Abstract
Mice carrying a truncated form of cAMP-responsive element binding protein (CREB)-binding protein (CBP) show several developmental abnormalities similar to patients with Rubinstein-Taybi syndrome (RTS). RTS patients suffer from mental retardation, whereas long-term memory formation is defective in mutant CBP mice. A critical role for cAMP signaling during CREB-dependent long-term memory formation appears to be evolutionarily conserved. From this observation, we reasoned that drugs that modulate CREB function by enhancing cAMP signaling might yield an effective treatment for the memory defect(s) of CBP+/− mice. To this end, we designed a cell-based drug screen and discovered inhibitors of phosphodiesterase 4 (PDE4) to be particularly effective enhancers of CREB function. We extend previous behavioral observations by showing that CBP+/− mutants have impaired long-term memory but normal learning and short-term memory in an object recognition task. We demonstrate that the prototypical PDE4 inhibitor, rolipram, and a novel one (HT0712) abolish the long-term memory defect of CBP+/− mice. Importantly, the genetic lesion in CBP acts specifically to shift the dose sensitivity for HT0712 to enhance memory formation, which conveys molecular specificity on the drug's mechanism of action. Our results suggest that PDE4 inhibitors may be used to treat the cognitive dysfunction of RTS patients.
Rubinstein-Taybi syndrome (RTS) is a human genetic disorder characterized by mental retardation and physical abnormalities including broad thumbs, big and broad toes, short stature, and craniofacial anomalies (1-3). RTS occurs in about 1 in 125,000 births and accounts for as many as 1 in 300 cases of institutionalized mentally retarded people. In many patients, RTS has been mapped to chromosome 16p13.3, a genomic region containing cAMP-responsive element binding protein (CREB)-binding protein (CBP) (4). Many RTS patients are heterozygous for CBP mutations that yield truncations of the CBP C terminus, suggesting that a dominant-negative mechanism may contribute to the clinical symptoms (5).
CBP is a transcriptional coactivator that binds to the CREB transcription factor when the latter is phosphorylated (6). CREB-dependent gene expression has been shown to underlie long-term memory formation in several vertebrate and invertebrate species (7-12), leading to the intriguing speculation that mental retardation in RTS patients may derive from reduced CBP function during long-term memory formation (13, 14). To this end, Oike et al. (15) generated a C-terminal truncation mutation in mouse CBP, which appears to act in a dominant-negative fashion to recapitulate many of the abnormalities observed in RTS patients. Homozygous CBP−/− mutants are embryonic lethal, whereas heterozygous CBP+/− mice show reduced viability, growth retardation, retarded osseous maturation, and hypoplastic maxilla (15). Importantly, CBP+/− mice show normal learning and short-term memory (STM) but defective long-term memory (LTM) for two passive avoidance tasks, substantiating the notion that normal CBP function is required for memory formation (15).
These studies demonstrate that CREB/CBP likely function together as a molecular switch during long-term memory formation. Because CREB/CBP function is reduced but not eliminated in CBP+/− mutant mice, the possibility existed to improve long-term memory formation by enhancing upstream signaling onto CREB/CBP. Here, we demonstrate that inhibitors of phosphodiesterase 4 (PDE4) (i) enhance CREB-dependent gene expression and (ii) ameliorate the long-term memory defects of CBP+/− mutant mice in a dose-dependent manner, which differs between normal and mutant mice.
Methods
The Mice. Generation of CBP+/− mice was described by Oike et al. (15). For these studies, we bred animals by crossing CBP+/− males to C57BL/6 females (The Jackson Laboratory). The mice were genotyped with a PCR protocol as described (15). We found CBP+/− mutants to be subviable. Only 25 of 168 progeny were heterozygous; this observed frequency (15%) is appreciably below that expected (50%) from the cross if wild-type and heterozygous animals were equally viable. Age-matched (12-14 weeks old by the time of handling) and gender-matched mutant mice and wild-type littermates were used for all experiments.
The mice were kept on a 12:12 light/dark cycle, and the experiments were conducted during the light phase of the cycle. With the exception of training and testing times, the mice had ad lib access of food and water. The experiments were conducted according to Animal Welfare Assurance no. A3280-01, and animals were maintained in accordance with the Animal Welfare Act and the Department of Health and Human Service guide.
