchial epithelial cultures (8). Thus, in human bronchial epithelia, WNT7a may function, at least in part, to induce or maintain E-cadherin expression.
Further support for the role of β-catenin was obtained from infection of H661 cells with an adenovirus expressing activated β-catenin. This was complicated, however, because even a low multiplicity of infection (1-2) led to widespread apoptosis by 24 h. Overexpressed β-catenin causes apoptosis in other contexts, even in cell lines harboring β-catenin mutations (47). At 6 h, which precedes the frank apoptotic response, activated β-catenin produced an 8.7-fold increase in WNT7a mRNA and a 2.7-fold increase in E-cadherin mRNA (Table 1). By 12 h and 18 h, WNT7a mRNA had diminished considerably, presumably reflecting β-catenin toxicity. This was reproducible in separate experiments. In contrast, WNT3a expression was largely unaffected (1.4-fold), whereas a control adenovirus expressing GFP had no significant effects (not shown). We also combined β-catenin overexpression with low-dose TSA (41 nM). Again, reproducible increases over TSA alone were detected for WNT7a and E-cadherin, whereas WNT3a showed little change. Thus, we conclude that canonical WNT/β-catenin signaling leads to elevations in WNT7a and E-cadherin in lung cancer cell lines.
Table 1. Activated β-catenin induces E-cadherin and WNT7a.
|
NCl-H661 treatments
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
|
β-Catenin
|
TSA
|
Both
|
|||||||
| 6 h | 12 h | 18 h | 6 h | 12 h | 18 h | 6 h | 12 h | 18 h | |
| WNT7a | 8.7 | 6.7 | 3.8 | 121.1 | 32.4 | 5.4 | 167.3 | 31.5 | 5.1 |
| E-CAD | 2.7 | 2.0 | 1.1 | 10.0 | 8.2 | 6.5 | 12.2 | 8.9 | 5.0 |
| WNT3a | 1.4 | 1.4 | 1.0 | 1.0 | 2.2 | 2.5 | 1.2 | 2.2 | 1.7 |
Real-time RT-PCR analysis of E-cadherin, WNT7a, and WNT3a mRNAs in H661 cells at 6, 12, and 18 h after activated (S37A) β-catenin adenovirus, low-dose (41 nM) TSA, or treatment with both. The Ct values were normalized to GAPDH and 18S RNA with identical results. Fold induction over untreated control cells was calculated by using the estimate that every PCR cycle difference, during exponential amplification, represents a 2-fold change in mRNA levels.
Transcriptional Suppression of E-Cadherin in Lung Cancer Cell Lines Is Caused by ZEB1. Snail, Slug, ZEB, and E12/E47 have been shown to bind E-box sites in the E-cadherin promoter and repress transcription (19-23). To determine whether any of these was associated with E-cadherin suppression in lung cancer, we quantified their expression in cell lines with real-time RT-PCR along with E-cadherin, β-catenin, and WNT7a. Reductions in E-cadherin protein were significantly correlated with levels of E-cadherin mRNA (Table 2), indicating that transcriptional repression is an important mechanism of E-cadherin dysfunction in lung cancer. ZEB1 levels were significantly negatively correlated with E-cadherin protein and mRNA. In addition, both ZEB1 and Snail levels were significantly negatively correlated with WNT7a, whereas neither Slug nor E12/E47 expression demonstrated any significant correlations (not shown).
Table 2. Statistical analysis of quantitative mRNA and protein expression levels.
| Data set (n = 18) | E-Cadherin mRNA | WNT7A mRNA | ZEB-1 mRNA | Snail mRNA | β-Catenin mRNA | β-Catenin protein |
|---|---|---|---|---|---|---|
| E-cadherin protein | 0.83 | 0.8 | −0.82 | −0.3 | 0.33 | 0.74 |
| <0.0001 | <0.0001 | <0.0001 | 0.23 | 0.18 | 0.0004 | |
| E-cadherin mRNA | 0.76 | −0.52 | −0.29 | 0.15 | 0.72 | |
| 0.0003 | 0.028 | 0.24 | 0.56 | 0.0007 | ||
| WNT7A mRNA | −0.71 | −0.57 | 0.40 | 0.76 | ||
| 0.0011 | 0.013 | 0.097 | 0.0003 | |||
| ZEB mRNA | 0.21 | −0.55 | 0.54 | |||
| 0.41 | 0.017 | 0.022 | ||||
| Snail mRNA | 0.01 | −0.42 | ||||
| 0.97 | 0.086 | |||||
| β-Catenin mRNA | 0.42 | |||||
| 0.083 |
Normalized real-time quantitative RT-PCR data were generated from 10 NSCLC and 3 mesothelioma lines for the indicated genes (see Methods). Relative E-cadherin protein expression levels were determined by Western blot and densitometric scanning. Correlations were evaluated by using the Spearman statistic (upper number) and significance (P, lower number). Significant correlations are shown in bold.
