Abstract
Dynamic nuclear polarization has enabled hyperpolarization of nuclei such as 13C and 15N in endogenous substances. The resulting high nuclear polarization makes it possible to perform subsecond 13C MRI. By using the dynamic nuclear polarization hyperpolarization technique, 10% polarization was obtained in an aqueous solution of 100 mM 13C-labeled urea, ready for injection. The in vivo T1 relaxation time of 13C in the urea solution was determined to 20 ± 2 s. Due to the long relaxation time, it is possible to use the hyperpolarized substance for medical imaging. A series of high-resolution (≈1-mm) magnetic resonance images were acquired, each with a scan time of 240 ms, 0-5 s after an i.v. injection of the hyperpolarized aqueous [13C]urea solution in a rat. The results show that it is possible to perform 13C angiography with a signal-to-noise ratio of ≈275 in ≈0.25 s. Perfusion studies with endogenous substances may allow higher spatial and/or temporal resolution than is possible with current proton imaging techniques.
The diagnostic quality of magnetic resonance (MR) images depends on the signal-to-noise ratio (SNR) as well as the contrast-to-noise ratio (CNR). The SNR is a measure of what resolution and/or scan time is needed to separate the NMR signal generated from a given region of interest from the background noise in the image. Every √2-fold improvement in SNR allows a doubling of the spatial resolution in one direction, and has a tremendous effect on the perceived image quality (1). The CNR is a measurement of how well a region of interest can be separated from surrounding regions, which in turn is the real measure of the clinical usefulness of an MR image. Above a given receiver frequency (when the dominating noise source is the subject to be imaged), the SNR will increase linearly with the main field (2). Consequently, the manufacturers of MRI scanners have directed considerable efforts toward the development of higher field instruments with the aim of improving the diagnostic quality of the images. In recent years, 3.0-T instruments have been introduced for clinical whole-body imaging (3).
The use of dynamic nuclear polarization (DNP) techniques such as the Overhauser effect (4) has been suggested as an alternative to increase SNR without the need for high magnetic field strengths (5-7), and a signal enhancement factor of ≈60 has been achieved at a main field of 0.01 T (8). This enhancement is smaller than the theoretically expected factor of 329, which is based on dipolar interaction between the electron spins of the administered free radical and the 1H nuclear spins. The main reason for this discrepancy is that the electron spin system cannot be fully saturated in vivo, because excessive radio frequency irradiation will lead to unacceptable heating of the subject. This problem can be circumvented by performing the polarization process outside the subject and the MR scanner. Nuclear polarizations close to 100% for protons and ≈50% for 13C in various organic molecules have been reported when DNP is performed in a strong magnetic field and at cryogenic temperatures (9, 10). These polarization levels are several orders of magnitude larger than the thermal equilibrium polarizations, which are in the parts-per-million range at clinically available field strengths (≈5 and ≈1 ppm at 1.5 T for 1H and 13C, respectively).
To comply with medical diagnostic needs, two challenges of the DNP technique must be solved.
The hyperpolarized, solid substance must be transferred from a temperature close to 1 K inside a several-Tesla magnet to an injectable solution having body temperature while preserving the nuclear polarization. A solution to this challenge has been described by Ardenkjær-Larsen in ref. 11, where it was shown that various endogenous and exogenous molecules labeled with 13C or 15N could be hyperpolarized at temperatures <2 K and brought to room temperature without significant loss of polarization.
The longitudinal relaxation rate, T1, of the entity in question must be sufficiently long in biological fluids to allow the substance to be distributed to the target organ before the hyperpolarization has vanished.
