Abstract
Conjugative coupling proteins (CPs) are proposed to play a role in connecting the relaxosome to a type IV secretion system (T4SS) during bacterial conjugation. Here we present biochemical and genetic evidence indicating that the prototype CP, TrwB, interacts with both relaxosome and type IV secretion components of plasmid R388. The cytoplasmic domain of TrwB immobilized in an affinity resin retained TrwC and TrwA proteins, the components of R388 relaxosome. By using the bacterial two-hybrid system, a strong interaction was detected between TrwB and TrwE, a core component of the conjugative T4SS. This interaction was lost when the transmembrane domains of either TrwB or TrwE were deleted, thus suggesting that it takes place within the membrane or periplasmic portions of both proteins. We have also analyzed the interactions with components of the related IncN plasmid pKM101. Its CP, TraJ, did not interact with TrwA, suggesting a highly specific interaction with the relaxosome. On the other side, CPs from three different conjugation systems were shown to interact with both their cognate TrwE-like component and the heterologous ones, suggesting that this interaction is less specific. Mating experiments among the three systems confirmed that relaxosome components need their cognate CP for transfer, whereas T4SSs are interchangeable. As a general rule, there is a correlation between the strength of the interaction seen by two-hybrid analysis and the efficiency of transfer.
Bacterial conjugation is the prevailing mechanism of horizontal gene transfer among prokaryotes. It is a complex process that involves at least two steps: conjugative DNA processing and DNA transport. DNA processing is conducted by a protein-DNA complex called relaxosome. In Gram-negative bacteria, a set of proteins constitute a type IV secretion system (T4SS) that forms a transmembrane channel. The nature of the molecular machine used for DNA transport is still under debate. The T4SS might just be needed to transport a pilot protein that would drive the displaced replicating DNA strand through the secretion pore (1), or it may be the device that directly pumps the DNA through the membranes and into the recipient cell (2). In both cases, a protein is needed to couple the relaxosome to the transport site.
The protein generally accepted to play this role is called coupling protein (CP), whose prototype is plasmid R388 protein TrwB. Evidence for its coupling role comes from several facts (reviewed in ref. 1): (i) genetic data suggest that it interacts with both the relaxosome and the transporter; (ii) its cellular localization is mainly cytoplasmic, anchored to the inner membrane; (iii) functionally, it does not fit in any of the conjugation moieties [it is neither required for initial DNA processing nor for pilus production (a characteristic of T4SS components)]; and (iv) many T4SSs involved in protein secretion do not have an associated CP. It has been proposed that CPs could also play an active role in DNA transport during conjugation, based on similarities to the DNA pumps FtsK and SpoIIIE (1, 3, 4). However, no direct physical proof for the coupling role has been reported. A key aspect of the coupling activity is the predicted interaction that the CP has to undertake, both with the relaxosome and with the T4SS. This issue was previously addressed by several laboratories working on different conjugation systems. Some evidences for interactions between CPs and relaxosome components were reported. Whereas in plasmid RP4 the CP TraG interacts directly with the relaxase (5, 6), in plasmid F the CP TraD interacts in vitro with the accessory nicking protein TraM (7). On the other hand, no interaction has ever been shown between a CP and a T4SS component, an analysis probably hampered by the fact that these are membrane proteins and thus most interactions probably take place within the membrane or at the periplasm. A recent work in the Agrobacterium Vir system used a peptide library that included the putative CP in addition to all T4SS components in an extensive search for protein peptides that would interact (8). Several interactions were shown but none involved the CP.
Previous work with a cytoplasmic soluble domain (TrwBΔN70) of plasmid R388 CP TrwB showed that it binds DNA and ATP (9). The crystal structure of this domain was determined. A model of the full-length, integral membrane protein displays a hexamer structurally similar to the ATP-synthase complex (10, 11). The full-length protein was recently characterized, and its structural characteristics confirm the previous model (12). In this work we analyze the interactions of TrwB with other R388 conjugation proteins in search of further evidence of its coupling role. The TrwB cytoplasmic domain interacts in vitro with the two R388 protein components of the relaxosome, TrwA and TrwC. Most interestingly, a strong interaction is shown in vivo between TrwB and TrwE, a core component of the conjugative T4SS that is highly conserved in other T4SSs that play a direct role in the virulence of plant and animal pathogens (2). We extended our analysis to other conjugative systems to gain further evidence that proves the significance of these interactions. It is shown that the specificity of the interaction lies mostly in contacting the relaxosome, whereas CPs can efficiently use heterologous T4SSs.
Methods
Bacterial Strains. Escherichia coli lacIq strain D1210 (13) was used for plasmid storage and Plac- or Ptac-driven expression. Strain BL21::DE3 (14) was used for protein overproduction from pET plasmids. Plasmid pLysS was introduced in this strain when over-producing the C-terminal His-tagged TrwA protein (TrwAh). For conjugation experiments, strains D1210 or DH5α (15) were used as donors, and strains DH5α and UB1637 (16) were the respective recipients. Strain DHM1 (F-, cya-854, recA1, endA1, gyrA96, thi1, hsdR17, spoT1, rfbD1, glnV44(AS); G. Karimova, Institut Pasteur, Paris) was used as a host in two-hybrid assays.
Plasmids. Plasmids used are shown in Tables 1, 2, 3 and were constructed by using standard recombinant DNA technology (25). A detailed description of their construction is provided in Table 6, which is published as supporting information on the PNAS web site, www.pnas.org. Expression vectors pGEX-3X (Pharmacia) and pET series (Novagen) were used for protein overproduction, and vectors pT18, pUT18, pUT18C, and pT25 (26) were used for two-hybrid assays.