Object Recognition Training and Testing. Mice were handled for 3-5 min for 5 days. The day before training, an individual mouse was placed into a training apparatus (a Plexiglas box 48 cm long, 38 cm wide, and 20 cm high, located in a dimly lit room) and allowed to habituate to the environment for 15 min (also see ref. 16). Training was initiated 24 h after habituation. A mouse was placed back into the training box, which contained two identical objects (e.g., a small conus-shape object), and was allowed to explore these objects. The objects were placed into the central area of the box, and the spatial position of objects (left-right sides) was counterbalanced between subjects. Among experiments, training times varied from 3.5 to 20 min.
We used four separate, and otherwise experimentally naïve, sets of animals. The first set was used for the experiment summarized in Fig. 2 A (wild-type mice, n = 20). The second set was used for experiments summarized in Fig. 2 B and C (n = 10 per genotype). The third set was used for the experiment summarized in Fig. 3A (wild-type mice, n = 20). The fourth set was used for the experiment summarized in Fig. 3B (n = 8 per genotype). For each experiment, the same set of animals was used repeatedly with different (new) sets of objects for each repetition. Five to nine repetitions were performed on each set of mice. Each mouse was trained and tested no more than once per week and with a 1-week interval between testing. In experiments with drug-injections (see below), vehicle-injected mice and high/low-dose-injected mice were counterbalanced. All experiments were designed and performed in a balanced fashion, meaning that (i) for each experimental condition (e.g., a specific dose-effect and/or genotype-memory effect) we used two to six experimental mice and two to six control mice; (ii) experiments with HT 0712 injections always consisted of the vehicle-injected mice and mice injected with two to three different doses of HT 0712; (iii) each experimental condition was replicated two to four independent times, and replicate days were added to generate final number of subjects. In each experiment, the experimenter was unaware (blind) to the treatment of the subjects during training and testing.
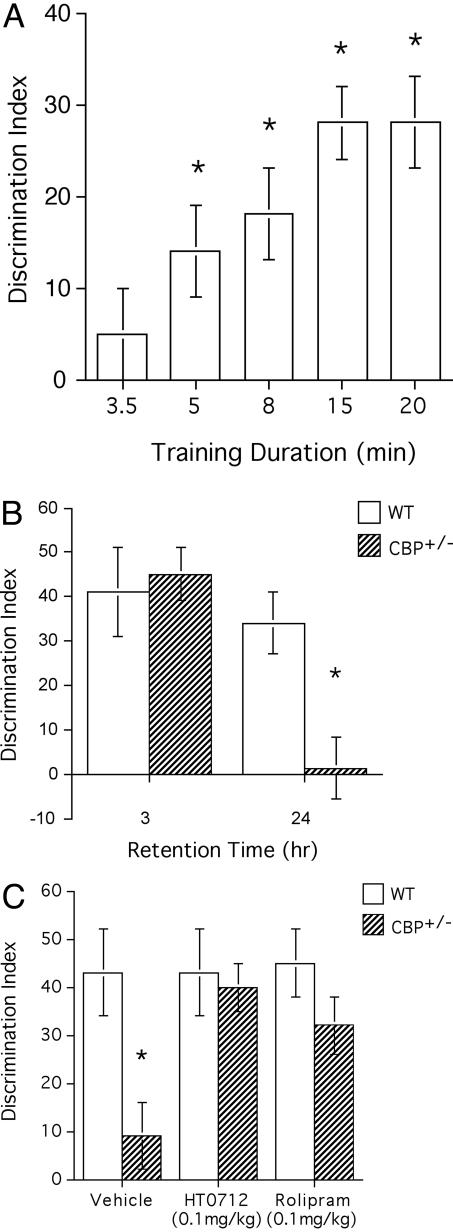
Fig. 2.
