Abstract
Chromatin remodeling is a major event that occurs during mammalian spermiogenesis, the process of spermatid maturation into spermatozoa. Nuclear condensation during spermiogenesis is accomplished by replacing somatic histones (linker histone H1 and core histones) and the testis-specific linker histone, H1t, with transition proteins and protamines. It has long been thought that H1t is the only testis-specific linker histone, and that all linker histones are replaced by transition proteins, and subsequently by protamines during spermiogenesis. Here, we report the identification and characterization of a spermatid-specific linker histone H1-like protein (termed HILS1) in the mouse and human. Both mouse and human HILS1 genes are located in intron 8 of the α-sarcoglycan genes. HILS1 is highly expressed in nuclei of elongating and elongated spermatids (steps 9-15). HILS1 displays several biochemical properties that are similar to those of linker histones, including the abilities to bind reconstituted mononucleosomes, produce a chromatosome stop during micrococcal nuclease digestion, and aggregate chromatin. Because HILS1 is expressed in late spermatids that do not contain core histones, HILS1 may participate in spermatid nuclear condensation through a mechanism distinct from that of linker histones. Because HILS1 also belongs to the large winged helix/forkhead protein superfamily, HILS1 may also regulate gene transcription, DNA repair, and/or other chromosome processes during mammalian spermiogenesis.
Keywords: spermatogenesis, transition nuclear proteins, in silico subtraction, genome database mining
Histones are a family of basic proteins that organize eukaryotic DNA into a compact chromatin. There are five major classes of histones, the core histones H2A, H2B, H3, and H4, and the linker histones, H1. Two of each of the four core histones constitute an octamer unit of the nucleosome particle. H1 histones bind to DNA in the nucleosome and to the linker DNA between nucleosomes, thereby facilitating the compaction of nucleosomes into a 30-nm chromatin fiber and higher-order chromatin structures (1). These interactions between histones and DNA modulate gene activity, and both core and H1 histones have profound effects on transcription (2, 3). Among the five histone classes, H1 histones exhibit the most diversity. In mice and humans, there are eight previously described H1 subtypes, including the five somatic subtypes H1a-H1e, the replacement subtype H1o, the testis-specific linker-histone H1t (4-6), and the oocyte-specific H1 linker histone, H1foo (7). The genomic organizations of these histone H1 genes are conserved (8, 9).
Spermatogenesis is the process of development of male germ cells from spermatogonia to highly differentiated spermatozoa. During spermatogenesis, chromatin is restructured, and dramatic changes in histone gene expression are observed (10). In spermatogonia and preleptotene spermatocytes (cells undergoing DNA replication), the H1a and H1b linker histones are expressed (11). During meiotic prophase, striking alterations in chromatin structure occur, and the somatic linker histones are replaced by testis-specific subtype H1t (12-14). H1t is first detected in mid-pachytene spermatocytes, where it rapidly integrates into the chromatin, replaces most of the other somatic H1 subtypes, and persists until the elongating spermatid stage (15). Surprisingly, knockout of H1t does not affect chromatin remodeling, indicating that other known or unknown linker histones may compensate for its absence (16-18).
The development of spermatids into spermatozoa, termed spermiogenesis, is characterized by striking morphological and molecular transformations. In elongating and elongated spermatids, major restructuring of the somatic chromatin occurs; this process involves the replacement of somatic histones and H1t by transition nuclear proteins (TNP1 and TNP2), which are subsequently replaced by protamines (PRM1 and PRM2). During this process, transcription ceases, nucleosomal-type chromatin is transformed into a smooth fiber, and chromatin condensation initiates (10). Decreasing levels of either PRM1 or PRM2 disrupts nuclear condensation, processing of PRM2, and normal sperm function (19). In contrast, lack of either TNP1 or TNP2 has only subtle effects on spermiogenesis and fertility (20, 21). Thus, H1t and transition proteins are not acting alone during chromatin restructuring and before PRMs are required.
We have identified a number of germ cell-specific genes by using in silico subtraction (22-26). Here, we report the identification and characterization of a gene encoding a spermatid-specific linker histone-like protein, histone H1-like protein in spermatids 1 (termed HILS1). Based on the spatiotemporal expression pattern of HILS1, we suggest that this linker histone-like protein participates in chromatin remodeling and/or transcriptional regulation during spermiogenesis.
Methods and Materials
In Silico Subtraction and Genomic Database Mining. ESTs (49,064) from three libraries [Lib.6786 (round spermatids), Lib.2549 (adult testis), and Lib.2511 (adult testis)] were downloaded from the mouse Unigene database (www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=Mm) at the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov). In silico subtraction and genomic database mining were performed as described (24).