Table 1. Plasmids used for protein production.
| Plasmid | Construction | Overproduced proteins | Ref. |
|---|---|---|---|
| pMTX501 | pGEX-3X::trwBΔN75 | GST-TrwBΔN75 | This work |
| pMTX515 | pGEX-3X::trwA | GST-TrwA | This work |
| pMTX609 | pGEX3X::traJΔN76 | GST-TraJΔN76 | This work |
| pSU1501 | pKK223-3::trwC | TrwC | 17 |
| pSU1547 | pET22b::trwA | TrwAh | G. Moncalián and F.d.l.C., unpublished |
| pSU1548 | pET22b::trwAN73 | TrwAhN73 | G. Moncalián and F.d.l.C., unpublished |
| pSU1550 | pET22b::trwAΔN35 | TrwAhΔN35 | G. Moncalián and F.d.l.C., unpublished |
| pSU1588 | pET3a::trwCN293 | TrwCN293 | This work |
| pSU4637 | pET3a::trwBΔN70 | TrwBΔN70 | 9 |
Table 2. Plasmids used for complementation assays.
| Plasmid | Construction | Relevant protein products | Ref. |
|---|---|---|---|
| pKM101 | Natural IncN plasmid | All pKM101 proteins | 18 |
| pMTX681 | pSU19::oriT, traK, traI | pKM101 TraK and TraI | This work |
| pSU1092 | ω insertion in trwA | All R388 proteins except TrwA | 19 |
| pSU1404 | pSU4051(MobW)::Tn5tac1 in trwB | R388 TrwA and TrwC | 20 |
| pSU1423 | pSU18::mobW | R388 TrwA, TrwB, and TrwC | 19 |
| pSU1425 | R388 without EcoRI site | All R388 proteins | 20 |
| pSU1443 | pSU1425::Tn5tac1 in trwB | All R388 proteins except TrwB | 21 |
| pSU1445 | pSU1425::Tn5tac1 in trwC | All R388 proteins except TrwC | 21 |
| pSU2007 | R388 KmR | All R388 proteins | 22 |
| pSU4132 | pSU1425::Tn5tac1 in trwD | All R388 proteins except TrwD | 23 |
| pSU4133 | pSU1425::Tn5tac1 in trwK | All R388 proteins except TrwK | This work |
| pSU4134 | pSU1425::Tn5tac1 in trwE | All R388 proteins except TrwE | This work |
| pSU4280 | pSU19::mobN | pKM101 TraK, TraJ, and TraI | This work |
| R6K-drd | Derepressed R6K plasmid | All R6K transfer proteins | 24 |
Table 3. Plasmid constructions used in two-hybrid assays.
| Plasmid | Construction | Complementation of (mutant)* |
|---|---|---|
| pMTX502 | pT25::trwBΔN75 | Not tested |
| pMTX503 | pT18::trwBΔN75 | Not tested |
| pMTX504 | pT25::trwC | <10−4 (pSU1445) |
| pMTX505 | pT18::trwC | <10−4 (pSU1445) |
| pMTX506 | pT25::trwA | Not tested |
| pMTX507 | pUT18::trwA | 0.01 (pSU1092) |
| pMTX508 | pUT18C::trwA | 0.003 (pSU1092) |
| pMTX512 | pUT18::trwB | 2.0 (pSU1443) |
| pMTX513 | pUT18C::trwB | 5.5 (pSU1443) |
| pMTX514 | pT25::trwB | 1.8 (pSU1443) |
| pMTX583 | pUT18::trwK | 0.003 (pSU4133) |
| pMTX584 | pT25::trwK | <10−4 (pSU4133) |
| pMTX585 | pUT18C::trwK | 0.005 (pSU4133) |
| pMTX631 | pUT18C::trwE | 0.7 (pSU4134) |
| pMTX632 | pT25::trwE | 0.8 (pSU4134) |
| pMTX634 | pT25::trwD | 0.005 (pSU4132) |
| pMTX635 | pT18::trwD | 0.01 (pSU4132) |
| pMTX643 | pUT18C::trwEΔN64 | Not tested |
| pMTX644 | pUT18C::traJ | Not tested |
| pMTX651 | pT25::traJ | Not tested |
| pMTX667 | pUT18C::traF | Not tested |
| pMTX668 | pT25::trwEN174 | Not tested |
| pMTX669 | pUT18C::trwEN174 | Not tested |
| pMTX674 | pT25::traF | Not tested |
| pMTX677 | pUT18C::pilX10 | Not tested |
| pMTX679 | pUT18C::taxB | Not tested |
| pMTX680 | pT25::taxB | Not tested |
Plasmids in boldface were introduced with the plasmid indicated in parentheses in strain D1210. These cells as donors were mated with strain DH5α. Figures show the percentage of transconjugants as compared to the transfer level of pSU2007 (100%). Plasmids in parentheses are all transfer-deficient except for pSU1092, which has a residual 1% conjugation frequency.
Protein Purification. Proteins TrwC (17), TrwCN293 (27), and TrwBΔN70 (9) were purified as described. His-tagged proteins were purified from soluble extracts by affinity to a Ni-NTA agarose column (Qiagen, Valencia, CA). Proteins fused to GST were purified by mixing the soluble lysates (17) from induced cells with glutathione-Sepharose resin (Pharmacia). Bound proteins were eluted from the resin either with 10 mM glutathione or by factor Xa digestion.