Object recognition memory in CBP+/− mutant mice and their normal siblings. (A) One-day memory retention in object recognition depends on duration of training. Wild-type mice were subjected to a 3.5-min (n = 24), 5-min (n = 24), 8-min (n = 16), 15-min (n = 46), or 20-min (n = 12) training duration and then tested 24 h later. Memory retention was quantified as a DI (see Methods). Maximal memory retention was reached with a training duration of 15 min. In contrast, 1-day memory was not detected after a 3.5-min training duration. (B) CBP+/− mutant mice have impaired long-term memory of an object recognition task. Wild-type mice and CBP+/− mutant mice were trained for 15 min and tested 3h or 24h later. Three-hour memory levels were similar for normal mice and CBP+/− mutants (P = 0.76; n = 6 for each genotype), but 24-h memory was significantly lower than normal in mutant mice (P < 0.01; n = 10 for each genotype). (C) PDE4 inhibitors ameliorate the long-term memory defect of CBP+/− mutant mice. Wild-type mice and CBP+/− mutant mice received i.p. injections of 0.1 mg/kg HT 0712 or Rolipram 20 min before training. Animals were trained with a 15-min training session, and memory retention was tested 24 h later (see Methods). In vehicle-injected CBP+/− mutants, memory was significantly lower than in vehicle-injected wild-type mice (P < 0.01; n = 12 and n = 6, respectively). In HT0712-injected CBP+/− mutants, memory was significantly higher than in vehicle-injected mutants (P < 0.05, n = 10 and n = 12, respectively). Similarly, memory retention in Rolipram-injected animals was significantly higher than that in vehicle-alone-treated animals (P < 0.05; n = 8 and n = 12, respectively). There was no significant difference between HT0712-treated CBP+/− mutants and HT0712-treated wild-type mice (P = 0.78) or between Rolipram-treated CBP+/− mutants and Rolipram-treated wild-type mice (P = 0.19). N values reflect number of observations (repetitions) per treatment.
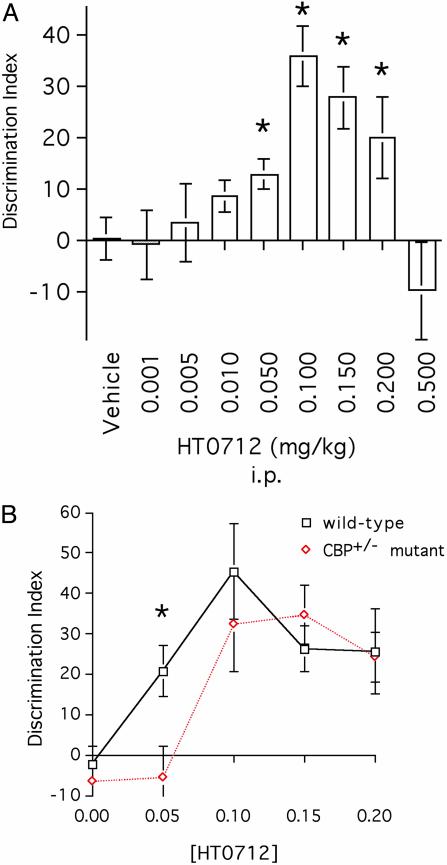
Fig. 3.
HT0712 dose sensitivity is decreased in mutant CBP+/− mice. (A) Dose-response curve for HT 0712 in wild-type mice. Mice received a single i.p. injection of drug or vehicle alone 20 min before training. Doses of 0.001, 0.005, 0.01, 0.05, 0.10, 0.15, 0.20, and 0.5 mg/kg were used. Animals experienced a 3.5-min training duration and were tested 24 h later (see Methods). Memory retention in drug-injected animals was significantly higher than that in vehicle-alone treated animals (n = 35) at doses of 0.05 mg/kg (n = 20; P < 0.05), 0.10 mg/kg (n = 22; P < 0.0001), 0.15 mg/kg (n = 18; P < 0.001), and 0.20 mg/kg (n = 17; P < 0.05). There were no significant effects at doses 0.001 mg/kg (P = 0.91; n = 6 for HT0712), 0.005 mg/kg (P = 0.72; n = 22 for HT0712), or 0.01 mg/kg (P = 0.34; n = 10 for HT0712). Because each session included vehicle-injected mice, the data for vehicle-injected mice were pooled. N values indicate a number of observations (repetitions) per treatment. (B) CBP+/− mutant mice are less sensitive than wild-type mice to the enhancing effects of HT 0712. CBP+/− mutants and wild-type mice received a single i.p. injection of vehicle or drug 20 min before training. They experienced a 3.5-min training duration and were tested 24-h later (see Methods). In wild-type mice, memory retention in drug-treated groups again was higher than in the vehicle-alone group (n = 26) at doses of 0.05 mg/kg (n = 12; P < 0.005), 0.10 mg/kg (n = 8; P < 0.0001), 0.15 mg/kg (n = 18; P < 0.005), and 0.2 mg/kg (n = 14; P < 0.005). In CBP+/− mutants, memory retention in drug-treated groups was higher than in the vehicle-alone group (n = 26) at doses of 0.10 mg/kg (n = 8; P < 0.0001), 0.15 mg/kg (n = 10; P < 0.0001), and 0.20 mg/kg (n = 14; P < 0.0001). In contrast to wild-type mice, a 0.05 mg/kg dose of HT0712 failed to enhance memory (n = 26; P = 0.79) in CBP+/− mutants. N values reflect number of observations (repetitions) per dose per genotype.