Protein Alignment, Phylogenetic Analysis, and Tertiary Structure Prediction. Protein alignment, sequence similarity, and the phylogenetic tree were determined by using the megalign program of the DNASTAR software package (DNASTAR, Madison, WI).
RNA and Protein Analyses. RNA and protein analyses were performed as described (24).
Preparation and Analysis of NPs. Nuclei and chromatin preparation, and extraction of basic NPs (BNPs) from total testes or sonication-resistant spermatids (SRS) were performed as described (20, 21, 27).
Linker Histone Binding Assay. Core histone preparation, mononucleosome and polynucleosome reconstitution, and binding assays were performed as reported (28), except that all DNA fragments were labeled by using digoxigenin-11-ddUTP (Roche Diagnostics, Indianapolis) as described (29). A comprehensive description of the methods and materials is available in Supporting Methods and Materials, which is published as supporting information on the PNAS web site, www.pnas.org.
Results and Discussion
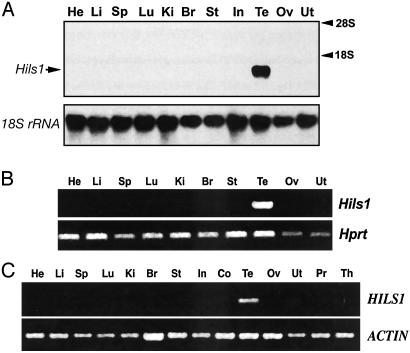
Identification of Mammalian Testis-Specific Transcripts. Using in silico subtraction, we identified seven similar mouse ESTs from testis or spermatid libraries. These ESTs had been grouped into Unigene cluster Mm.30482, which contains 100 ESTs derived solely from testis libraries. Three of the 100 ESTs (NM_108792, AB045724, and AK005718) contain an ORF predicted to encode a mouse protein of 170 amino acids, which had been deposited into GenBank and named as HILS1. To confirm the accuracy of these sequences and verify the 5′ end of the cDNA, we performed RT-PCR and 5′ RACE by using mouse testis total RNA; the complete full-length cDNA sequence is shown in Fig. 6, which is published as supporting information on the PNAS web site. Northern blot, RT-PCR, and RACE analyses are all consistent with a single ≈1.0-kb Hils1 transcript expressed exclusively in testis (Figs. 1 A and B and 6).
Fig. 1.
Analysis of Hils1 mRNA in mouse and human tissues. (A) Northern blot analysis of Hils1 mRNA expression in mouse tissues. Total RNA (15 g) from heart (He), liver (Li), spleen (Sp), lung (Lu), kidney (Ki), brain (Br), stomach (St), intestine (In), testis (Te), ovary (Ov), and uterus (Ut) was electrophoresed, transferred, and probed with Hils1 or 18S rRNA cDNAs. (B) RT-PCR analysis of Hils1 and Hprt mRNA in multiple mouse tissues. (C) RT-PCR analysis of HILS1 and ACTIN mRNA in human tissues. The abbreviations are similar to A and include colon (Co), prostate (Pr), and thymus (Th).
To identify the human HILS1 ortholog, we used the mouse Hils1 cDNA sequence to search the human genome database at the Sanger Center (Hinxton, Cambridge, U.K.), and at the Washington University School of Medicine (St. Louis). A 384-bp genomic fragment from human chromosome 17 was shown to share 66% nucleotide identity. RT-PCR analysis confirmed that HILS1 was testis-specific in humans (Fig. 1C). The full-length human cDNA was then isolated by using 5′ RACE and 3′ RACE; it is predicted to encode a 231-aa protein (see Fig. 7, which is published as supporting information on the PNAS web site). Based on our in silico subtraction, Northern blot, and RT-PCR analyses, as well as database searches, Hils1 is, to our knowledge, a novel mammalian testis-specific gene.