Protein-Protein Interactions by Affinity Chromatography. Fusion proteins containing either a N-terminal GST or a C-terminal His-tag were bound to glutathione-Sepharose or Ni-NTA agarose, respectively. After extensive wash, 20 μg of the purified proteins of interest, or BSA as a control, were added in buffer A (50 mM Tris, pH 7.6/50 mM NaCl/5 mM MgCl2) plus BSA 1 μg/ml and incubated at room temperature for 1 h. The resin was washed again, proteins were eluted with glutathione/Xa or imidazol, and eluates were loaded on SDS/PAGE gels stained with Coomassie brilliant blue.
Quantitative Mating Assays. One hundred microliters of overnight cultures of donor and recipient strains were mixed, and cells were collected and placed on filters on prewarmed LB plates for 1 h at 37°C. Plating was done in selective medium for both donor cells and transconjugants.
Two-Hybrid Assay. Strain DHM1 was grown at 30°C and cotransformed with plasmids bearing T25 and T18 fusions. Three independent transformants were grown overnight in liquid medium at 30°C. β-galactosidase levels were measured on 100-μl samples as described (28). All experiments included positive and negative controls. Plasmid pSU4111 (29), which carries lacZ under the control of the lactose promoter, produced ≈6,000 Miller units in this system.
Results
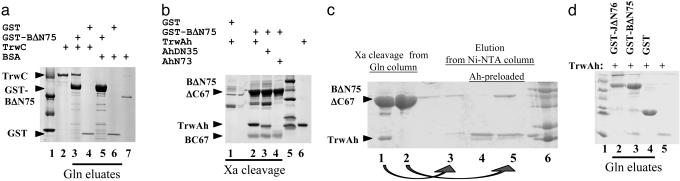
TrwB in Vitro Interactions. The soluble domain of TrwB (TrwBΔN75) was fused to the C-terminal end of GST. The fusion protein (GST-TrwBΔN75) was bound to glutathione-Sepharose resin and checked for specific retention of added proteins after extensive washing. Fig. 1 shows that TrwBΔN75 retains both protein TrwC (Fig. 1a) and protein TrwAh (Fig. 1b), which are the components of the R388 relaxosome. Both interactions were lost when the salt concentration was raised to 250 mM (data not shown). It was found that factor Xa protease cleaves off a peptide of TrwB, as can be observed in Fig. 1b. This peptide was gel-extracted, and its N terminus was sequenced and found to correspond to the C-terminal 67 aa of TrwB.
Fig. 1.
Protein-protein interactions detected by affinity chromatography. Soluble lysates containing GST fusion proteins were bound to glutathione-Sepharose resin, incubated with 20 μg of the indicated added proteins, and either eluted with glutathione (a and d) or cleaved with factor Xa protease (b and c). The figure shows 10% (a) or 12% (b-d) SDS/PAGE Coomassie-stained gels of eluted proteins. Molecular weight markers shown are 97.4 (not seen in b and c), 66, 45, 31, 21.5 (not seen in a), and 14.4 kDa (not seen in a). Lanes in a: 1, markers; 2, 2 μg of purified TrwC; 3-6, resin eluates; 3 and 5, lysates from pMTX501 (containing GST-TrwBΔN75); 4 and 6, lysates from pGEX-3X (containing GST); 3 and 4, bound proteins incubated with TrwC; 5 and 6, bound proteins incubated with BSA; 7, 1 μg of purified BSA. Lanes in b:1-4, Xa cleavage products; 1, lysates from pGEX-3X; 2-4, lysates from pMTX501; 1 and 2, incubation with TrwAh; 3, incubation with TrwAhΔN35; 4, incubation with TrwAhN73; 5, markers; 6, 2 μg of purified TrwAh. Arrowheads point to the two TrwB fragments obtained by factor Xa digestion. (c) TrwB-TrwA reciprocal interaction by affinity chromatography. Eluates from either glutathione-Sepharose resin by factor Xa digestion (lanes 1 and 2) or from Ni-NTA columns (lanes 3-5). Lanes: 1, TrwBΔN75 + TrwAh; 2, TrwBΔN75 + BSA; 3, eluate from lane 1 loaded on a Ni-NTA column; 4 and 5, Ni-NTA columns preloaded with TrwAh and then eluates from Xa digestions added (lane 4, from GST + TrwAh; lane 5, from TrwBΔN75 + BSA); 6, markers. Lanes in d: 1, markers; 2-4, eluates from lysates incubated with TrwAh; 2, pMTX609 (containing GST-TraJΔN76); 3, pMTX501; 4, pGEX-3X; 5, 1 μg of purified TrwAh.
To dissect the protein domains involved in the interactions, we assayed polypeptides containing the different domains separately. Protein TrwA can be separated into two domains obtained by partial trypsin treatment. The C-terminal domain (represented by TrwAΔN35) contains the tetramerization determinant, whereas the N-terminal domain (TrwAN73) retains the oriT-binding ability (G. Moncalián and F.d.l.C., unpublished observations). As seen in Fig. 1b, TrwAhΔN35 was efficiently retained by TrwBΔN75, whereas TrwAhN73 lost most interaction capacity. TrwC can be separated into relaxase and DNA helicase domains (27). The relaxase domain (TrwCN293) fully retained its ability to interact with TrwBΔN75 (data not shown).
The TrwBΔN75-TrwAh interaction was also detected by immobilizing protein TrwAh in a Ni-NTA column through its C-terminal His-tag and adding protein TrwBΔN75. The TrwBΔN75ΔC67-TrwAh complex obtained by factor Xa cleavage was also retained by the Ni-NTA column (Fig. 1c). This approach could not be used to detect the TrwBΔN75-TrwC interaction because the relaxase on its own binds the Ni-NTA column strongly and irreversibly (G. Moncalián, unpublished observations), and a GST fusion to either the C or the N terminus of the relaxase rendered fusion proteins insoluble (data not shown). Finally, a GST-TrwA fusion was constructed that retained purified TrwBΔN70 as expected but did not retain TrwC (data not shown).