The transit time from the injection site (i.v., vena cubiti) to the aorta and the coronary arteries is ≈15 s. From the aorta to the peripheral arteries, a further time delay of 10-30 s occurs. In a magnetic field of 0.1-3.0 T, the T1 of tissue protons is in the range of 0.1-2.0 s. Hyperpolarization of protons ex vivo is therefore less interesting for medical applications, because most of the hyperpolarization would have vanished before the molecule reaches the target organ. However, the T1 of 13C in small molecules, in general, is much longer than the T1 of protons. A large number of low molecular weight substances with 13C T1 in excess of 10 s is available. Some of these substances are endogenous and therefore are likely to have lower toxicity than drugs and exogenous contrast media. Urea, a metabolic product of proteins, is an example of such an endogenous substance, with a natural concentration of 1-10 mM in the blood and body fluids. An additional advantage of using 13C rather than 1H as the target nucleus is that the body tissues will be virtually invisible. Only regions where the 13C-labeled, hyperpolarized substance is present will appear in the generated images. Angiographic imaging thus can be performed without background signal from surrounding tissues.
The purpose of the present work was to demonstrate the feasibility of injecting an endogenous substance containing in vitro highly polarized 13C into an animal and visualizing the vascular system and interstitial space with high temporal and spatial resolution in a standard MRI instrument.
Materials and Methods
The Endogenous Substance. [13C]Urea (isotopically enriched to 99%) was used as the imaging agent. The polarization and subsequent dissolution of the substance were performed as described in ref. 11. In each experiment, a syringe containing 2 ml of water with a urea concentration of 100 mM was used for i.v. injection into an animal. At the end of the dissolution process, the polarization and temperature of the urea solution were 25% and ≈30°C, respectively. The solution was transported to the MRI instrument within 40 s. The T1 of the 13C in the solution was 40 s, resulting in a polarization of ≈10% at the moment of injection.
MRI Equipment. The imaging experiments were performed on a 2.35-T animal MR scanner (BioSpec, Bruker Biospin, Ettlingen, Germany). Used was a double-tuned birdcage coil with a 72-mm diameter and 110-mm length and operating at the frequencies of 1H (100.1 MHz) and 13C (25.1 MHz) (Bruker Biospin).
Spectroscopic Experiments. Six female NMRI mice (body weights, 20-25 g) were used in a series of experiments to measure the in vivo T1 relaxation time. The animals were anaesthetized i.v. with a mixture of Hypnorm and Dormicum before placing them in the scanner. All animal experiments were approved by the local ethics committee. The hyperpolarized urea solution was injected i.v. (2 mmol/kg, 0.1 ml/s) through a tail vein. A series of broadband radio frequency (RF) excitation pulses were applied with a time spacing of 4 s. No field gradients were applied; consequently all 13C nuclei within the animal contributed to the generated NMR signal. A total number of 40 RF pulses with a small flip angle of 3o was applied in each experiment. After each RF pulse, the free induction decay signal was acquired and Fourier-transformed. The T1 relaxation was calculated from a monoexponential fit to the amplitudes of the spectral peaks after correction for the signal decay caused by the excitation pulses.
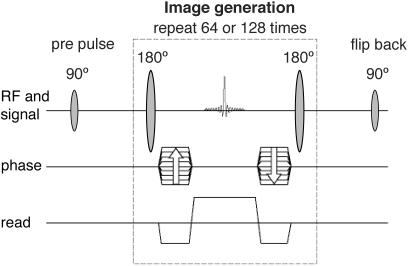
Imaging Experiments. Imaging experiments were performed and included six female and three male Wistar rats (body weights, 250-300 g). The rats were anaesthetized with a mixture of Hypnorm and Dormicum before they were placed in the scanner. In a first series of experiments, angiographic 13C images were generated after an i.v. injection of 2 ml of [13C]urea solution in a tail vein (injection rate, 0.5 ml/s), resulting in a dose of ≈0.7 mmol/kg. The pulse sequence applied in the 13C-imaging experiments was a fully balanced, true fast imaging with steady-state precession sequence (12), as outlined in Fig. 1. The sequence parameters were repetition time/echo time/flip angle = 3.8 ms/1.9 ms/180o, field of view = 7 × 7 cm2, and matrix = 64 × 64 (interpolated to 128 × 128), resulting in a scan time of 240 ms. No slice selection was used. Thus, all generated images corresponded to a coronal projection of the in vivo urea concentration.
Fig. 1.
