Abstract
The polyomavirus coat protein viral protein 1 (VP1) has the intrinsic ability to self-assemble in vitro into polymorphic capsid-like structures on addition of calcium. In contrast, polyomavirus assembly in vivo is rigorously controlled, such that virions of uniform size are formed only in the cell nucleus. During viral infection, the 72 kDa cellular chaperone heat shock cognate protein (hsc70) binds VP1 posttranslation and colocalizes with VP1 to the nucleus, thereby suggesting a role for ≈70-kDa heat shock protein (hsp70) family chaperones in regulating the quality and location of capsid assembly. We found that, after expression of recombinant VP1 in Escherichia coli, the prokaryotic hsp70 chaperone DnaK copurified with the VP1 C-terminal domain that links pentamers in an assembled capsid. When stably bound to VP1, DnaK inhibited in vitro assembly induced by calcium. However, in the presence of ATP, the hsp70 chaperone system comprised of DnaK, DnaJ, and GrpE assembled VP1 into uniform capsids without requiring calcium. Chaperone-mediated assembly was similarly catalyzed by the eukaryotic hsc70 protein, in combination with the J-domain function of the simian virus 40 large T-antigen protein. Thus, polyomavirus capsid assembly can be recapitulated with high-fidelity in vitro using either prokaryotic or eukaryotic hsp70 chaperone systems, thereby supporting a role for cellular chaperones in the in vivo regulation of virion assembly.
The Polyomaviridae family of small, nonenveloped, icosahedral DNA viruses includes murine polyomavirus and simian virus 40 (SV40). Like most DNA viruses, polyomavirus capsid proteins are synthesized in the cytosol, whereas assembly of virions occurs only in the nucleus. Polyomavirus capsids are comprised of 72 pentamers (capsomeres) of the major capsid viral protein (VP1), which is arranged in a T = 7 icosahedral lattice ≈50 nm in diameter (1, 2). One minor capsid protein, either VP2 or VP3, binds in the central 5-fold cavity of each VP1 pentamer (3). The atomic structure of the virion reveals that the C-terminal domain of each VP1 monomer “invades” a neighboring pentamer to form the principal interpentamer contacts, and these contacts are stabilized by calcium ions (2). Consistent with the structural data, a recombinant VP1 protein lacking 63 amino acids from the C terminus forms pentamers that are incapable of assembly (4). In vitro capsid assembly of full-length recombinant VP1 pentamers incubated with calcium or at high ionic strength is robust but frequently yields polymorphic assemblies of sizes that are disparate from authentic capsids (5, 6). In contrast to calcium-mediated assembly in vitro, expression of recombinant VP1 in eukaryotic cells results in the formation of uniform virus-like particles (VLPs), exclusively in the cell nucleus (7).
Cellular ≈70-kDa heat shock protein (hsp70) family chaperones bind newly synthesized proteins and escort the unfolded domains until protein folding is completed (reviewed in ref. 8). The constitutively expressed eukaryotic chaperone ≈70-kDa heat shock cognate protein (hsc70) associates with VP1 posttranslation and then colocalizes with VP1 in the nucleus of virus-infected mouse cells (9). Similar to this interaction in eukaryotic cells, the bacterial hsp70 family member, DnaK, associates with VP1 that is recombinantly expressed in Escherichia coli (9). Hence, hsp70 chaperones could be regulatory factors that control the fidelity and subcellular location of polyomavirus capsid assembly in vivo.
Hsp70 family chaperones assist in protein folding through a catalytic cycle that depends on cochaperones and ATP hydrolysis. Hsp70 chaperones typically exhibit rapid binding and release of the substrate in the presence of ATP but are locked in a stable complex with the substrate in the presence of ADP (10, 11). The E. coli hsp40 cochaperone DnaJ contains a highly conserved J-domain that binds DnaK and stimulates ATP hydrolysis (12), and the GrpE cochaperone serves as a nucleotide exchange factor to replace ADP with ATP in the rate-limiting step (13, 14). The eukaryotic hsp70 chaperone system differs in that the rate-limiting step of the catalytic cycle is completed by the hsp70 family member and the hsp40 cochaperone.
In this article, we show that hsp70 chaperones interact with the C-terminal domain of VP1 to inhibit calcium-mediated capsid assembly in vitro. We also demonstrate that both prokaryotic and eukaryotic hsp70 chaperones can assemble VP1 capsomeres in vitro into uniform capsids in an energy-dependent reaction.