To test the specificity of the TrwB-TrwA interaction, the soluble domain of TraJ (the CP of the related IncN plasmid pKM101) was equally fused to GST and assayed for interaction with TrwAh. TraJΔN76 did not retain TrwAh under the same conditions in which TrwBΔN75 did (Fig. 1d).
TrwB in Vivo Interactions. The in vitro approach used to test for interactions between TrwB and cytoplasmic components of the R388 conjugation machinery could not be used to search for interacting partners in the membrane. T4SSs are formed by 10-11 interacting proteins spanning inner and outer membranes and forming a transmembrane complex. To analyze protein-protein interactions in T4SSs, recent strategies were based on the isolation of protein subcomplexes from the membrane (30) and on the use of two-hybrid methods (8, 31-35). None of the interactions shown by these approaches involved the CP. We used a bacterial two-hybrid method (36) because, unlike the yeast two-hybrid system, it should detect interactions that involve the bacterial membranes and periplasm.
The DNA sequence of the R388 T4SS coding region was determined (GenBank accession no. X81123). It contains 11 genes, named trwD to trwN, with high similarity both in DNA sequence and genetic organization to the Agrobacterium tumefaciens VirB operon. We were particularly interested in proteins TrwD, TrwE, and TrwK, homologous to VirB11, VirB10, and VirB4, respectively, because they are the three best conserved components in all T4SSs and, like TrwB, are associated with the inner membrane. Both C- and N-terminal fusions to the T18 and T25 domains of adenylate cyclase were constructed whenever possible. To confirm their functionality the fused proteins were tested for complementation of corresponding R388 mutations. Results shown in Table 3 indicate that, except for TrwC and TrwK, significant levels of complementation were detected.
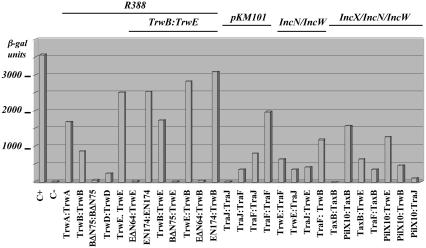
Fig. 2 shows the results of the two-hybrid analysis. Proteins TrwA, TrwB, TrwD, and TrwE interact with themselves, as expected, because TrwA is known to be a tetramer (29), TrwB and TrwD form hexamers (12, 37), and protein VirB10, the TrwE homolog in the Vir system, interacts with itself and forms high-molecular-weight structures (32, 35, 38). We also detected a weak (but consistent) interaction of TrwBΔN75 with itself, only visible on plates after long incubation periods. This result is consistent with the fact that TrwBΔN75 behaves as a monomer in solution but forms hexamers under crystallization conditions (10). TrwC and TrwK did not show signs of self-interaction, but because the corresponding fusion proteins produced a negative result in the in vivo complementation test, the negative result here could be due to a lack of general functionality. In addition, TrwK behaves as a monomer in solution (39).
Fig. 2.
In vivo protein-protein interactions measured by using the bacterial two-hybrid assay. The indicated plasmid pairs were introduced in strain DHM1 in the absence of other conjugation functions, and β-galactosidase units were measured. Only the most representative results are shown. All other combinations between R388 proteins were tested, and they gave background levels (<30 β-galactosidase units). Tested pairs are cited by the proteins fused to T18-T25. C+, positive control [pT18zip + pT25zip (36)]; C-, negative control (pT18zip + pT25).
When plasmids expressing two different R388 proteins were tested, a single pair gave a positive result: TrwB-TrwE. The interaction was as strong as that of TrwE interacting with itself. All protein pairs were also tested in the presence of pSU2007 (providing all Trw proteins) in case a fully assembled conjugation machinery were required to elicit the interactions, but the same results were obtained.
From its sequence it is predicted that TrwE contains a single N-terminal transmembrane segment (residues 46-64). This finding would suggest that TrwE is located in the periplasm, anchored to the inner membrane by its amino terminus, as determined for VirB10 (40). We fused TrwEΔN64 to the T18 domain for two-hybrid assays. Results show that interactions with both full-length TrwE and TrwB are lost. The TrwB-TrwE interaction is also lost when the N-terminal segments of TrwB are deleted (in TrwBΔN75). On the other hand, the N-terminal half of TrwE (TrwEN174) interacts both with itself and with TrwB with the same strength as the full-length protein.
The significance of this interaction was tested by searching for similar interactions between TrwB and TrwE homologs of other conjugative plasmids. These are proteins TraJ and TraF in the related IncN plasmid pKM101 (41, 42) and proteins TaxB and PilX10 in IncX plasmid R6K (ref. 43; B. Núñez and F.d.l.C., unpublished observations; GenBank accession number AJ006342). Results (Fig. 2) confirmed that there was also a strong interaction between TraJ and TraF, and between TaxB and PilX10; that is to say, this specific CP-T4SS interaction can be reproduced in three diverse transfer systems (with proteins that are ≈30% identical in their amino acid sequences). This fact underscores the broad significance of this interaction. In addition, Fig. 2 shows that the VirB10 homologs (TrwE, TraF, and PilX10) interact strongly with each other, suggesting that they could form heterologous multimers despite sharing modest levels of amino acid identity (23% for TraF-PilX10, 25.8% for TrwE-PilX10, and 33.6% for TrwE-TraF). In turn, CP can either interact with its cognate or with the heterologous VirB10-like partners. For example, TrwB interacts not only with its partner TrwE, but also with TraF and PilX10. It should be noted that all of these interactions are detected in the absence of other T4SS components.