To directly test the relationship between ZEB1 and E-cadherin, we used small interfering RNA (siRNA) to inhibit ZEB1 expression in H661 (Table 3). Compared with mock-transfected cells, ZEB-188 siRNA lowered ZEB1 mRNA by 2.7 PCR cycles (∼6.5-fold), whereas E-cadherin expression increased by ∼2-fold. Several additional siRNAs designed to target ZEB1 were ineffective and caused no change in E-cadherin levels. When combined with Li+ and TSA, ZEB1 inhibition led to further increases in E-cadherin mRNA. In contrast, WNT7a levels were unaffected by anti-ZEB1 siRNA alone (not shown), suggesting that Snail may contribute to WNT7a suppression.
Table 3. Inhibition of ZEB1 by RNAi.
|
Fold difference vs. mock-transfected cells
|
||
|---|---|---|
| Treatment | ZEB1 mRNA | E-cadherin mRNA |
| Mock | 1.0 | 1.0 |
| ZEB-188 | −6.5 | 2.2 |
| Mock + lithium | 1.0 | 15 |
| ZEB-188 + lithium | −4.0 | 23 |
| Mock + TSA | −2.3 | 64 |
| ZEB-188 + TSA | −7.0 | 223 |
H661 cells were mock-transfected, transfected with the indicated siRNA as described in the text, or treated with siRNA and either TSA or Li+. The Ct values were normalized to GAPDH, and the fold induction was calculated as in Table 1.
No significant correlation was observed between β-catenin protein and mRNA levels (Table 2), consistent with its predominantly posttranscriptional mode of regulation (11). β-catenin protein, however, was significantly correlated with E-cadherin mRNA, E-cadherin protein, WNT7a and, to a lesser extent, with ZEB1. These findings support the experimental data that WNT/β-catenin signaling affects E-cadherin transcription in lung cancers and also suggest that multiple regulatory interactions might occur among these genes. Thus, at least in lung cancer cell lines, ZEB1 appears to be the predominant suppressor of E-cadherin, whereas WNT7a, at least on the basis of correlative expression data, may be negatively regulated by both ZEB1 and Snail.
Discussion
This investigation stemmed from a 3p21 homozygous deletion confined to the β-catenin gene in H28, which also lacked E-cadherin expression (8, 9). Although that deletion was rare, loss of 3p encoded WNT7a expression occurred in most lung cancer cell lines and many direct tumors (8). In resected lung tumors, β-catenin and E-cadherin protein levels were significantly related, which may explain why reduction of either protein is a poor prognostic feature (17, 34, 48). In our series, the 2-year survival rate for individuals with squamous cancer was 78% with high E-cadherin expression but only 14% with low expression (17).
Our results indicate that WNT7a leads to increased free β-catenin and up-regulation of E-cadherin in lung cancer cells. Based on quantitative analyses, E-cadherin was significantly correlated with WNT7a mRNA and β-catenin protein. Li+, a GSK3 inhibitor (36), led to substantial increases in E-cadherin and WNT7a. Because Li+ also inhibits IMPase (37), it was necessary to exclude that these results were caused by inositol metabolism. Inclusion of myo-inositol did not block the effects of Li+ and L690,330, a specific IMPase inhibitor (38), also had no significant effects. More importantly, mWnt1 and mWnt7a induced E-cadherin mRNA and protein, and both ligands increased levels of free β-catenin. The mechanism of Wnt7a signaling varies in different contexts. In C57MG mammary cells, where Wnt transformation is strongly correlated with free β-catenin induction, Wnt7a effects vary from weak to highly transforming (25, 26). In cerebellar neurons, WNT7a inhibits GSK3β (27), whereas in limb bud development, Wnt7a functions in a non-β-catenin pathway (28). Our finding that mWnt7a induces free β-catenin suggests that endogenous WNT7a in bronchial epithelial cells functions in canonical WNT/β-catenin signaling. In Drosophila, E-cadherin is under the control of Wnt/β-catenin (18). In the mouse, a LEF-1 binding site was identified in the E-cadherin promoter, although no functional studies were described (49). In Xenopus, cadherin-11 is upregulated by Wnt/β-catenin (50). Thus, there is precedent for Wnt/β-catenin signaling to up-regulate cadherin expression.