The fully balanced true fast imaging with steady-state precession sequence used in all 13C-imaging experiments. The sequence starts with a prepulse and is followed by an imaging loop, which is repeated 64 or 128 times depending on the number of phase-encoding steps. The flip-back pulse brings the remaining xy magnetization back to the longitudinal direction.
To compare the 13C angiograms with conventional proton angiograms, a second series of experiments was performed, where representative state-of-the-art proton images were generated after injection of a blood-pool contrast agent (Feruglose, Amersham Health) at a dosage of 4 mg Fe/kg. The proton images were acquired in both 2D and 3D modes by using a first-order flow-compensated gradient echo pulse sequence with low flip angle. The pulse-sequence parameters were repetition time/echo time/flip angle = 26 ms/2.6 ms/40° and field of view = 7 × 7 cm2. The matrix size was 64 × 64 (interpolated to 128 × 128) in 2D mode and 64 × 64 × 32 (interpolated to 128 × 128 × 64) in 3D mode. The slice thickness was 10 mm in the 2D image. In the 3D image set, the total slab thickness was 32 mm, resulting in a slice thickness of 1 mm. The total scan times of the 2D and 3D image sets were 1.7 and 53 s, respectively. The 3D data set was postprocessed to generate a maximum-intensity projection (MIP).
Results and Discussion
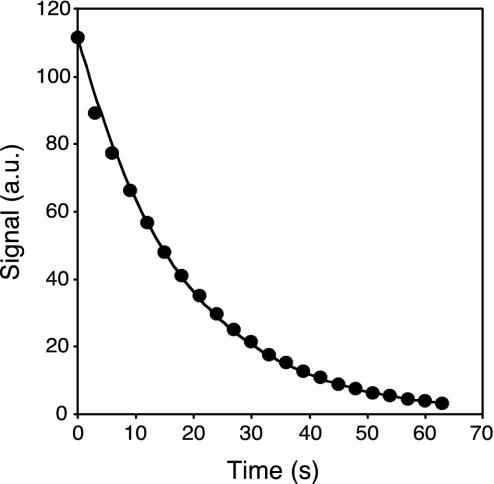
To determine the T1 relaxation time of 13C in vivo after administration of the hyperpolarized 13C-labeled urea, 13C-NMR spectroscopy experiments were carried out in mice. The peak amplitude values of spectra acquired after a train of low flip-angle RF pulses demonstrated a monoexponential decay as function of time after injection (Fig. 2). From these experiments, an in vivo T1 value of 20 ± 2 s was obtained for [13C]urea in the mouse. When a hyperpolarized contrast agent is used for imaging, the magnetization components never reach a true steady-state situation. Instead, the polarization irreversibly decays toward its thermal equilibrium value. However, the long relaxation time of the [13C]urea, compared with the duration of the imaging sequence, ensures that a pseudo-steady-state situation is maintained long enough to allow for collection of all image data. The flip-back RF pulse after the imaging part of the pulse sequence (see Fig. 1) restores the magnetization along the longitudinal direction (13). Due to the long T1, the prepulse, the imaging part, and the flip-back pulse may be repeated for the generation of additional images.
Fig. 2.
Peak amplitude of the NMR spectrum from mouse as a function of time after injection. Circles show the measured values after correction for the signal decay caused by the excitation pulses. The line indicates a monoexponential decay with a T1 of 20 s.