To test for memory retention, mice were observed for 10 min 3 and 24 h after training. Mice were presented with two objects, one of which was used during training, and thus was “familiar,” and the other of which was novel (e.g., a small pyramid-shape object). The test objects were divided into 10 sets of two “training” plus “testing” objects, and a new set of objects was used for each training session. To ensure that the discrimination targets did not differ in smell, after each experimental subject, the apparatus and the objects were thoroughly cleaned with 90% ethanol, dried, and ventilated for a few minutes.
Drug Compound Administration. Twenty minutes before training, mice were injected in their home cages with the indicated doses of HT 0712, Rolipram (in 1% DMSO/PBS) or with vehicle alone (1% DMSO/PBS). HT0712 was administered i.p. at doses of 0.001, 0.005, 0.01, 0.05, 0.10, 0.15, 0.20, and 0.50 mg/kg. Rolipram was administered i.p. at 0.10 mg/kg. Drug compounds were injected with 1-week intervals to allow for sufficient wash-out time (the half-life for HT 0712 and Rolipram <3 h). In addition, vehicle- and drug-injected mice were counterbalanced from experiment to experiment. Such design allowed at least a 2-week wash-out time between repeated usages of high doses. We have not observed any dose-accumulating effects with repeated injections between/within the groups (data not shown).
Data Analysis. The experiments were videotaped via an overhead video camera system. Types were reviewed by a blinded observer and the following behavioral parameters were determined: time of exploration of each object, the total time of exploration of the objects, number of approaches to the objects, and time (latency) to first approach an object. The discrimination index (DI) was determined as the difference in exploration time expressed as a ratio of the total time spent exploring the two objects in the choice (testing) phase (17). This ratio serves to normalize for individual or group differences in the total amount of exploration time. Data were analyzed by Student's unpaired t test using a software package (STATWIEW 5.0.1, SAS Institute, Cary, NC). All values in the text and figures are expressed as mean ± SEM.
CRE-Luciferase Assay. A cell-based screening system was developed with a functional read-out of CREB-dependent transcription. Ten thousand SK-N-MC neuroblastoma cells, stably transfected with a reporter construct consisting of the VIP promoter containing two CRE elements together with two additional CRE elements from the tyrosine aminotransferase gene driving expression of luciferase, were plated in a 96-well plate format overnight at 37°C. The next day, cells were incubated with drug (5 μM) or vehicle (0.2% DMSO) for 2 h, after which forskolin (5 μM) was added to the cells. After a further 6-h incubation at 37°C, cells were washed briefly with PBS and lysed, and luciferase activity was measured by using a Victor 5 luminometer.
Real-Time Quantitative PCR. Drugs were tested for their ability to superinduce the endogenous CREB-responsive gene Somatostatin in the neuroblastoma SK-N-MC cell line. A total of 500,000 cells (in triplicate for each condition) were plated in a 96-well plate format and incubated overnight at 37°C. Cells were incubated for 2 h with drug (5 μM) or vehicle (0.2% DMSO), followed by a 6-h incubation with forskolin (5 μM). Cells were washed twice with cold PBS, and RNA was isolated. Somatostatin RNA levels were examined by real-time quantitative PCR using an Applied Biosystems 7900 real-time PCR instrument for rapid RNA analysis. Somatostatin RNA levels were normalized against TFIID (TATA-binding protein) RNA levels.