Our results appear to vary from reports in colon cancer. For example, activating β-catenin mutations in lung tumors are rare except in the unusual fetal-type adenocarcinoma (51-53). In colon cancer, downregulation of nuclear β-catenin induced by dominant-negative integrin-linked kinase leads to up-regulation of E-cadherin (54). However, Snail promoter activity was also inhibited (54), suggesting that E-cadherin changes might be the result of reduced Snail. In lung cancer cells, our results indicate that ZEB1 is the predominant E-cadherin suppressor. Another important difference between lung and colon cancer may involve the transcription factors with which β-catenin interacts. Although TCF/LEFs have been strongly implicated in colon cancer, this relationship has not been established in lung cancer, and we were unable to demonstrate up-regulation of reported TCF/LEF targets (i.e., MYC, Cyclin-D, PPARδ, and fibronectin) in cells treated with Li+. In breast cancer, β-catenin was shown to interact with the retinoic acid receptor (12). Of note, abnormalities in retinoic acid receptor responses have been frequently reported in lung cancer (55-57). Similarly, β-catenin can bind ligand-activated vitamin D receptor and induce E-cadherin (13). Thus, it is possible that the effects of WNT7a and β-catenin in lung cancer might involve transcription factors other than TCF/LEF.
DNA methylation is a well recognized component of gene silencing and affects E-cadherin (58). Methylation at CpG sites is part of a larger regulatory mechanism linked to histone deacetylation. The methyl-CpG binding protein MBD2 is part of a complex (MeCP1) containing HDACs (59). Similarly, the Mi-2 complex, suggested to be the most abundant form of HDAC in mammalian cells, contains the methylcytosine binding protein MBD3 (60). Specific gene silencing, however, requires directed transcriptional repressors. A minimal E-cadherin promoter, capable of epithelial specific expression was shown to be dependent on E-box elements (CANNTG) (61). Subsequent studies identified three zinc-finger proteins and E12/E47 that bind these sites and suppress E-cadherin transcription (19-21, 23, 62). We found that E-cadherin protein levels were significantly correlated with its mRNA (Table 1) in lung cancer cell lines, indicating that a major point of regulation is transcriptional. Among these repressors, only ZEB1 was significantly correlated with E-cadherin downregulation, and inhibition of ZEB1 by siRNA resulted in elevated levels of E-cadherin. Both ZEB1 and Snail were correlated with WNT7a repression (P = 0.0011 and 0.0132, respectively), whereas Slug and E12/E47 showed no significant associations. Because WNT7a was not induced by ZEB1 inhibition, Snail may contribute to WNT7a repression. We note that there is an E-box (CACCTG) within 500-bp upstream of the translation start as well as other sites upstream and within the first intron.
In summary, our results indicate that transcriptional repression of E-cadherin in lung cancer cells may be twofold: by direct action of the repressor(s) on the E-cadherin promoter as reported (62), and by indirect effects on WNT7a, which is a positive regulator of E-cadherin. Thus, loss of WNT7a may contribute to the pathogenesis of lung cancer through effects on E-cadherin. Although GSK3β and HDACs affect many pathways (63-66), our results suggest that inhibitors of these molecules may have a role in the treatment or chemoprevention of lung cancer.
Acknowledgments
We thank Drs. C. Korch and P. Bunn for their comments, J. Jacobsen for tissue culture, and the University of Colorado Cancer Center DNA Sequencing Core (supported by National Institutes of Health Grant CA46934). This work was supported by Lung Cancer Specialized Program of Research Excellence Grant CA58187 and Early Detection Research Network Grant CA85070. J.R. and S.K. were supported by Association pour la Recherche sur le Cancer and Ligue Nationale Contre le Cancer, comités de la Vienne et de la Charente.
Abbreviations: NSCLC, non-small-cell lung cancer; TSA, trichostatin A; Ct values, cycle threshold values; IMPase, inositol monophosphatase; HDAC, histone deacetylase; siRNA, small interfering RNA.