The above results suggested a lack of specificity in the CP-T4SS interaction that could give rise to functional interchangeability among the different conjugative systems. To test whether this observation was functionally significant, we constructed several plasmids containing either the relaxosomal components or the complete mobilization region (relaxosomal components plus CP) of each system (Table 2). Thus, RLXW contains oriTW plus trwA and trwC, whereas MOBW contains the RLXW components plus trwB. Similarly, RLXN contains oriTN plus traK and traI, whereas MOBN contains RLXN components plus traJ. We carried out mobilization experiments using the conjugal machineries (TRAW, TRAN, and TRAX) of these plasmids. Results are shown in Table 4. These experiments confirm the fact that relaxosomal components of each plasmid need their cognate CP for transfer (RLXW and RLXN are not mobilized by heterologous TRA systems), whereas each CP can operate with either T4SS with high efficiency (MOBW and MOBN are mobilized by the three TRA systems).
Table 4. Conjugation assays.
|
TRA*
|
MOB†
|
RLX‡
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Helper | Mobilizable | W | N | X | W | N | W | N | Transfer |
| pSU2007 | pSU1423 | + | + | 5,2 × 10−1 | |||||
| pSU2007 | pSU4280 | + | + | 1,8 × 10−2 | |||||
| pSU2007 | pSU1404 | + | + | 6,1 × 10−1 | |||||
| pSU2007 | pMTX681 | + | + | <9 × 10−8 | |||||
| pKM101 | pSU1423 | + | + | 1,3 × 10−1 | |||||
| pKM101 | pSU4280 | + | + | 2,8 × 10−1 | |||||
| pKM101 | pSU1404 | + | + | <6 × 10−7 | |||||
| pKM101 | pMTX681 | + | + | 1,0 × 10−1 | |||||
| R6K-drd | pSU1423 | + | + | 1,6 × 10−3 | |||||
| R6K-drd | pSU4280 | + | + | 3,4 × 10−5 | |||||
Donors were derivatives of strain DH5α harboring the indicated helper and mobilizable plasmids (first two columns). The recipient was strain UB 1637. Transfer frequencies are indicated as the number of transconjugants per donor. The table indicates the parts of the conjugal machinery that are present in each tested plasmid pair. W, N, and X refer to the specific mating system of the parental conjugal plasmid.
TRA = complete transfer system = MOB + T4SS.
MOB = RLX + CP; MOBW = oriTW + trwA + trwB + trwC; MOBN = oriTN + traK + traJ + traI.
RLX = oriT + relaxase + accessory nicking protein; RLXW = oriTW + trwA + trwC; RLXN = oriTN + traK + traI.
Significantly, we observed that the efficiency of heterologous mobilization correlates with the strength of the CP-T4SS interaction as measured in the two-hybrid assay. For instance, MOBW is mobilized at a frequency of 5.2 × 10-1 by TRAW, at 1.3 × 10-1 by TRAN, and only at 1.6 × 10-3 by TRAX. These figures can be compared with 1,740, 1,190, and 470 β-galactosidase units for the corresponding TrwB interactions with TrwE, TraF, and PilX10, respectively. A scheme summarizing the most significant data from Fig. 2 and Table 4 is shown in Table 5, where the correlation between CP interactions in the two-hybrid system and performance in the in vivo assays becomes obvious.
Table 5. Correlation between the CP-T4SS interaction and conjugal efficiency.
| TrwB (TRAW) | TraJ (TRAN) | TaxB (TRAX) | MOBW | MOBN | RLXW | RLXN | ||
|---|---|---|---|---|---|---|---|---|
| TrwE (TRAW) | ++++ | + | + | TRAW | 100 | 10−2 | 100 | 0 |
| TraF (TRAN) | ++ | ++ | + | TRAN | 10−1 | 10−1 | 0 | 10−1 |
| PilX10 (TRAX) | + | + | +++ | TRAX | 10−3 | 10−5 | nt | nt |
The most relevant results from Fig. 2 and Table 4 are represented as follows. Left, two-hybrid assays between proteins on the first column fused to T18 and proteins in the first row fused to T25. β-Galactosidase units are represented as follows: ++++, ≥2,500; +++, 1,500−2,500; ++, 500−1,500; +, 30−500. Right, mobilization by different transfer systems (TRA) of plasmids containing cognate or heterologous MOB and RLX regions (see legend to Table 3). W, N, and X refer to the incompatibility group. Data have been rounded to the logarithmic range to facilitate comparisons.
Discussion
The TrwB-Relaxosome Interaction. Results show that TrwB interacts in the cytoplasm with proteins TrwA and TrwC, the two protein components of the R388 relaxosome. Protein GST-TrwBΔN75 bound to an affinity matrix retained proteins TrwC and TrwA (Fig. 1). TrwC dissection indicated that the interaction domain lies in the relaxase moiety. A TrwB-TrwC interaction was expected because an analogous result was previously shown for plasmid RP4 CP TraG, both with its cognate relaxase and with that of a mobilizable plasmid (5, 6). In addition, TrwB enhances TrwC-mediated cleavage of supercoiled DNA containing plasmid R388 oriT (9). Because TrwB is a nonspecific DNA binding protein (9), nic-cleavage enhancement could also result from an indirect effect by which TrwB might affect the supercoiling negative density of the DNA around nic and thus help TrwC to separate the DNA strands at the nic site and cleave it. Overall, the TrwB-TrwC interaction shown is relatively weak, and its physiological relevance remains uncertain.