Results
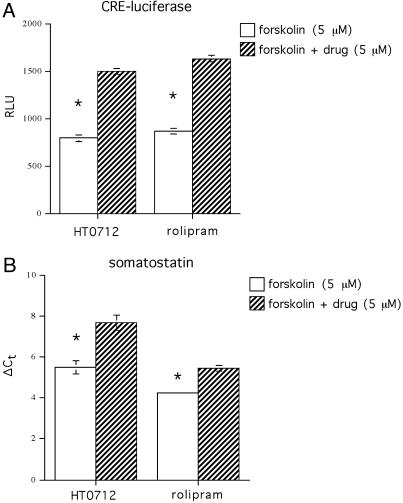
PDE4 Inhibitors Increase CREB-Dependent Gene Expression. A high-throughput drug screen was conducted by using human SKN-MC neuroblastoma cells, which were stably transfected with a luciferase reporter gene driven by a CRE promoter (18). Cells were exposed to drug for 2 h and then stimulated with a suboptimal dose of forskolin for further 6 h. Compounds were identified that had no effect on their own but that significantly increased forskolin-induced CRE-luciferase expression. Among the dozens of confirmed active compounds for several molecular targets identified from this screen, inhibitors of PDE4 were numerous. Consequently, we focused our attention on a novel PDE4 inhibitor, HT 0712. In addition, we also studied Rolipram, a prototypical PDE4 inhibitor that has been shown previously to affect performance in animal models of memory (19-21). Both compounds produced robust effects on CRE-luciferase expression and on expression of a CRE-dependent endogenous gene, somatostatin (Fig. 1).
Fig. 1.
PDE4 inhibitors enhance forskolin-induced gene expression in human neuroblastoma cells. (A) Cells were stably transfected with a CRE-luciferase reporter gene and exposed to vehicle alone or drug (HT0712 or rolipram) for 2 h before stimulation by a suboptimal dose of forskolin. Relative light units (RLU) emitted from luciferase were quantified. Both drugs increased forskolin-induced CRE-luciferase expression 1.9-fold above forskolin alone (P < 0.001 in each case). (B) Real-time PCR was used to quantify expression of somatostatin, an endogenous cAMP-responsive gene. Expression levels induced by forskolin or by forskolin plus drug are quantified as differences in threshold cycle number (ΔCt) above vehicle alone control groups (not shown). HT0712 and rolipram produced 4.6-fold (P < 0.001) and 2.3-fold (P < 0.001) increases, respectively, in forskolin-induced expression of somatostatin.
Long-Term Memory for Object Recognition Is Defective in CBP+/−
Mutant Mice. Our initial experiments on normal, young-adult mice established that long-term memory formation after contextual fear conditioning was enhanced by HT0712 and rolipram, delivered directly to the hippocampus, intraventricularly, or i.p. (refs. 7 and 18 and data not shown). Specifically, these drugs enhance memory formation by reducing the amount of training required to produce maximal long-term memory. We also were interested to determine whether these drugs could ameliorate memory defects caused by molecular lesions in the CREB pathway. The mouse model of RTS was particularly attractive because (i) the molecular lesion (truncated protein) in mice was similar to those known for some RTS patients, (ii) CBP function in CBP+/− heterozygous mice was reduced but not blocked, and (iii) long-term memory formation, but not learning or short-term memory, appeared specifically to be disrupted in these mutant animals (15).
Long-term memory defects in CBP+/− mutant mice have been reported only for fear-based tasks (15). Hence, we first wanted to know whether CBP+/− mutant mice also had defective long-term memory for a different type of experience. Object recognition is a nonaversive task that relies on a mouse's natural exploratory behavior. During training for this task, mice are presented with two identical novel objects, which they explore for some time by orienting toward, sniffing, and crawling over. Normal mice then will remember having explored that object. To test for this memory, mice are presented at a later time with two different objects, one of which was presented previously during training and thus is “familiar,” and the other of which is novel. If the mouse remembers the familiar object, it spends more time exploring the novel object. By analogy to an object recognition-based “nonmatching to sample” task in monkeys and rats (22, 23), this task can be performed repeatedly on the same animals by exposing them serially to different sets of novel objects.
Compared with fear-based tasks, which use relatively strong punishment as reinforcement, object recognition is considered to be a poorly motivated task. One consequence is appreciable interstrain variation in performance. Hence, we calibrated our training protocol for the genetic background represented by the wild-type (normal) littermates of CBP+/− mutant mice. Initially, we subjected these wild-type mice to various durations of training, ranging from 3.5 to 20 min and then quantified 1-day memory thereafter. Maximal 1-day memory was reached with a 15- or 20-min training duration, whereas no 1-day memory was detected with a 3.5-min training duration (Fig. 2 A).