Materials and Methods
Coexpression and Purification of Recombinant Capsid Proteins in E. coli. Recombinant polyomavirus capsid proteins were coexpressed in E. coli as described (15). Recombinant SV40 VP3 was generated by PCR amplification from SV40 viral DNA, strain 776, and cloning into the XmaI/XhoI site of pXABN (16). The recombinant SV40 VP3 construct was coexpressed with SV40 VP1 (17) in the BL21(DE3) strain of E. coli.
The recombinant polyomavirus or SV40 capsid proteins were purified as described (15), with the following modifications. For VP1 + VP3 with copurified chaperones, the thrombin-cleaved, glutathione-Sepharose eluate was further purified by using gel filtration chromatography with a Superdex 200 column (Amersham Pharmacia Biotech, Piscataway, NJ) to obtain pentamers. For VP1 pentamer with VP3 (VP1 + VP3) (without chaperones), the concentrated eluate in 0.2 M NaCl was bound to a phosphocellulose column (P11 cellulose phosphate, Whatman), eluted with a salt gradient, and dialyzed into 0.2 M NaCl buffer by centrifugation (Ultra, Amicon). Pentamers were isolated by gel filtration chromatography.
Chaperone Protein and Virus Purification. DnaK was isolated as a by-product of capsid protein purification and was further purified by using gel filtration chromatography with a Superdex 200 column (Amersham Pharmacia Biotech). Recombinant YDJ-1 and recombinant avian hsp70 were expressed and purified as described (18). Recombinant DnaJ (SPP-640), GrpE (SPP-650), GroELS (SPP-610 and SPP-620), bovine hsc70 (SPP-751), and purified recombinant human hsp40 (SPP-400) were obtained from StressGen Biotechnologies (Victoria, BC, Canada). Large T antigen (LgT) proteins were purified from recombinant baculovirus constructs (19). Polyomavirus virions and VP1 VLPs were purified as described (20, 21).
Immunoblots. Purified capsid protein pentamers with and without copurified chaperones were analyzed by SDS/PAGE, and were transferred to a poly(vinylidene difluoride) membrane. The membranes were blotted by using primary antibodies for VP1 (I58) (7, 22), VP3 (482) (7, 22), DnaK (AXL623, Accurate Chemical and Scientific, Westbury, NY), DnaJ (SPA-410, StressGen Biotechnologies), GrpE (SPA-240, StressGen Biotechnologies), and GroEL (SPA-870, StressGen Biotechnologies) at 1:1,000 dilution, and secondary anti-rabbit or anti-mouse alkaline phosphatase conjugates (S373B or S372B, Promega) at 1:5,000 dilution. The blots were developed using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate.
Coimmunoprecipitation. Purified proteins were mixed to a final concentration of VP1 + VP3 pentamers at 0.016 μM (based on monomeric VP1), 0.064 μM DnaK, and 0.008 μM YDJ-1 in coimmunoprecipitation buffer (50 mM Tris·Cl/15 mM EDTA/100 mM NaCl/0.1% (wt/vol) Triton X-100/1 mM DTT/1 mM PMSF/5 μg/ml pepstatin A/5 μg/ml leupeptin, pH 7.5). The indicated reactions were treated with 1 mM MgATP, and all reactions were incubated for 15 min at 22°C. All reactions then were stored at 4°C, except for one reaction treated with an additional 100 mM ADP and incubated at 22°C, and one reaction treated with 15 units of apyrase (Grade VI, Sigma) and incubated at 30°C for 15 min. All reactions were coimmunoprecipitated by using anti-VP1 rabbit polyclonal antibody (I58) (7, 22) and protein A-Sepharose (Sigma). Proteins bound to protein A-Sepharose were analyzed by SDS/PAGE and Coomassie blue staining.
In Vitro Assembly Reactions. Calcium-mediated assembly reactions were performed as described (5). Recombinant capsid proteins (standardized to 150 μg/ml for the monomeric form of VP1) with and without copurified chaperones are added to minidialysis units (molecular weight cutoff of 3,500; Pierce), and are dialyzed against calcium buffer (50 mM Tris·Cl/150 mM NaCl/0.5 mM CaCl2/5% glycerol, pH 7.2) at 22°C for 1 h.
E. coli chaperone-mediated assembly reactions were performed by using recombinant capsid proteins with and without copurified chaperones (150 μg/ml for VP1). The capsid proteins were added to minidialysis units (molecular weight cutoff of 3,500; Pierce), and the reactions were dialyzed against dissociation buffer (50 mM Tris·Cl/200 mM NaCl/5% glycerol/1 mM EDTA/5 mM DTT, pH 7.2) at 22°C for 1 h. For the reconstituted E. coli system, the purified proteins were added to minidialysis units with capsid proteins at 150 μg/ml or 3.75 μM (for VP1), 2 μM DnaK, 0.2 μM DnaJ, 0.2 μM GrpE, and 2 μM GroELS, and the reactions were dialyzed against dissociation buffer containing 20 mM MgATP at 22°C for 1 h.