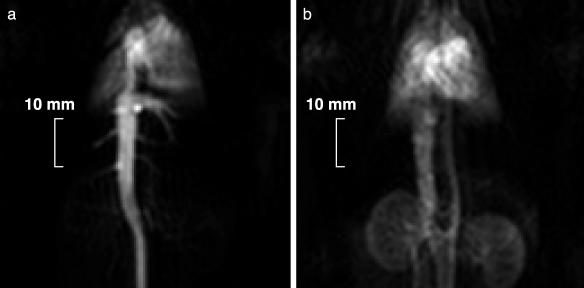
In vivo angiographic imaging with 13C-labeled, hyperpolarized urea was performed in anesthetized rats. The [13C]urea was administered at a dose of 0.7 mmol/kg and an injection rate of 0.5 ml/s via a cannula inserted in a tail vein. Immediately after the injection of the [13C]urea, vena cava, the right side of the heart, and the vascular tree in the lung were visualized (Fig. 3a, image acquired 1 s after the injection). The heart muscle was visualized as well immediately after the injection. Because the molecular weight of urea is low (60 g/mol) and the substance is nonionic, it rapidly diffuses from the intravascular to the extravascular space. Thus, urea may constitute an excellent marker of perfusion in tissue (14). Two seconds later, the urea solution was present in the aorta and had reached the vascular system of the kidneys (Fig. 3b). Distinct angiographic images without tissue background were observed. In these images, acquired with a scan time of 0.25 s each, maximum SNR values of 275 and 180 were measured in vena cava and the cardiac region, respectively. These results demonstrate the feasibility of using hyperpolarization techniques to image 13C-enriched endogenous molecules in vivo. The high degree of nuclear polarization remaining after the sample has been brought from cryogenic to physiological temperatures and the redistribution of the hyperpolarized solution in the circulatory system after i.v. injection suggest that this can be a generally applicable imaging modality with a judicious choice of endogenous tracers.
Fig. 3.
13C coronal projection images of a rat. The image acquisitions were started immediately (a) and 2 s after completing the injection of the contrast agent (b). The scan time of each image was 0.24 s. For other pulse-sequence parameters see Imaging Experiments.
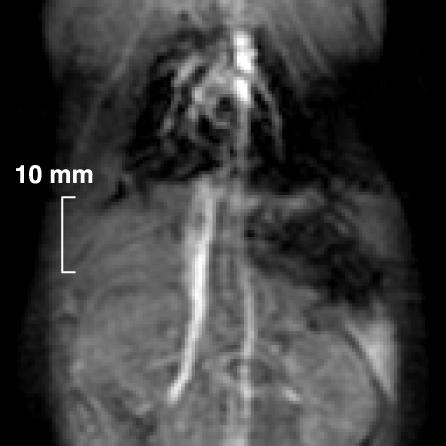
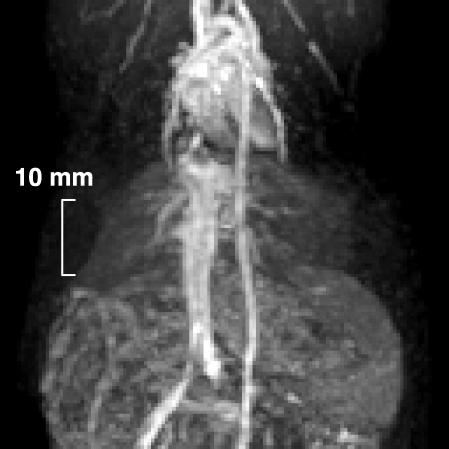
To compare the 13C images obtained by using hyperpolarized urea with conventional proton images, a blood-pool agent (Feruglose, 4 mg/kg Fe) was injected i.v. Contrast-enhanced 1H imaging with 3D acquisition has been established as the method of choice for MR angiography (15, 16). 2D imaging with thick slices generally gives an unsatisfactory result, because the signal from vessels parallel to the slice is obscured by the large background signal coming from the surrounding tissues. This effect is demonstrated in the 2D 1H image (Fig. 4), where e.g., the aorta is poorly visible despite its high signal intensity, and small vessels are not discernible at all. By using 3D imaging sequences in which the slice thickness is comparable to the vessel diameters, combined with MIP postprocessing, also tiny vessels can be visualized, as shown in the MIP of the 3D 1H data set (Fig. 5). Here the heart, the vena cava, the aorta, and part of the lung vascular tree were visualized. In this image, a maximum SNR value of 36 was measured in the heart. The penalty for the increased image quality with 3D sequences, however, is a significantly prolonged scan time, because the scan time increases proportionally with the number of slices. For example, the 32 slices used for generation of the image in Fig. 5 were acquired in 53 s, which is 32 times longer than for the single-slice proton image (Fig. 4), and >200 times longer than for the [13C]urea images with a scan time of 0.25 s (Fig. 3).