Compared with TrwC, TrwB shows a clearly stronger interaction with protein TrwA, which was demonstrated when either interacting protein was immobilized in a solid support (Fig. 1). The interaction was further dissected to specific protein domains. Results indicate that the TrwA C-terminal domain, which contains the tetramerization domain, is involved in the interaction (Fig. 1). In the case of the related F-like conjugative plasmids, the roles of TrwA seem to be performed by the conjunction of the oriT-binding proteins TraY and TraM, which are functionally redundant to some extent (44). Both proteins, like R388 TrwA, are involved in transcriptional regulation of transfer genes and in activating the relaxase nicking activity (29, 45-47). Protein TraY is homologous only to the N-terminal DNA-binding domain of TrwA. Protein TraM is a tetramer, as is the TrwA C-terminal domain (29, 48). The CP of F-like plasmids, TraD, interacts with TraM (7). It is tempting to speculate that the N- and C-terminal domains of TrwA carry the activities of TraY and TraM, respectively.
No interaction was seen in the two-hybrid assays between TrwB (or TrwBΔN75) and TrwA despite the in vitro evidence. This might be significant and suggest that TrwB is unable to bind TrwA when both proteins adopt their standard in vivo conformation, which may change during the conjugation process. Alternatively, the adenylate cyclase fusions could sterically prevent the two partner proteins from interacting.
We also tested protein TraJ (the CP of TRAN plasmid pKM101) for interaction with TrwA, and it was shown (Fig. 1d) that the cytoplasmic domain of TraJ does not retain TrwA. Because TrwB and TraJ are functionally equivalent, share 40% identity, and have similar molecular weight, pI values, and overall secondary structure, this result suggests that the TrwB-TrwA interaction is specific and may indicate that CPs interact specifically only with components of their cognate relaxosomes.
We would like to emphasize that, despite its interactions with both relaxosome components, TrwB is not permanently associated with the R388 relaxosome. Processing at oriT takes place in the absence of CP. Neither electron microscopy nor electrophoresis or gel filtration experiments indicate a stable and specific association of TrwB with oriT, either naked or bound by relaxosome components (unpublished results).
The TrwB-T4SS Interaction. Previous efforts to identify CP-interacting partners within the T4SS may have failed because these interactions might involve extracytoplasmic locations, and the approaches used did not deal with this complication. By using the bacterial two-hybrid system, we show a strong interaction between TrwB and TrwE (Fig. 2) that occurs independently of the presence of other T4SS components. TrwE is predicted to localize to the periplasmic space. Thus, it is not surprising that previous work using the yeast two-hybrid system did not detect such interactions in the Vir system, because TrwE (or its homolog VirB10) cannot properly localize in the yeast cell. While this work was under review, a similar finding has been reported between the homologous proteins TraG and TrhB of conjugative plasmid R27 (49).
The bulk of TrwB and TrwE proteins belong to different cell compartments: cytoplasm and periplasm, respectively. Both proteins are anchored to the inner membrane by their N-terminal transmembrane segments. Not surprisingly, the TrwB-TrwE interaction is lost when the TrwB transmembrane domain is deleted (in TrwBΔN75). It is important to note that TrwBΔN75 is a properly folded protein that displays the biochemical features of the native protein, such as DNA and ATP binding (9). Thus, the result suggests that the TrwB-TrwE interaction takes place at the inner membrane or in the periplasm. With respect to the TrwE interaction domain, we show that TrwEΔN64 interacts neither with TrwB nor with TrwE, although this could be explained simply by TrwEΔN64 being located in the wrong cell compartment (cytoplasm instead of periplasm). Finally, we show that the N-terminal 174 residues of TrwE fully retain the interaction domain, both with itself and with TrwB. This is in agreement with the recent results by Gilmour et al. (49), who show that the N-terminal half of R27 protein TrhB contains the domain for self-interaction and CP interaction.
In an effort to gain further proof for the significance of the TrwB-TrwE interaction as a way of connecting CP with T4SSs in conjugation, we extended the two-hybrid analysis to other conjugative systems that code for similar T4SSs: the related TRAN system of plasmid pKM101 and the more distant TRAX system of plasmid R6K (Fig. 2). We determined that the same CP-VirB10-like interaction occurs in the three systems. In addition, we observed that although CPs interact most strongly with their cognate T4SS component, they also interact with heterologous T4SSs with considerable strength. This lack of specificity could explain the functionality of hybrid TRA systems previously observed for TRAW and TRAN plasmids (19). To confirm and extend these results, mating experiments were carried out to test for mobilization by heterologous T4SSs (Table 4). It is shown that a given MOB region (that is, the relaxosome with its cognate CP) can efficiently use different T4SSs for its transfer. Moreover, there is a correlation between the strength of the CP-T4SS interaction seen by the two-hybrid analysis and the efficiency of mobilization, as outlined in Table 5.