For the reconstituted mammalian chaperone system, the purified proteins were added to minidialysis units (molecular weight cutoff of 3,500) with purified SV40 capsid proteins to 150 μg/ml or 3.75 μM (for VP1), hsc70 to 4 μM, and LgT to 0.4 μM for the J-domain, and were dialyzed into dissociation buffer containing 20 mM MgATP at 22°C for 1 h.
Transmission Electron Microscopy (TEM). Assembly reactions were absorbed to glow-discharged, carbon and formvar-coated copper grids (G400 copper, EM Science), rinsed with buffer lacking phosphates, stained with uranyl acetate, and examined by TEM (Philips CM10) at 80 kV. The number of 50-nm particles was determined by direct count of the indicated number (n = 3 or n = 10) of grid squares (1,369 μm2) at ×46,000 total magnification.
Results
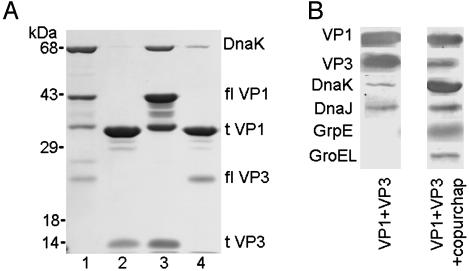
Coexpression of VP1 and GST-VP3 (a fusion of GST to VP3) in E. coli generates a pentamer of VP1 bound to one molecule of VP3 (15). After capsid protein coexpression in E. coli, we found that the prokaryotic hsp70 chaperone DnaK initially purified with pentamer complexes of full-length VP1 and VP3, but not with truncated proteins lacking the N and C termini of VP1 (Fig. 1A). Similar results were obtained when full-length and truncated VP1 proteins were expressed in E. coli without VP3 (data not shown). Capsid protein complexes that were isolated by using only glutathione affinity and gel filtration chromatography contained the E. coli chaperones DnaK, DnaJ, GrpE, and GroEL (referred to as VP1 + VP3 + copurified chaperones; Fig. 1B). Complexes of full-length VP1 and VP3 that were purified by glutathione affinity chromatography, followed by ion exchange chromatography and gel filtration chromatography, resulted in pentamer complexes (VP1 + VP3) without detectable GrpE and GroEL (Fig. 1B). DnaK and and DnaJ were present in this VP1 + VP3 preparation; however, the relative amounts were decreased 10- to 30-fold. Thus, DnaK and other bacterial chaperones associate with the C-terminal assembly domain of VP1 after capsid protein expression in E. coli, and they remain associated unless further purification steps are used.
Fig. 1.
E. coli chaperones interact with the C-terminal domain of the polyomavirus capsid protein VP1. (A) Recombinant VP1 and a GST-VP3 fusion protein were coexpressed in E. coli and were purified by using glutathione Sepharose chromatography, followed by thrombin cleavage as described (15). Eluates were analyzed by SDS/PAGE and Coomassie blue staining. Lane 1, full-length VP1 (flVP1) coexpressed with full-length VP3 (flVP3); lane 2, residues 32-316 of VP1 (tVP1) coexpressed with the C-terminal 105 residues of VP3 (tVP3); lane 3, flVP1 coexpressed with tVP3; and lane 4, tVP1 coexpressed with flVP3. (B) Immunoblots of VP1 + VP3 pentamers purified by glutathione affinity, ion exchange, and gel filtration chromatography (VP1 + VP3), or only glutathione affinity and gel filtration chromatography (VP1 + VP3 + copurified chaperones).