Fig. 4.
A 2D 1H coronal image obtained from a rat. The total scan time was 1.7 s. A slice thickness of 10 mm was used. For other pulse-sequence parameters see Imaging Experiments.
Fig. 5.
A coronal MIP of a rat. The MIP was calculated from a 3D 1H data set. The total scan time was 53 s. For other pulse-sequence parameters see Imaging Experiments.
The introduction of a hyperpolarized contrast agent based on an endogenous substance opens up a field of MRI examinations. The function of the injected substance is no longer to modify the amplitude of the signal generated by the protons. Instead, the hyperpolarized nuclei, contained in each molecule, are the source of the NMR signal. When the scanner is tuned to the NMR frequency of the 13C nuclei, the generated images will only show areas where the hyperpolarized substance is present. Because the 13C images completely lack any background signal, the technique offers the possibility for angiographic imaging without the traditional tradeoff between spatial resolution and scan time encountered in 1H MR angiography. A single 13C projection image, as shown in Fig. 3, directly provides the same information as obtained after 3D imaging and MIP postprocessing but with an acquisition time corresponding to the much faster 2D acquisition scheme. A short scan time will reduce motion artifacts and is imperative when imaging rapidly moving vessels, e.g., the coronary arteries. In particular, it is desirable to be able to acquire a full data set within a single breath-hold, because respiratory motion may cause severe image degradation otherwise. With recent advances in scanner technology, proton 3D data sets can be acquired as fast as 7-10 s (17), i.e., at a tolerable duration of a breath-hold. This is achieved, however, at the expense of a fairly coarse image resolution. By using the 2D 13C projection technique, as proposed in the present work, a comparable or even higher spatial resolution may potentially be obtained within a subsecond time frame, thereby minimizing image artifacts associated with both voluntary and involuntary motions of the object.
In contrast-enhanced MRI, the goal is to visualize the different structures in the body with as high SNR and CNR as possible for a given voxel volume and with lowest possible toxic reaction. Traditional MR-contrast media, based on iron, gadolinium, or manganese chelates, interact with the water protons in the tissues (through dipolar interaction) and shortens the relaxation times. The NMR signal originates from the protons and not the contrast molecule per se. The ultimate SNR that can be achieved in proton imaging will depend mainly on the magnetic field strength and the relaxation times of the protons combined with the pulse-sequence parameters. When the contrast agent is present only in the vascular system, short repetition times between excitation pulses then may be exploited to suppress the signal from surrounding tissue, making it possible to generate angiographic images with high SNR and high CNR. However, the contrast agent leaks out fast into the surrounding tissue (with the exception of certain blood-pool agents), and the high CNR is lost after the first passage of the bolus through the capillary bed. This reduces the obtainable CNR when the contrast substance is used to visualize the perfusion of the myocardium (18) and where trapped contrast molecules reduce the relaxation times of the whole heart muscle.
The SNR and CNR in 13C images obtained by using the hyperpolarization concept, will, in addition to the T1 and T2 relaxation times of the hyperpolarized nucleus, be a function of the local tissue concentration of the substance and its polarization level. It is possible to theoretically calculate the image SNR that can be achieved by using the hyperpolarized 13C concept and compare it with the SNR in conventional proton images. Table 1 shows a comparison of thermal equilibrium proton imaging at 3 T, hyperpolarized 13C imaging as performed in the present work, and hyperpolarized 13C imaging under foreseeable optimized conditions. In the table, the concentration (c), the polarization (P), and the gyromagnetic ratio (γ) of the respective nuclei are listed. The theoretically achievable SNR (ignoring differences in RF coil design and loading conditions between the two nuclei) is proportional to the product of these quantities. The product thus may serve as a “figure of merit” when comparing the relative SNR of the different imaging scenarios. For the hyperpolarized urea injection in the present work, the figure of merit is a factor 3 higher than the one obtained for proton imaging at 3 T. As shown in Table 1, if the polarization and concentration of the [13C]urea solution could be increased to 50% and 1 M, respectively, the SNR (from an undiluted bolus) may potentially be ≈100 times higher than the SNR in proton images from a 3-T scanner. The latter estimation is only relevant for an arterial injection or during interventional work where the dilution is insignificant. During an i.v. injection, the bolus will be diluted in the heart and lungs before reaching the arteries. Taking into account both the bolus dilution during a fast i.v. injection and the signal attenuation due to relaxation, it should still be possible, however, to improve the SNR in the heart and the brain by a factor of ≈5, relative to 3 T proton imaging, by using a contrast agent based on hyperpolarized [13C]urea. With respect to the injection of high-osmolality solutions, e.g., iodinated x-ray contrast agents can be injected in concentrations of 1 M (in doses up to 2.5 mmol/kg). We therefore foresee that an injection of 1 M urea should be acceptable, considering its low toxicity (19). Consequently, the polarization method in the present work, combined with fast imaging pulse sequences, should open the possibility of improved image quality in MRI.