The Coupling Model. Bacterial conjugation is a complex process that brings about the efficient transfer of long DNA chains between bacteria. In terms of mechanism, we know some details of the DNA processing steps in the donor bacteria, principally catalyzed by the relaxase, but little else. Analysis of the 3D structure of TrwB suggested that it may work as a DNA transporter (1), effectively pumping DNA from donor to recipient bacteria once a channel has been formed. Apart from their direct role in DNA transport, genetic evidence (50) suggested that TrwB family proteins may have an additional coupling role, being responsible for the interactions between the relaxosome and the transport apparatus. Our results identify and characterize the two-handed interactions that govern the function of the CP. On one hand, TrwB interacts with the T4SS. The experiments reported here suggest that this interaction occurs primarily at the inner membrane or in the periplasm between TrwB and TrwE, the VirB10-like component of the T4SS. VirB10-like proteins are key elements of the transporter, as shown by their presence in all T4SS-like systems, and by their interactions with other components. The “core complex” of the Agrobacterium T4SS is formed by proteins VirB8, VirB9, and VirB10. This has been shown by their interactions in vivo and in vitro (32, 35, 38, 51), and by their presence together in a protein subassembly in the membrane (30). The CP-VirB10 interaction that we describe in this work occurs in the three conjugative systems that we have studied and plasmid R27 (49), so we presume it is a general feature of conjugative systems. On the other hand, the interaction with the relaxosome is mediated by different relaxosome components, depending on the type of transfer system. In F-type transfer systems, the interaction takes place with the “nicking accessory protein” (TrwA in R388 or TraM in F-like plasmids). In P-type transfer systems, this interaction may be mediated directly by the relaxase.
Conjugation experiments show that the specificity lies preferentially in the cytoplasmic side of the CP interactions (Table 4). Thus, relaxosomes of related conjugation systems, such as R388 and pKM101, can efficiently use heterologous T4SSs if they connect with them via their cognate CP, whereas they are unable to do so with the heterologous CP. This situation supports the view that T4SSs are separate biological entities that recruit (or are recruited by) different biological machineries for the secretion of diverse macromolecules. For instance, highly homologous T4SSs are present in the pathogenic bacterium Bartonella (presumably for secretion of virulence factors) and in conjugative plasmid R388 (52).
In summary, the two sets of interactions described in this work illustrate how the relaxosome may come into close contact with the core complex of the T4SS, which probably spans both bacterial membranes (53). The mechanism by which the processed DNA (or the pilot protein heading the T-DNA) now exits through the membrane transporter remains to be elucidated. It is our view that this event is followed by the active pumping of the T-strand by the CP, as postulated by the two-step conjugation model (1).
Supplementary Material
Acknowledgments
We thank Dr. Daniel Ladant and Hybrigenics for providing strains and plasmid vectors for the two-hybrid assay, María Lucas and Itsaso Hormaeche for providing purified proteins TrwCN293 and TrwBΔN70, respectively, and M. Hernando, M. V. Mendiola, and S. Bolland for various plasmid constructions. This work was supported by Ministerio de Ciencia y Tecnología Grants BMC2002-00379 (to F.d.l.C.) and BIO2002-00063 (to M.L.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: T4SS, type IV secretion system; CP, coupling protein; TrwAh, TrwA with a C-terminal histidine tag.
References
- 1.Llosa, M., Gomis-Rüth, F.-X., Coll, M. & de la Cruz, F. (2002) Mol. Microbiol. 45, 1-8. [DOI] [PubMed] [Google Scholar]
- 2.Christie, P. J. (2001) Mol. Microbiol. 40, 294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egelman, E. H. (2001) Nature 409, 573-575. [DOI] [PubMed] [Google Scholar]
- 4.Errington, J., Bath, J. & Wu, L. J. (2001) Nat. Rev. Mol. Cell Biol. 2, 538-545. [DOI] [PubMed] [Google Scholar]
- 5.Schroder, G., Krause, S., Zechner, E. L., Traxler, B., Yeo, H. J., Lurz, R., Waksman, G. & Lanka, E. (2002) J. Bacteriol. 184, 2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szpirer, C. Y., Faelen, M. & Couturier, M. (2000) Mol. Microbiol. 37, 1283-1292. [DOI] [PubMed] [Google Scholar]
- 7.Disque-Kochem, C. & Dreiseikelmann, B. (1997) J. Bacteriol. 179, 6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward, D. V., Draper, O., Zupan, J. R. & Zambryski, P. C. (2002) Proc. Natl. Acad. Sci. USA 99, 11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moncalián, G., Cabezón, E., Alkorta, I., Valle, M., Moro, F., Valpuesta, J. M., Goñi, F. M. & de la Cruz, F. (1999) J. Biol. Chem. 274, 36117-36124. [DOI] [PubMed] [Google Scholar]
- 10.Gomis-Rüth, F. X., Moncalián, G., Pérez-Luque, R., González, A., Cabezón, E., de la Cruz, F. & Coll, M. (2001) Nature 409, 637-641. [DOI] [PubMed] [Google Scholar]
- 11.Gomis-Rüth, F. X., Moncalián, G., de la Cruz, F. & Coll, M. (2002) J. Biol. Chem. 277, 7556-7566. [DOI] [PubMed] [Google Scholar]
- 12.Hormaeche, I., Alkorta, I., Moro, F., Valpuesta, J. M., Goñi, F. M. & de la Cruz, F. (2002) J. Biol. Chem. 277, 46456-46462. [DOI] [PubMed] [Google Scholar]
- 13.Sadler, J. R., Tecklenburg, M. & Betz, J. L. (1980) Gene 8, 279-300. [DOI] [PubMed] [Google Scholar]
- 14.Studier, F. W. & Moffatt, B. A. (1986) J. Mol. Biol. 189, 113-130. [DOI] [PubMed] [Google Scholar]
- 15.Grant, S. G., Jessee, J., Bloom, F. R. & Hanahan, D. (1990) Proc. Natl. Acad. Sci. USA 87, 4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Cruz, F. & Grinsted, J. (1982) J. Bacteriol. 151, 222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandoso, G., Llosa, M., Zabala, J. C. & de la Cruz, F. (1994) Eur. J. Biochem. 226, 403-412. [DOI] [PubMed] [Google Scholar]
- 18.Langer, P. J. & Walker, G. C. (1981) Mol. Gen. Genet. 182, 268-272. [DOI] [PubMed] [Google Scholar]
- 19.Bolland, S., Llosa, M., Avila, P. & de la Cruz, F. (1990) J. Bacteriol. 172, 5795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llosa, M., Bolland, S. & de la Cruz, F. (1994) J. Mol. Biol. 235, 448-464. [DOI] [PubMed] [Google Scholar]
- 21.Llosa, M., Bolland, S., Grandoso, G. & de la Cruz, F. (1994) J. Bacteriol. 176, 3210-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez, E. & de la Cruz, F. (1988) Mol. Gen. Genet. 211, 320-325. [DOI] [PubMed] [Google Scholar]
- 23.Rivas, S., Bolland, S., Cabezón, E., Goñi, F. M. & de la Cruz, F. (1997) J. Biol. Chem. 272, 25583-25590. [DOI] [PubMed] [Google Scholar]
- 24.Avila, P., Núñez, B. & de la Cruz, F. (1996) J. Mol. Biol. 261, 135-143. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 26.Karimova, G., Ullmann, A. & Ladant, D. (2001) J. Mol. Microbiol. Biotechnol. 3, 73-82. [PubMed] [Google Scholar]
- 27.Llosa, M., Grandoso, G., Hernando, M. A. & de la Cruz, F. (1996) J. Mol. Biol. 264, 56-67. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria (Cold Spring Harbor Lab. Press, Plainview, NY).