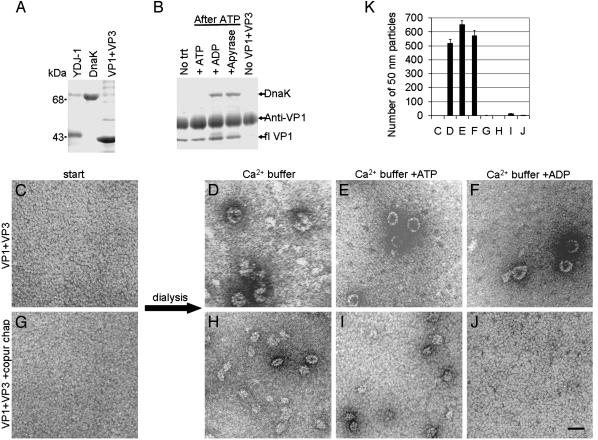
Previous studies (23) have demonstrated that hsp70 chaperones can be rebound in vitro to a substrate such as the progesterone receptor, by using an hsp40 cochaperone and ATP. To determine whether chaperones would reassociate with capsid proteins, we incubated a recombinant hsp40 cochaperone from yeast (YDJ-1) and DnaK with VP1 + VP3 pentamers in vitro (Fig. 2A). A stable interaction between DnaK and VP1 was not detected without the addition of ATP to initiate binding of DnaK to the substrate, or if ATP was maintained in the reaction (Fig. 2B). After the subsequent addition of either excess ADP or apyrase, an enzyme that hydrolyses ATP, a stable interaction between DnaK and VP1 could be detected by coimmunoprecipitation with anti-VP1 antibodies (refs. 7 and 22 and Fig. 2B). No interaction was detected between DnaK and the antibody in the absence of capsid proteins (Fig. 2B). Thus, hsp40 cochaperones and ATP facilitate addition of DnaK to purified VP1 + VP3 in vitro, and excess ADP stabilizes this interaction.
Fig. 2.
A stable interaction between E. coli chaperones and VP1 inhibits calcium-mediated assembly. (A) Purified proteins YDJ-1, DnaK, and VP1 + VP3 analyzed by SDS/PAGE and Coomassie blue staining. (B) Anti-VP1 (I58) (7, 22) coimmunoprecipitation of purified proteins (YDJ-1 at 0.5× molar concentration, DnaK at 4× molar concentration, and VP1 + VP3 at 1× molar concentration) incubated without treatment (No trt), which is incubated briefly in ATP, followed by either ATP, excess ADP, or apyrase; or incubation of YDJ-1 and DnaK in the absence of VP1 + VP3 (No VP1 + VP3). (C-G) Purified pentamers of recombinant VP1 + VP3 without chaperones (C) or with copurified E. coli chaperones (G), in dissociating buffer visualized by negative stain and TEM. (D-F and H-J) In vitro assembly reactions of VP1 + VP3 pentamers without (D-F) or with (H-J) copurified chaperones, after dialysis into indicated calcium buffers, and visualized by negative stain and TEM. (Scale bar, 50 nm.) (K) Quantitation of the mean number of 50-nm particles per grid square (±SEM, n = 3) from the in vitro assembly reactions shown in C-J.
A stable interaction between DnaK and the C-terminal domain of VP1 might sterically inhibit assembly of pentamers. We performed calcium-mediated in vitro assembly reactions to test this hypothesis. Pentamer complexes of either VP1 + VP3 (Fig. 2C)orVP1 + VP3 + copurified chaperones (Fig. 2G) were used as the starting material. Assembly was observed when VP1 + VP3 complexes were dialyzed into buffers containing calcium (Fig. 2 D-F). Although assembly of smaller (25 nm) particles was observed when VP1 + VP3 + copurified chaperones were dialyzed into calcium-containing buffer (Fig. 2H) or a calcium buffer with ATP (Fig. 2I), no assembly occurred when the interaction between DnaK and the capsid proteins was stabilized by ADP (Fig. 2 J). Quantitation of 50-nm assembled particles demonstrated that calcium-mediated assembly was inhibited in the presence of chaperones (Fig. 2K). The small, irregular aggregates seen in the presence of chaperones in buffers without ADP (Fig. 2 H and I) may be unproductive assembly, perhaps because of GroEL in the copurified chaperone preparation. Similar results were seen for recombinant VP1 (not coexpressed with VP3) purified with and without associated chaperones (data not shown). Thus, a stable interaction between DnaK and the C-terminal assembly domain of VP1 inhibits calcium-mediated in vitro assembly.