Table 1. The concentration (c), gyromagnetic ratio (γ), and polarization (P) of protons at 3 T main magnetic field compared with the corresponding values when injecting solutions of hyperpolarized [13C]urea.
| 1H, 3 T | [13C]Urea | [13C]Urea optimized | |
|---|---|---|---|
| c, M | 80 | 0.1 | ≈1.0 |
| γ, MHz/T | 42.5 | 10.7 | 10.7 |
| P, polarization | 1·10−5 | 0.1 | ≈0.5 |
| c·γ·P | 0.034 | 0.11 | ≈3.2 |
The middle column corresponds to the conditions during the performed experiments, and the right column corresponds to foreseen optimized conditions. The relative SNR is indicated by the product of c·γ·P.
The presented DNP method is not only applicable to 13C but also to other NMR-active nuclei (e.g., 15N). Thus, hyperpolarized injectables based on other endogenous substances such as alanine, glutamine, and acetate may be generated (see ref. 11) and imaged. Depending on the metabolic rates, some of these endogenous substances may allow real-time metabolic mapping to be carried out. The MR field may be expanded to use [13C]acetate as a tracer of regional myocardial oxygen consumption, much in the same way as has been proven feasible recently by using [11C]acetate and positron emission tomography (14, 20).
The possibility of using 13C-labeled d-[1-13C]glucose to perform an in vivo study of the glucose metabolism has been reported in the literature (21-23). In those studies, scan times have been 3-10 min, depending on the main magnetic field. If the metabolic turnover rates are sufficiently high compared with the T1 relaxation time, the hyperpolarization of 13C-labeled endogenous substances may allow mapping of the metabolites with scan times on the order of seconds. Due to the large chemical shift of 13C and 15N, it should be possible to separate the metabolite image from that of the originally injected molecule. Molecular imaging with MRI thus may become a reality.
Abbreviations: MR, magnetic resonance; SNR, signal-to-noise ratio; CNR, contrast-to-noise ratio; DNP, dynamic nuclear polarization; RF, radio frequency; MIP, maximum-intensity projection.
References
- 1.Haacke, E. M., Brown, R. W., Thompson, M. R. & Venkatesan, R. (1999) Magnetic Resonance Imaging—Physical Principles and Sequence Design (Wiley-Liss, New York), pp 331-380.