We then gave CBP+/− mutants and their wild-type littermates 15 min to explore novel objects during training and tested their memory retention 3 and 24 h later (Fig. 2B). Three-hour memory appeared normal, but 1-day memory was abnormally low, in CBP+/− mutants (DI = 34 ± 7 and 1 ± 7 for wild-type mice and CBP+/− mutants, respectively; P < 0.01]. These results indicate that CBP+/− mutant mice have impaired long-term memory, but normal short-term memory, for object recognition under training conditions that yield maximal 1-day memory in normal mice of the same genetic background. These findings extend the observations of Oike et al. (15) to an ethologically relevant, nonaversive behavior and confirm the notion that loss-of-function mutations in CBP can yield specific defects in long-term memory formation.
PDE4 Inihibitors Ameliorate the Long-Term Memory Deficit of CBP+/−
Mutant Mice in a Dose-Dependent Manner. To evaluate the PDE4 inhibitors, drug or vehicle alone were administered i.p. to normal mice and CBP+/− mutants 20 min before a 15-min training session (Fig. 2C). As in the previous experiment, 1-day memory retention was significantly reduced in CBP+/− mutants treated with vehicle alone (DI = 9 ± 7; P < 0.01). In striking contrast, however, a single administration of 0.10 mg/kg HT0712 or Rolipram restored 24-h memory to normal levels in CBP+/− mutants (DI = 40 ± 5 and 32 ± 6 for HT 0712 and Rolipram treated mice, respectively).
To address whether the drugs' effects were specific to the molecular lesion in CBP, we changed our training protocol and determined dose sensitivity curves for mutant and wild-type animals. Our 15-min training protocol produces maximum 1-day retention in the wild-type mice used here (Fig. 2 A). Consequently, we did not see any drug-induced memory enhancement in wild-type mice (Fig. 2C). By reducing training to a 3.5-min protocol, however, 1-day retention was near zero in wild-type mice, thereby allowing an evaluation of the enhancing effects of the PDE4 inhibitors. We then reasoned that, because CBP+/− mutants had less functional CBP than wild-type animals, a higher concentration of drug might be required in the mutants than in wild-type mice to produce equivalent levels of memory enhancement. In essence, the molecular lesion in CBP would act specifically to shift the dose sensitivity for a PDE4 inhibitor to enhance memory formation, a concept advanced by Silva and coworkers (24).
We first quantified the dose-response curve for wild-type mice (Fig. 3A). In mice treated with vehicle alone, the 3.5-min training protocol did not produce any appreciable 1-day memory. At concentrations below 0.05 mg/kg or above 0.20 mg/kg, HT 0712 failed to produce any memory enhancement. One-day memory retention was significantly increased, however, at HT0712 concentrations of 0.05, 0.10, 0.15, and 0.20 mg/kg. Next, we compared memory retention between CBP+/− mutants and wild-type animals at selected concentrations of HT0712 (Fig. 3B). We found that the initial effective dose differed between mutant and wild-type animals. Again, at a dose of 0.05 mg/kg, wild-type animals showed significant enhancement of 1-day memory (21 ± 6 vs. −2 ± 4; HT0712- and vehicle-treated mice, respectively; P < 0.005), but CBP+/− mutants did not. Memory enhancement was first seen in CBP+/− mutants at the next higher dose of HT0712 (0.10 mg/kg; 32 ± 12 vs. −7 ± 4 for HT0712- and vehicle-treated mice, respectively, P < 0.0001). Similarly, the peak effective dose appears shifted to a higher concentration in mutants (0.15 mg/kg) than in wild-type mice (0.10 mg/kg).
We considered whether HT 0712 might be increasing performance in our task nonspecifically by affecting perception of the training context (objects) or the motivation to explore objects during training or testing. We analyzed the latency to first approach an object during training, the total number of approaches to an object, and the total exploration time. In all experiments, no differences between genotypes and/or drug treatments were observed in the latency to first approach (data not shown). CBP+/− mutant mice showed increases in total exploration time and in the total number of object-approaches, but drug treatments did not change these measures, and these behavioral responses were not correlated with DIs (data not shown). Similarly, we did not observe any behavioral side effects (lethargy, nausea, etc.) from any doses of HT0712.
We also administered HT0712 immediately after training to dissociate further any possible nonspecific effects on performance from its more specific effect on memory consolidation. Mutant and wild-type mice were trained and then injected with 0.15 mg/kg HT0712 or vehicle immediately thereafter. Again, we observed significant memory enhancement 1 day after training in both CBP+/− mutant mice (23 ± 5 vs. −5 ± 3 for HT0712- vs. vehicle-treated mice; n = 12 and n = 8, respectively; P < 0.001) and wild-type mice (22 ± 5 vs. −9 ± 9 for HT0712- vs. vehicle-treated mice; n = 12 and n = 8, respectively; P < 0.01).