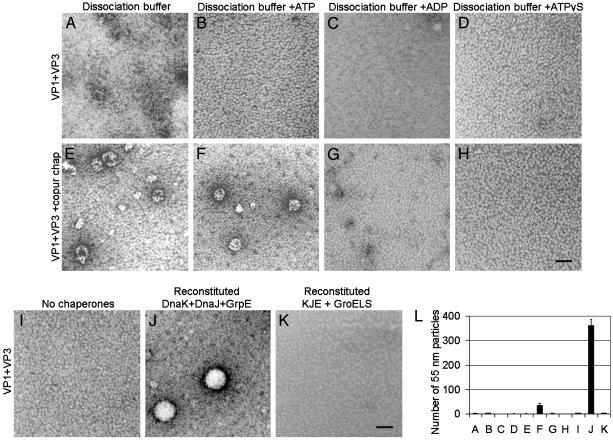
The interaction of DnaK with the C-terminal assembly domain of VP1 suggested an active role for this chaperone in assembly. Recombinant VP1 purified in a buffer containing chelating and disulfide-reducing agents (i.e., dissociation buffer) yields only VP1 pentamers (6). Similarly, the purified VP1 + VP3 complexes (Fig. 2C) remained pentameric (Fig. 3 A-D) after dialysis against the indicated dissociation buffers, except for minor aggregation (Fig. 3A). In contrast, VP1 + VP3 + copurified chaperones (Fig. 2G) assembled into both 25-nm and 50-nm particles after dialysis into dissociation buffer or a dissociation buffer with ATP (Fig. 3 E and F). The variation in size of the assemblies again may be because of the presence of GroEL in the copurified chaperones. Chaperone-mediated assembly did not occur in a dissociation buffer with ADP (Fig. 3G) and depended on ATP hydrolysis (Fig. 3H). Assembly was not induced by the addition of magnesium ions in any buffer (Fig. 3 B-D and data not shown). These in vitro reactions suggested that DnaK was actively assembling pentamers into capsid-like particles with the assistance of cochaperones and ATP.
Fig. 3.
E. coli chaperones DnaK, DnaJ, and GrpE assemble capsid proteins in vitro. (A-H) In vitro assembly reactions of VP1 + VP3 pentamers without (A-D) or with (E-H) copurified chaperones dialyzed into indicated dissociation buffers and visualized by negative stain and TEM. (I-K) In vitro assembly reactions of VP1 + VP3 pentamers at 1× molar concentration in dissociating buffer with ATP, using no chaperones (I), a reconstituted chaperone system with purified DnaK (0.5×), DnaJ (0.05×), and GrpE (0.05×)(J), or a reconstituted chaperone system with purified DnaK (0.5×), DnaJ (0.05×), GrpE (0.05×), and GroELS (1×) visualized by negative stain and TEM. (Scale bar, 50 nm.) (L) Quantitation of the mean number of 55-nm particles per grid square (±SEM, n = 3) from the in vitro assembly reactions shown in A-K.
To identify the essential protein components for chaperone-mediated assembly, the assembly reaction was reconstituted with purified chaperones. E. coli chaperones were added to VP1 + VP3 complexes, were dialyzed into dissociation buffer with ATP, and were assayed for in vitro assembly. No assembly occurred without chaperones (Fig. 3I) or when purified DnaK, DnaJ, or GrpE alone (data not shown) were added to VP1 + VP3 pentamers. When DnaK, DnaJ, and GrpE were simultaneously present (without GroEL contamination), uniform capsids of ≈55 nm were observed (Fig. 3J). Assembly by DnaK, DnaJ, and GrpE was inhibited by the addition of GroEL to the reaction (Fig. 3K). Capsids formed by chaperones remained stable in the absence of ATP (data not shown). Quantitation of in vitro assembly shown in Fig. 3 confirmed that the reconstituted E. coli chaperone system with DnaK, DnaJ, and GrpE was sufficient to promote assembly of uniform capsids (Fig. 3L). The reaction with reconstituted chaperones was ≈10-fold more efficient at assembly than the reaction using copurified chaperones (Fig. 3L, F versus J), perhaps because of a suboptimal stoichiometry of chaperones in the latter mixture.
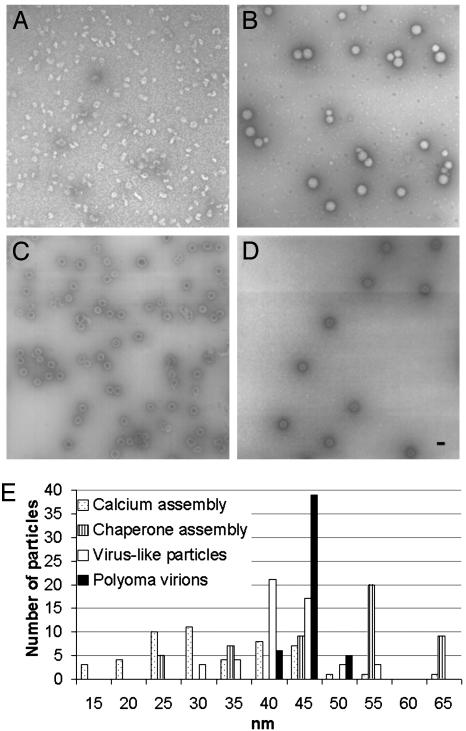
The particles assembled by the reconstituted bacterial chaperones were more uniform in size and appearance than were particles formed in the presence of calcium (Fig. 4 A and B). Moreover, the chaperone-assembled capsids lacked the internal staining typically seen in empty particles such as VP1 VLPs (Fig. 4C), and appeared more similar to “full” virions (Fig. 4D). The minor capsid protein VP3 may not allow stain penetration into the interior of the capsid, although this possibility requires further study. The size distribution (Fig. 4E) of chaperone assembly products resembled that of polyoma virions and VLPs more closely than the wide distribution seen in calcium assembly of VP1 + VP3 (Fig. 4E). Calcium-mediated assembly of VP1 alone is more uniform (6). A similar size distribution was observed by using density gradient centrifugation (data not shown). The chaperone-assembled products averaged 55 nm and appeared larger than virus, possibly because DNA is not present to contribute to particle compaction. Thus, the products assembled by chaperones are uniform, and more closely resemble virions than do products obtained in other in vitro reactions.