References
- 1.Wistuba, I. I., Behrens, C., Virmani, A. K., Mele, G., Milchgrub, S., Girard, L., Fondon, J. W., III, Garner, H. R., McKay, B., Latif, F., et al. (2000) Cancer Res. 60, 1949-1960. [PubMed] [Google Scholar]
- 2.Burbee, D. G., Forgacs, E., Zochbauer-Muller, S., Shivakumar, L., Fong, K., Gao, B., Randle, D., Kondo, M., Virmani, A., Bader, S., et al. (2001) J. Natl. Cancer Inst. 93, 691-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomizawa, Y., Kohno, T., Kondo, H., Otsuka, A., Nishioka, M., Niki, T., Yamada, T., Maeshima, A., Yoshimura, K., Saito, R., et al. (2002) Clin. Cancer Res. 8, 2362-2368. [PubMed] [Google Scholar]
- 4.Tomizawa, Y., Sekido, Y., Kondo, M., Gao, B., Yokota, J., Roche, J., Drabkin, H., Lerman, M. I., Gazdar, A. F. & Minna, J. D. (2001) Proc. Natl. Acad. Sci. USA 98, 13954-13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tse, C., Xiang, R. H., Bracht, T. & Naylor, S. L. (2002) Cancer Res. 62, 542-546. [PubMed] [Google Scholar]
- 6.Brambilla, E., Constantin, B., Drabkin, H. & Roche, J. (2000) Am. J. Pathol. 156, 939-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agathanggelou, A., Dallol, A., Zochbauer-Muller, S., Morrissey, C., Honorio, S., Hesson, L., Martinsson, T., Fong, K. M., Kuo, M. J., Yuen, P. W., et al. (2003) Oncogene 22, 1580-1588. [DOI] [PubMed] [Google Scholar]
- 8.Calvo, R., West, J., Franklin, W., Erickson, P., Bemis, L., Li, E., Helfrich, B., Bunn, P., Roche, J., Brambilla, E., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 12776-12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shigemitsu, K., Sekido, Y., Usami, N., Mori, S., Sato, M., Horio, Y., Hasegawa, Y., Bader, S. A., Gazdar, A. F., Minna, J. D., et al. (2001) Oncogene 20, 4249-4257. [DOI] [PubMed] [Google Scholar]
- 10.Huelsken, J. & Birchmeier, W. (2001) Curr. Opin. Genet. Dev. 11, 547-553. [DOI] [PubMed] [Google Scholar]
- 11.Bienz, M. & Clevers, H. (2000) Cell 103, 311-320. [DOI] [PubMed] [Google Scholar]
- 12.Easwaran, V., Pishvaian, M., Salimuddin & Byers, S. (1999) Curr. Biol. 9, 1415-1418. [DOI] [PubMed] [Google Scholar]
- 13.Palmer, H. G., Gonzalez-Sancho, J. M., Espada, J., Berciano, M. T., Puig, I., Baulida, J., Quintanilla, M., Cano, A., de Herreros, A. G., Lafarga, M. & Munoz, A. (2001) J. Cell Biol. 154, 369-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, F., Li, X., Sharma, M., Sasaki, C. Y., Longo, D. L., Lim, B. & Sun, Z. (2002) J. Biol. Chem. 277, 11336-11344. [DOI] [PubMed] [Google Scholar]
- 15.Truica, C. I., Byers, S. & Gelmann, E. P. (2000) Cancer Res. 60, 4709-4713. [PubMed] [Google Scholar]
- 16.Christofori, G. & Semb, H. (1999) Trends Biochem. Sci. 24, 73-76. [DOI] [PubMed] [Google Scholar]
- 17.Bremnes, R. M., Veve, R., Gabrielson, E., Hirsch, F. R., Baron, A., Bemis, L., Gemmill, R. M., Drabkin, H. A. & Franklin, W. A. (2002) J. Clin. Oncol. 20, 2417-2428. [DOI] [PubMed] [Google Scholar]
- 18.Yanagawa, S., Lee, J. S., Haruna, T., Oda, H., Uemura, T., Takeichi, M. & Ishimoto, A. (1997) J. Biol. Chem. 272, 25243-25251. [DOI] [PubMed] [Google Scholar]
- 19.Cano, A., Perez-Moreno, M. A., Rodrigo, I., Locascio, A., Blanco, M. J., del Barrio, M. G., Portillo, F. & Nieto, M. A. (2000) Nat. Cell Biol. 2, 76-83. [DOI] [PubMed] [Google Scholar]