- 2.Edelstein, W. A., Glover, G. H., Hardy, C. J. & Redington, R. W. (1986) Magn. Reson. Med. 3, 604-618. [DOI] [PubMed] [Google Scholar]
- 3.Campeau, N. G., Huston, J., Bernstein, M. A., Lin, C. & Gibbs, G. F. (2001) Top. Magn. Reson. Imaging 12, 183-204. [DOI] [PubMed] [Google Scholar]
- 4.Overhauser, A. W. (1953) Phys. Rev. 92, 411-412. [Google Scholar]
- 5.Lurie, D. J., Bussell, D. M., Bell, L. H. & Mallard, J. R. (1988) J. Magn. Reson. 76, 366-370. [Google Scholar]
- 6.Grucker, D. (1990) Magn. Reson. Med. 4, 140-147. [DOI] [PubMed] [Google Scholar]
- 7.Golman, K., Leunbach, I., Ardenkjær-Larsen, J. H., Ehnholm, G. J., Wistrand, L.-G., Petersson, J. S., Järvi, A. & Vahasalo, S. (1998) Acta Radiol. 39, 10-17. [DOI] [PubMed] [Google Scholar]
- 8.Golman, K., Petersson, J. S., Ardenkjær-Larsen, J.-H., Leunbach, I., Wistrand, L.-G., Ehnholm, G. & Liu, K. (2000) J. Magn. Reson. Imaging 12, 929-938. [DOI] [PubMed] [Google Scholar]
- 9.de Boer, W., Borghini, M., Morimoto, K., Niinikoski, T. O. & Udo, T. (1974) J. Low Temp. Phys. 15, 249-267. [Google Scholar]
- 10.de Boer, W. & Niinikoski, T. O. (1974) Nucl. Instrum. Methods 114, 495-498. [Google Scholar]
- 11.Ardenkjær-Larsen, J. H., Fridlund, B., Gram, A., Hansson, G., Hansson, L., Lerche, M. H., Servin, R., Thaning, M. & Golman, K. (2003) Proc. Natl. Acad. Sci. USA 100, 10158-10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oppelt, A., Graumann, R., Barfuss, H., Fisher, H., Hartl, W. & Schajor, W. (1986) Electromedica 54, 15-18. [Google Scholar]
- 13.Scheffler, K., Heid, O. & Hennig, J. (2001) Magn. Reson. Med. 45, 1075-1080. [DOI] [PubMed] [Google Scholar]
- 14.Klein, L. J., Visser, F. C., Knaapen, P., Peters, J. H., Teule, G. J. J., Visser, C. A. & Lammertsma, A. A. (2001) Eur. J. Nucl. Med. 28, 651-668. [DOI] [PubMed] [Google Scholar]
- 15.Rubin, G. D., Herfkens, R. J., Pelc, N. J., Foo, T. K. Napel, S., Shimakawa, A., Steiner, R. M. & Bergin, C. J. (1994) Invest. Radiol. 29, 766-792. [DOI] [PubMed] [Google Scholar]
- 16.Gilfeather, M., Holland, G. A., Siegelman, E. S., Schnall, M. D., Axel, L., Carpenter, J. P. & Golden, M. A. (1997) Radiographics 17, 423-432. [DOI] [PubMed] [Google Scholar]
- 17.Masunaga, H., Takehara, Y., Isoda, H., Igarashi, T., Sugiyama, M., Isogai, S., Kodaira, N., Takeda, H., Nozaki, A. & Sakahara, H. (2001) Am. J. Roentgenol. 176, 1213-1219. [DOI] [PubMed] [Google Scholar]
- 18.Bertschinger, K. M., Nanz, D., Buechi, M., Luescher, T. F., Marincek, B., von Schulthess, G. K. & Schwitter, J. (2001) J. Magn. Reson. Imaging 14, 556-562. [DOI] [PubMed] [Google Scholar]
- 19.Gratton, J. A., Lightman, S. L. & Bradbury, M. W. (1993) J. Physiol. (London) 470, 651-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sciacca, R. R., Akinboboye, O., Chou, R. L., Epstein, S. & Bergmann, S. R. (2001) J. Nucl. Med. 42, 63-70. [PubMed] [Google Scholar]
- 21.Skehira, H., Hashimoto, T., Fukuda, H., Ueshima, Y., Yamai, S., Maki, T., Iinuma, T. A. & Tateno, Y. (1990) Am. J. Physiol. Imaging 5, 50-54. [PubMed] [Google Scholar]
- 22.Morishita, S., Sumi, M., Nishimura, R., Takahashi, M. & Iriguchi, N. (1992) Magn. Reson. Med. 10, 94-100. [PubMed] [Google Scholar]
- 23.Bulsing, J. M., Brooks, W. M., Field, J. & Doddrell, D. M. (1984) J. Magn. Reson. 56, 167-172. [Google Scholar]