Fig. 4.
Capsids assembled by chaperones in vitro are uniform. (A-D) Negative stain and TEM of calcium assembly reaction (A), chaperone assembly reaction (B), VP1 VLPs (C), and polyoma virions (D). (Scale bar, 50 nm.) (E) Size polymorphism, based on 50 random assembled particles for the reactions shown in A-D.
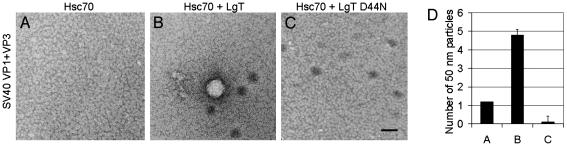
To determine whether chaperone-mediated capsid assembly could be catalyzed by eukaryotic chaperones, in vitro assembly reactions were performed by using purified recombinant eukaryotic chaperones reconstituted with SV40 VP1 + VP3 complexes. SV40 protein complexes behaved identically to the murine polyomavirus proteins in both calcium and bacterial chaperone-mediated capsid assembly (data not shown). We did not observe assembly by using either bovine hsc70 or avian hsp70 with either YDJ-1 or human hsp40 (data not shown). However, we observed the assembly of SV40 VP1 + VP3 complexes, by using bovine hsc70 and SV40 LgT. The viral protein LgT encodes for a J-domain at its N terminus that functions as a molecular chaperone in cellular transformation and viral genome replication in the nucleus (24). Moreover, the LgT J-domain can functionally substitute for the J-domain of E. coli DnaJ to support phage λ growth (25). Assembly did not occur when pentamers of SV40 VP1 + VP3 alone were dialyzed into dissociation buffer with ATP (data not shown). Limited assembly was observed when bovine hsc70 was added to pentamers of SV40 VP1 + VP3 (Fig. 5A). Similar results were obtained when only SV40 LgT was added to pentamers (data not shown). However, when both bovine hsc70 and SV40 LgT were added to SV40 VP1 + VP3 pentamers, assembly of uniform 50-nm capsids was observed (Fig. 5B). Importantly, hsc70 + LgT-mediated assembly was inhibited by the LgT J-domain mutant protein (D44N), which does not bind hsc70 (ref. 26 and Fig. 5C), indicating that the J-domain was responsible for the assembly reaction. Assembly by eukaryotic chaperones was ≈50-fold less efficient than with bacterial chaperones, possibly because of contaminating bacterial proteins or because of a requirement for additional eukaryotic cofactors and buffer optimization. Nevertheless, quantitation of the assembled particles (Fig. 5D) verifies that the J-domain of LgT is required for in vitro capsid assembly by bovine hsc70, and that the background basal level of assembly with hsc70 alone is abolished when the reaction is inhibited by the D44N protein.
Fig. 5.
Mammalian hsc70 requires the J-domain of large T antigen to assemble capsid proteins in vitro.(A-C) In vitro assembly reactions of purified recombinant SV40 capsid proteins VP1 + VP3 at 1× molar concentration with mammalian hsc70 (1×) (A), mammalian hsc70 (1×) + LgT (at 0.1× for the J-domain) (B), or mammalian hsc70 (1×) + LgT D44N, a J-domain mutant (0.1×)(C), visualized by negative stain and TEM. (Scale bar, 50 nm.) (D) Quantitation of the mean number of 50-nm particles per grid square (±SEM, n = 10) from the in vitro assembly reactions shown in A-C.
Discussion
First identified as essential in bacteriophage assembly (27-32), hsp70 family chaperones have now been implicated in the life cycle of a number of eukaryotic DNA and RNA viruses. The use of chaperones by viruses is perhaps not surprising, given the requirements for virion quality control and for the regulation of structural proteins intrinsically poised for assembly. However, a mechanistic role for chaperones has not yet been described for controlling either the quality or the subcellular location of viral capsid assembly. Along with the previous finding (9) that hsc70 complexes with VP1 in virus-infected mouse cells, the current observation that hsp70 chaperones bind the C terminus of VP1 and inhibit capsid assembly in vitro strongly suggests an active role for this chaperone in polyomavirus assembly.