- 20.Batlle, E., Sancho, E., Franci, C., Dominguez, D., Monfar, M., Baulida, J. & Garcia De Herreros, A. (2000) Nat. Cell Biol. 2, 84-89. [DOI] [PubMed] [Google Scholar]
- 21.Hajra, K. M., Chen, D. Y. & Fearon, E. R. (2002) Cancer Res. 62, 1613-1618. [PubMed] [Google Scholar]
- 22.Postigo, A. A. & Dean, D. C. (1999) Proc. Natl. Acad. Sci. USA 96, 6683-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Moreno, M. A., Locascio, A., Rodrigo, I., Dhondt, G., Portillo, F., Nieto, M. A. & Cano, A. (2001) J. Biol. Chem. 276, 27424-27431. [DOI] [PubMed] [Google Scholar]
- 24.Chinnadurai, G. (2002) Mol. Cell 9, 213-224. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu, H., Julius, M. A., Giarre, M., Zheng, Z., Brown, A. M. & Kitajewski, J. (1997) Cell Growth Differ. 8, 1349-1358. [PubMed] [Google Scholar]
- 26.Wong, G. T., Gavin, B. J. & McMahon, A. P. (1994) Mol. Cell. Biol. 14, 6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucas, F. R., Goold, R. G., Gordon-Weeks, P. R. & Salinas, P. C. (1998) J. Cell Sci. 111, 1351-1361. [DOI] [PubMed] [Google Scholar]
- 28.Kengaku, M., Capdevila, J., Rodriguez-Esteban, C., De La Pena, J., Johnson, R. L., Belmonte, J. C. & Tabin, C. J. (1998) Science 280, 1274-1277. [DOI] [PubMed] [Google Scholar]
- 29.Sharma, M., Chuang, W. W. & Sun, Z. (2002) J. Biol. Chem. 277, 30935-30941. [DOI] [PubMed] [Google Scholar]
- 30.Herman, J. G., Graff, J. R., Myohanen, S., Nelkin, B. D. & Baylin, S. B. (1996) Proc. Natl. Acad. Sci. USA 93, 9821-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graff, J. R., Herman, J. G., Myohanen, S., Baylin, S. B. & Vertino, P. M. (1997) J. Biol. Chem. 272, 22322-22329. [DOI] [PubMed] [Google Scholar]
- 32.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 33.Oie, H. K., Russell, E. K., Carney, D. N. & Gazdar, A. F. (1996) J. Cell. Biochem., 24, Suppl., 24-31. [DOI] [PubMed] [Google Scholar]
- 34.Kase, S., Sugio, K., Yamazaki, K., Okamoto, T., Yano, T. & Sugimachi, K. (2000) Clin. Cancer Res. 6, 4789-4796. [PubMed] [Google Scholar]
- 35.Sulzer, M. A., Leers, M. P., van Noord, J. A., Bollen, E. C. & Theunissen, P. H. (1998) Am. J. Respir. Crit. Care Med. 157, 1319-1323. [DOI] [PubMed] [Google Scholar]
- 36.Klein, P. S. & Melton, D. A. (1996) Proc. Natl. Acad. Sci. USA 93, 8455-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atack, J. R., Broughton, H. B. & Pollack, S. J. (1995) Trends Neurosci. 18, 343-349. [DOI] [PubMed] [Google Scholar]
- 38.Sconzo, G., Cascino, D., Amore, G., Geraci, F. & Giudice, G. (1998) Cell Biol. Int. 22, 91-94. [DOI] [PubMed] [Google Scholar]
- 39.Hidalgo, A. (1991) Mech. Dev. 35, 77-87. [DOI] [PubMed] [Google Scholar]
- 40.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Science 281, 1509-1512. [DOI] [PubMed] [Google Scholar]
- 41.Tetsu, O. & McCormick, F. (1999) Nature 398, 422-426. [DOI] [PubMed] [Google Scholar]
- 42.Gradl, D., Kuhl, M. & Wedlich, D. (1999) Mol. Cell. Biol. 19, 5576-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He, T. C., Chan, T. A., Vogelstein, B. & Kinzler, K. W. (1999) Cell 99, 335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizushima, T., Nakagawa, H., Kamberov, Y. G., Wilder, E. L., Klein, P. S. & Rustgi, A. K. (2002) Cancer Res. 62, 277-282. [PubMed] [Google Scholar]