- 29.Moncalián, G., Grandoso, G., Llosa, M. & de la Cruz, F. (1997) J. Mol. Biol. 270, 188-200. [DOI] [PubMed] [Google Scholar]
- 30.Krall, L., Wiedemann, U., Unsin, G., Weiss, S., Domke, N. & Baron, C. (2002) Proc. Natl. Acad. Sci. USA 99, 11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das, A., Anderson, L. B. & Xie, Y. H. (1997) J. Bacteriol. 179, 3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das, A. & Xie, Y. H. (2000) J. Bacteriol. 182, 758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris, R. L., Sholl, K. A., Conrad, M. N., Dresser, M. E. & Silverman, P. M. (1999) Mol. Microbiol. 34, 780-791. [DOI] [PubMed] [Google Scholar]
- 34.Harris, R. L., Hombs, V. & Silverman, P. M. (2001) Mol. Microbiol. 42, 757-766. [DOI] [PubMed] [Google Scholar]
- 35.Ding, Z., Zhao, Z., Jakubowski, S. J., Krishnamohan, A., Margolin, W. & Christie, P. J. (2002) J. Bacteriol. 184, 5572-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karimova, G., Pidoux, J., Ullmann, A. & Ladant, D. (1998) Proc. Natl. Acad. Sci. USA 95, 5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krause, S., Pansegrau, W., Lurz, R., de la Cruz, F. & Lanka, E. (2000) J. Bacteriol. 182, 2761-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaupre, C. E., Bohne, J., Dale, E. M. & Binns, A. N. (1997) J. Bacteriol. 179, 78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabel, C., Grahn, A. M., Lurz, R. & Lanka, E. (2003) J. Bacteriol. 185, 1045-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward, J. E., Jr., Dale, E. M., Nester, E. W. & Binns, A. N. (1990) J. Bacteriol. 172, 5200-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pohlman, R. F., Genetti, H. D. & Winans, S. C. (1994) Mol. Microbiol. 14, 655-668. [DOI] [PubMed] [Google Scholar]
- 42.Paterson, E. S., More, M. I., Pillay, G., Cellini, C., Woodgate, R., Walker, G. C., Iyer, V. N. & Winans, S. C. (1999) J. Bacteriol. 181, 2572-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Núñez, B., Avila, P. & de la Cruz, F. (1997) Mol. Microbiol. 24, 1157-1168. [DOI] [PubMed] [Google Scholar]
- 44.Karl, W., Bamberger, M. & Zechner, E. L. (2001) J. Bacteriol. 183, 909-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fowler, T., Taylor, L. & Thompson, R. (1983) Gene 26, 79-89. [DOI] [PubMed] [Google Scholar]
- 46.Lahue, E. E. & Matson, S. W. (1990) J. Bacteriol. 172, 1385-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polzleitner, E., Zechner, E. L., Renner, W., Fratte, R., Jauk, B., Hogenauer, G. & Koraimann, G. (1997) Mol. Microbiol. 25, 495-507. [DOI] [PubMed] [Google Scholar]
- 48.Verdino, P., Keller, W., Strohmaier, H., Bischof, K., Lindner, H. & Koraimann, G. (1999) J. Biol. Chem. 274, 37421-37428. [DOI] [PubMed] [Google Scholar]
- 49.Gilmour, M. W., Gunton, J. E., Lawley, T. D. & Taylor, D. E. (2003) Mol. Microbiol. 49, 105-116. [DOI] [PubMed] [Google Scholar]
- 50.Cabezón, E., Sastre, J. I. & de la Cruz, F. (1997) Mol. Gen. Genet. 254, 400-406. [DOI] [PubMed] [Google Scholar]
- 51.Finberg, K. E., Muth, T. R., Young, S. P., Maken, J. B., Heitritter, S. M., Binns, A. N. & Banta, L. M. (1995) J. Bacteriol. 177, 4881-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seubert, A., Hiestand, R., de la Cruz, F. & Dehio, C. (2003) Mol. Microbiol., in press. [DOI] [PubMed]
- 53.Cao, T. B. & Saier, M. H., Jr. (2001) Microbiology 147, 3201-3214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.