Although calcium ions are found in polyoma virions (2), they are apparently not required for capsid assembly under our in vitro conditions for chaperone-mediated assembly. Perhaps the VP1 C-terminal strand insertion is accomplished by the chaperone reaction in such an ordered or shielded manner that other ions can be used to neutralize repulsive side groups [e.g., in vitro capsid assembly induced by high concentrations of sodium chloride in the absence of calcium (5, 6)]. Consistent with a well ordered coaxing of C-terminal domains into bonding positions, the assemblies generated by the chaperones are highly regular in appearance, which is in contrast to the variable morphologies generated by calcium-induced assembly.
The previous characterization (9) of the colocalization of hsc70 with VP1 from posttranslation into the nucleus of an infected cell, and the experiments described here allow us to hypothesize an in vivo regulatory role for hsp70 chaperones in polyomavirus assembly. Hsc70 binds the disordered C-terminal domains of VP1 immediately after translation to inhibit premature assembly in the cytosol, although other inhibitory factors may also be involved. For example, the E. coli chaperone GroEL inhibited assembly by the hsp70 chaperone system in these in vitro reactions, and homologous eukaryotic hsp60 family chaperones may inhibit assembly in the cytosol. Alternatively, karyopherins bound to the nuclear localization signal of the capsid proteins may inhibit assembly until they release the cargo in the nucleus. On arrival in the nucleus, hsc70 and a J-domain cochaperone complete the task of C-terminal strand invasion into a neighboring capsomere, to yield capsids in an energy-dependent fashion. Thus, hsp70 family chaperones and cochaperones may also control the location of assembly in vivo.
Exclusive assembly of virions within the eukaryotic cell nucleus necessarily implies a compartmentalized trigger that links capsid assembly to the location of viral genomes. Thus, a protein bound specifically to the viral genome would be an ideal candidate to properly initiate capsid protein assembly. LgT not only specifically binds the polyoma origin but also possesses a J-domain that can catalyze hsc70 chaperone assembly. Although the SV40 LgT D44N mutant virus fails to replicate the viral genome and cannot be used to measure capsid assembly in vivo, other viral mutants have demonstrated that the J-domain of LgT also participates in viral assembly (33, 34). The inability to induce assembly with several heterologous combinations of eukaryotic chaperones may be due to a more stringent requirement for appropriate cochaperones or an actual specificity between hsc70-bound VP1 and the J-domain (or other sites) of LgT. Further characterization of this reaction will be required before concluding that a unique interaction is occurring with LgT. Alternative J-domains must be present in eukaryotic cells used for recombinant VP1 expression, where nuclear capsid assembly occurs in the absence of other viral proteins or viral DNA (7). Nonetheless, the uniform construction of 15-MDa spheres from 210-kDa building blocks suggests that the hsp70 chaperone system is a robust molecular machine of unusual precision in the assembly of viral capsids.
Acknowledgments
We thank Xiaojiang Chen for the recombinant capsid protein coexpression constructs, Patricia Estes for initial expression of these constructs, and Avrom Caplan and David Toft for the YDJ-1 and hsp70 expression vectors. This work was supported by National Institutes of Health Grant CA 37667 (to R.L.G.), National Institutes of Health Grant CA 40586 (to J.M.P.), and National Institutes of Health Predoctoral Training Grant T32-FM08730 (to L.R.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: VP1, viral protein 1; VP1 + VP3, VP1 pentamer with VP3; VLP, virus-like particle; LgT, large T-antigen; TEM, transmission electron microscopy; SV40, simian virus 40; hsc70, ≈70-kDa heat shock cognate protein; hsp70, ≈70-kDa heat shock protein.