Finally, the 3.5-min training protocol did not produce significant short-term memory, tested 1 h after training, and HT 0712 did not have any appreciable memory-enhancing effects on short-term memory (n = 6 per genotype; data not shown). These observations support our previous findings (19), where Rolipram had no effect on short-term memory for fear conditioning. They also are in agreement with the functional role of CREB in memory formation, namely, its requirement for long-term, but not for short-term, memory.
Discussion
Our data indicate that the memory impairments observed for CBP+/− mutants in an object recognition task can be ameliorated by inhibitors of PDE4. These PDE inhibitors likely enhance signaling to CREB during memory formation by increasing cAMP levels in response to experience-dependent changes in neural activity (18-20, 25). This effect apparently is specific molecularly, because CBP+/− mutants require a higher dose of HT0712 than wild-type mice to produce an equivalent amount of enhanced memory. This result, in fact, might be expected because the genetic lesion of CBP+/− mutant mice likely reduces the amount of CREB/CBP available for functional activation via cAMP signaling. Hence, greater upstream enhancement is required for a comparable effect on the regulation of downstream genes (see ref. 8). Given that CBP interacts with several transcription factors (26-28), likely via an innate histone acetyltransferase activity (29, 30), our results strongly suggest CREB/CBP to be the relevant interaction for long-term memory formation. Because of the observed molecular and pathological similarities between these CBP+/− mice and patients with RTS, we hypothesize that the mental retardation of RTS patients likely results, at least in part, from defects in long-term memory formation. Further, PDE4 inhibitors may ameliorate this biochemical block on memory formation, thereby rendering the patient capable to benefit from training and experience.
Acknowledgments
We thank Jim Watson, Bruce Stillman, Walt Lovenberg, George Carmany, and John Tallman for their support and encouragement on this project, and John Tallman and Josh Dubnau for comments on the manuscript. T.T. is a St. Giles Foundation Professor of Neuroscience at the Cold Spring Harbor Laboratory.
Abbreviations: CREB, cAMP-responsive element binding protein; CBP, CREB-binding protein; RTS, Rubinstein-Taybi syndrome; PDE4, phosphodiesterase 4; DI, discrimination index.
References
- 1.Rubinstein, J. H. & Taybi, H. (1963) Am. J. Dis. Child. 105, 588-608. [DOI] [PubMed] [Google Scholar]
- 2.Hennekam, R. C., Baselier, A. C., Beyaert, E., Bos, A., Blok, J. B., Jansma, H. B., Thorbecke-Nilsen, V. V. & Veerman, H. (1992) Am. J. Mental Retard. 96, 645-660. [PubMed] [Google Scholar]
- 3.Cantani, A. & Gagliesi, D. (1998) Eur. Rev. Med. Pharmacol. Sci. 2, 81-87. [PubMed] [Google Scholar]
- 4.Petrij, F., Giles, R. H., Dauwerse, H. G., Saris, J. J., Hennekam, R. C., Masuno, M., Tommerup, N., van Ommen, G. J., Goodman, R. H., Peters, D. J., et al. (1995) Nature 376, 348-351. [DOI] [PubMed] [Google Scholar]
- 5.Petrij, F., Dorsman, J. C., Dauwerse, H. G., Giles, R. H., Peeters, T., Hennekam, R. C., Breuning, M. H. & Peters, D. J. (2000) Am. J. Med. Genet. 92, 47-52. [DOI] [PubMed] [Google Scholar]
- 6.Lonze, B. E. & Ginty, D. D. (2002) Neuron 35, 605-623. [DOI] [PubMed] [Google Scholar]