- 45.Munemitsu, S., Albert, I., Souza, B., Rubinfeld, B. & Polakis, P. (1995) Proc. Natl. Acad. Sci. USA 92, 3046-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slusarski, D. C., Yang-Snyder, J., Busa, W. B. & Moon, R. T. (1997) Dev. Biol. 182, 114-120. [DOI] [PubMed] [Google Scholar]
- 47.Kim, K., Pang, K. M., Evans, M. & Hay, E. D. (2000) Mol. Biol. Cell 11, 3509-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Retera, J. M., Leers, M. P., Sulzer, M. A. & Theunissen, P. H. (1998) J. Clin. Pathol. 51, 891-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huber, O., Korn, R., McLaughlin, J., Ohsugi, M., Herrmann, B. G. & Kemler, R. (1996) Mech. Dev. 59, 3-10. [DOI] [PubMed] [Google Scholar]
- 50.Hadeball, B., Borchers, A. & Wedlich, D. (1998) Mech. Dev. 72, 101-113. [DOI] [PubMed] [Google Scholar]
- 51.Ueda, M., Gemmill, R. M., West, J., Winn, R., Sugita, M., Tanaka, N., Ueki, M. & Drabkin, H. A. (2001) Br. J. Cancer 85, 64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sunaga, N., Kohno, T., Kolligs, F. T., Fearon, E. R., Saito, R. & Yokota, J. (2001) Genes Chromosomes Cancer 30, 316-321. [DOI] [PubMed] [Google Scholar]
- 53.Nakatani, Y., Masudo, K., Miyagi, Y., Inayama, Y., Kawano, N., Tanaka, Y., Kato, K., Ito, T., Kitamura, H., Nagashima, Y., et al. (2002) Mod. Pathol. 15, 617-624. [DOI] [PubMed] [Google Scholar]
- 54.Tan, C., Costello, P., Sanghera, J., Dominguez, D., Baulida, J., de Herreros, A. G. & Dedhar, S. (2001) Oncogene 20, 133-140. [DOI] [PubMed] [Google Scholar]
- 55.Kim, Y. H., Dohi, D. F., Han, G. R., Zou, C. P., Oridate, N., Walsh, G. L., Nesbitt, J. C., Xu, X. C., Hong, W. K., Lotan, R., et al. (1995) Cancer Res. 55, 5603-5610. [PubMed] [Google Scholar]
- 56.Moghal, N. & Neel, B. G. (1995) Mol. Cell. Biol. 15, 3945-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gebert, J. F., Moghal, N., Frangioni, J. V., Sugarbaker, D. J. & Neel, B. G. (1991) Oncogene 6, 1859-1868. [PubMed] [Google Scholar]
- 58.Zochbauer-Muller, S., Fong, K. M., Virmani, A. K., Geradts, J., Gazdar, A. F. & Minna, J. D. (2001) Cancer Res. 61, 249-255. [PubMed] [Google Scholar]
- 59.Ng, H. H., Zhang, Y., Hendrich, B., Johnson, C. A., Turner, B. M., Erdjument-Bromage, H., Tempst, P., Reinberg, D. & Bird, A. (1999) Nat. Genet. 23, 58-61. [DOI] [PubMed] [Google Scholar]
- 60.Wade, P. A., Gegonne, A., Jones, P. L., Ballestar, E., Aubry, F. & Wolffe, A. P. (1999) Nat. Genet. 23, 62-66. [DOI] [PubMed] [Google Scholar]
- 61.Giroldi, L. A., Bringuier, P. P., de Weijert, M., Jansen, C., van Bokhoven, A. & Schalken, J. A. (1997) Biochem. Biophys. Res. Commun. 241, 453-458. [DOI] [PubMed] [Google Scholar]
- 62.Comijn, J., Berx, G., Vermassen, P., Verschueren, K., van Grunsven, L., Bruyneel, E., Mareel, M., Huylebroeck, D. & van Roy, F. (2001) Mol. Cell 7, 1267-1278. [DOI] [PubMed] [Google Scholar]
- 63.Eldar-Finkelman, H. (2002) Trends Mol. Med. 8, 126-132. [DOI] [PubMed] [Google Scholar]
- 64.Marks, P., Rifkind, R. A., Richon, V. M., Breslow, R., Miller, T. & Kelly, W. K. (2001) Nat. Rev. Cancer 1, 194-202. [DOI] [PubMed] [Google Scholar]
- 65.Johnstone, R. W. (2002) Nat. Rev. Drug Discovery 1, 287-299. [DOI] [PubMed] [Google Scholar]
- 66.Vigushin, D. M. & Coombes, R. C. (2002) Anticancer Drugs 13, 1-13. [DOI] [PubMed] [Google Scholar]