References
- 1.Rayment, I., Baker, T. S., Caspar, D. L. & Murakami, W. T. (1982) Nature 295, 110-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liddington, R. C., Yan, Y., Moulai, J., Sahli, R., Benjamin, T. L. & Harrison, S. C. (1991) Nature 354, 278-284. [DOI] [PubMed] [Google Scholar]
- 3.Griffith, J. P., Griffith, D. L., Rayment, I., Murakami, W. T. & Caspar, D. L. (1992) Nature 355, 652-654. [DOI] [PubMed] [Google Scholar]
- 4.Garcea, R. L., Salunke, D. M. & Caspar, D. L. (1987) Nature 329, 86-87. [DOI] [PubMed] [Google Scholar]
- 5.Salunke, D. M., Caspar, D. L. & Garcea, R. L. (1989) Biophys. J. 56, 887-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salunke, D. M., Caspar, D. L. & Garcea, R. L. (1986) Cell 46, 895-904. [DOI] [PubMed] [Google Scholar]
- 7.Montross, L., Watkins, S., Moreland, R. B., Mamon, H., Caspar, D. L. & Garcea, R. L. (1991) J. Virol. 65, 4991-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartl, F. U. & Hayer-Hartl, M. (2002) Science 295, 1852-1858. [DOI] [PubMed] [Google Scholar]
- 9.Cripe, T. P., Delos, S. E., Estes, P. A. & Garcea, R. L. (1995) J. Virol. 69, 7807-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukau, B., Deuerling, E., Pfund, C. & Craig, E. A. (2000) Cell 101, 119-122. [DOI] [PubMed] [Google Scholar]
- 11.Bukau, B. & Horwich, A. L. (1998) Cell 92, 351-366. [DOI] [PubMed] [Google Scholar]
- 12.Kelley, W. L. (1998) Trends Biochem. Sci. 23, 222-227. [DOI] [PubMed] [Google Scholar]
- 13.Packschies, L., Theyssen, H., Buchberger, A., Bukau, B., Goody, R. S & Reinstein, J. (1997) Biochemistry 36, 3417-3422. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, C. J., Hayer-Hartl, M., Di Liberto, M., Hartl, F. & Kuriyan, J. (1997) Science 276, 431-435. [DOI] [PubMed] [Google Scholar]
- 15.Chen, X. S., Stehle, T. & Harrison, S. C. (1998) EMBO J. 17, 3233-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finnen, R. L., Erickson, K. D., Chen, X. S. & Garcea, R. L. (2003) J. Virol. 77, 4818-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cilmi, S. A. (1991) B.S. thesis (Harvard Univ., Cambridge, MA).
- 18.Caplan, A. J., Tsai, J., Casey, P. J. & Douglas, M. G. (1992) J. Biol. Chem. 267, 18890-18895. [PubMed] [Google Scholar]
- 19.Cantalupo, P., Saenz-Robles, M. T. & Pipas, J. M. (1999) Methods Enzymol. 306, 297-307. [DOI] [PubMed] [Google Scholar]
- 20.Schaffhausen, B. S. & Benjamin, T. L. (1976) Proc. Natl. Acad. Sci. USA 73, 1092-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcea, R. L. & Estes, P. A. (1998) in Cell Biology: A Laboratory Handbook (Academic, New York), pp. 521-527.
- 22.Moreland, R. B., Montross, L. & Garcea, R. L. (1991) J. Virol. 65, 1168-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosano, H., Stensgard, B., Charlesworth, M. C., McMahon, N. & Toft, D. (1998) J. Biol. Chem. 273, 32973-32979. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan, C. S. & Pipas, J. M. (2002) Microbiol. Mol. Biol. Rev. 66, 179-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley, W. L. & Georgopoulos, C. (1997) Proc. Natl. Acad. Sci. USA 94, 3679-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan, C. S., Gilbert, S. P. & Pipas, J. M. (2001) J. Virol. 75, 1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niewiarowska, J., D' Halluin, J. C. & Belin, M. T. (1992) Exp. Cell Res. 201, 408-416. [DOI] [PubMed] [Google Scholar]
- 28.Saphire, A. C., Guan, T., Schirmer, E. C., Nemerow, G. R. & Gerace, L. (2000) J. Biol. Chem. 275, 4298-4304. [DOI] [PubMed] [Google Scholar]
- 29.Prokhnevsky, A. I., Peremyslov, V. V., Napuli, A. J. & Dolja, V. V. (2002) J. Virol. 76, 11003-11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peremyslov, V. V., Hagiwara, Y. & Dolja, V. V. (1999) Proc. Natl. Acad. Sci. USA 96, 14771-14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satyanarayana, T., Gowda, S., Mawassi, M., Albiach-Marti, M. R., Ayllon, M. A., Robertson, C., Garnsey, S. M. & Dawson, W. O. (2000) Virology 278, 253-265. [DOI] [PubMed] [Google Scholar]
- 32.Zimmerman, C., Klein, K. C., Kiser, P. K., Singh, A. R., Firestein, B. L., Riba, S. C. & Lingappa, J. R. (2002) Nature 415, 88-92. [DOI] [PubMed] [Google Scholar]
- 33.Peden, K. W. & Pipas, J. M. (1992) Virus Genes 6, 107-118. [DOI] [PubMed] [Google Scholar]
- 34.Brodsky, J. L. & Pipas, J. M. (1998) J. Virol. 72, 5329-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]