- 7.Tully, T., Bourtchouladze, R., Scott, R. & Tallman, J. (2003) Nat. Rev. Drug Discovery 2, 267-277. [DOI] [PubMed] [Google Scholar]
- 8.Dubnau, J., Chiang, A. S., Grady, L., Barditch, J., Gossweiler, S., McNeil, J., Smith, P., Buldoc, F., Scott, R., Certa, U., et al. (2003) Curr. Biol. 13, 286-296. [DOI] [PubMed] [Google Scholar]
- 9.Poser, S. & Storm, D. R. (2001) Int. J. Dev. Neurosci. 19, 387-394. [DOI] [PubMed] [Google Scholar]
- 10.Bailey, C. H., Giustetto, M., Huang, Y. Y., Hawkins, R. D. & Kandel, E. R. (2000) Nat. Rev. Neurosci. 1, 11-20. [DOI] [PubMed] [Google Scholar]
- 11.Dubnau, J. & Tully, T. (1998) Annu. Rev. Neurosci. 21, 407-444. [DOI] [PubMed] [Google Scholar]
- 12.Menzel, R. (2001) Learn. Mem. 8, 53-62. [DOI] [PubMed] [Google Scholar]
- 13.D'Arcangelo, G. & Curran, T. (1995) Nature 376, 292-293. [DOI] [PubMed] [Google Scholar]
- 14.Weeber, E. J. & Sweatt, J. D. (2002) Neuron 33, 845-848. [DOI] [PubMed] [Google Scholar]
- 15.Oike, Y., Hata, A., Mamiya, T., Kaname, T., Noda, Y., Suzuki, M., Yasue, H., Nabeshima, T., Araki, K. & Yamamura, K. (1999) Hum. Mol. Genet. 8, 387-396. [DOI] [PubMed] [Google Scholar]
- 16.Pittenger, C., Huang, Y. Y., Paletzki, R. F., Bourtchouladze, R., Scanlin, H., Vronskaya, S. & Kandel, E. R. (2002) Neuron 34, 447-462. [DOI] [PubMed] [Google Scholar]
- 17.Ennaceur, A. & Aggleton, J. P. (1997) Behav. Brain Res. 88, 181-193. [DOI] [PubMed] [Google Scholar]
- 18.Scott, R., Bourtchuladze, R., Gossweiler, S., Dubnau, J. & Tully, T. (2002) J. Mol. Neurosci. 19, 171-177. [DOI] [PubMed] [Google Scholar]
- 19.Barad, M., Bourtchouladze, R., Winder, D. G., Golan, H. & Kandel, E. (1998) Proc. Natl. Acad. Sci. USA 95, 15020-15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagakura, A., Takagi, N. & Takeo, S. (2002) Neuroscience 113, 519-528. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, K., Farooqui, S. M. & O'Donnell, J. M. (1999) Brain Res. Dev. Brain Res. 112, 11-19. [DOI] [PubMed] [Google Scholar]
- 22.Mishkin, M. & Appenzeller, T. (1987) Sci. Am. 256, 80-89. [DOI] [PubMed] [Google Scholar]
- 23.Wood, E. R., Mumby, D. G., Pinel, J. P. & Phillips, A. G. (1993) Behav. Neurosci. 107, 51-62. [DOI] [PubMed] [Google Scholar]
- 24.Frankland, P. W., Ohno, M., Takahashi, E., Chen, A. R., Costa, R. M., Kushner, S. A. & Silva, A. J. (2003) Neuroscientist 9, 104-109. [DOI] [PubMed] [Google Scholar]
- 25.Vitolo, O. V., Sant'Angelo, A., Costanzo, V., Battaglia, F., Arancio, O. & Shelanski, M. (2002) Proc. Natl. Acad. Sci. USA 99, 13217-13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamei, Y., Xu, L., Heinzel, T., Torchia, J., Kurokawa, R., Gloss, B., Lin, S. C., Heyman, R. A., Rose, D. W., Glass, C. K. & Rosenfeld, M. G. (1996) Cell 85, 403-414. [DOI] [PubMed] [Google Scholar]
- 27.Horvai, A. E., Xu, L., Korzus, E., Brard, G., Kalafus, D., Mullen, T. M., Rose, D. W., Rosenfeld, M. G. & Glass, C. K. (1997) Proc. Natl. Acad. Sci. USA 94, 1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torchia, J., Rose, D. W., Inostroza, J., Kamei, Y., Westin, S., Glass, C. K. & Rosenfeld, M. G. (1997) Nature 387, 677-684. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides, T. (2000) EMBO J. 19, 1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korzus, E., Torchia, J., Rose, D. W., Xu, L., Kurokawa, R., McInerney, E. M., Mullen, T. M., Glass, C. K. & Rosenfeld, M. G. (1998) Science 279, 703-707. [DOI] [PubMed] [Google Scholar]