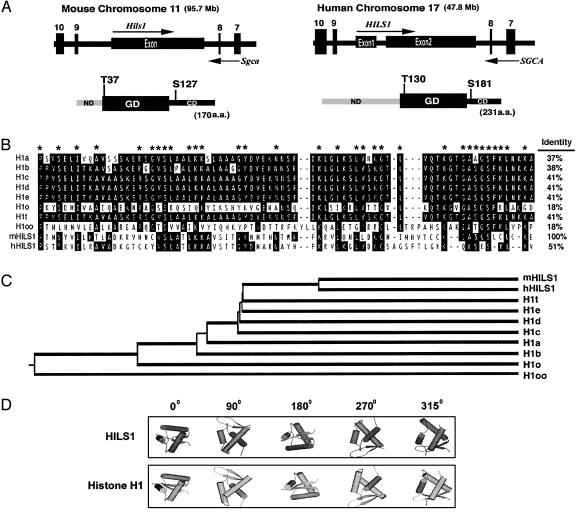
Taking advantage of the human genome and mouse genome databases at NCBI and Sanger Center, we identified the genomic location and structures of mouse Hils1 and its human ortholog. Mouse Hils1 is an intronless gene located on distal chromosome 11 at 95.7 Mb (Fig. 2A), whereas human HILS1 consists of two exons and a small 106-bp intron, mapping to a syntenic position on chromosome 17q21.33 (at 47.8 Mb; Fig. 2 A). Both mouse and human HILS1 genes are located within intron 8 of the α-sarcoglycan gene (SGCA, also called ADHALIN; Fig. 2 A). Mutations in SGCA cause autosomal recessive Duchenne-like muscular dystrophy type 2 (30-32).
Fig. 2.
HILS1 genomic and protein structures. (A) Genomic organization and domain structures of the mouse and human HILS1 proteins. The mouse Hils1 is located in intron 8 of the mouse α-sarcoglycan gene (SGCA), and a similar genomic arrangement is conserved in humans. Arrows represent the transcriptional orientations of the genes. Each protein contains a globular domain (GD), which is flanked by an N-terminal domain (ND) and a C-terminal domain (CD). The conserved putative phosphorylation sites are marked. (B) Alignment analysis of the globular domains of histone H1 family members. Identical residues are highlighted, and residues conserved in at least 8 of 10 members are marked with asterisks. Amino acid identity between the globular domains of the mouse HILS1 and other family members is shown. (C) Phylogenetic tree of linker histone H1 family proteins. (D) Predicted 3D structure of mouse HILS1 versus histone H1a. Cylinders represent α-helical bundles, and arrows represent β-sheets. The angles of rotation are shown.
HILS1 Structurally Resembles Linker Histone H1s. Histones of the H1 family interact at high affinity with linker DNA, and hence are known as linker histones (33). The histone H1 family in meta-zoans and other multicellular eukaryotes is a heterogeneous family of developmentally regulated histones that includes highly tissue-specific proteins, such as histone H5 from the nucleated erythrocytes of birds (33). In contrast to core histones, linker histones are less evolutionarily conserved (33). Whereas the central globular domains of these proteins are relatively well conserved through evolution in animals, plants, and fungi, the N- and C-terminal domains are extremely heterogeneous, both in length and amino acid composition (33). The C termini of H1 histones are enriched in serine, proline, and lysine repeats, which appear to be essential for the formation of higher-order structures (33).
Protein alignment analyses show that mouse and human HILS1 have 50% amino acid identity, whereas their globular domains share 51% amino acid identity (Fig. 2B). The N terminus of human HILS1 is longer than that of mouse HILS1, but the two globular domains and C termini are similar in size. Multiple alignment analyses demonstrated that the globular domains of the human and mouse HILS1 share ≈40% amino acid identity with globular domains of histone H1a-H1e and H1t, but only 18% with those of H1o and H1foo (Fig. 2B). Phylogenetic analysis suggests that HILS1 is evolutionarily more similar to histone H1t (Fig. 2C). Using the motif scan program (http://scansite.mit.edu), we identified multiple putative phosphorylation sites, among which two sites are conserved in human and mouse HILS1 (mouse T37 and S127, and human T130 and S180; Fig. 2 A). Twenty percent of HILS1 amino acids are strongly basic (K and R), and its isoelectric point (IP) is ≈10.0, whereas ≈28% of amino acids are strongly basic in other histone H1s, with an average IP of ≈10.9. Based on these data, HILS1 is, to our knowledge, a novel linker histone H1-like basic protein with testis specificity.
The crystallographic structure of the globular domain of H1 linker histones has been determined (34). This domain consists of a winged-helix motif containing 3-helical bundles, and interacts with the nucleosome at a region close to the pseudodyad axis of symmetry (34). A Pfam search (www.sanger.ac.uk/Software/Pfam/index.shtml) revealed that both the mouse and human HILS1 proteins contain a conserved globular domain. To predict the 3D structure of HILS1, three homologs [1GHC (globular domain of linker histone H1), 1BM9 (DNA-binding domain of replication terminator protein), and 1HST (globular domain of linker histone H5)], which have winged-helix DNA-binding domains with a DNA/RNA-binding 3-helical bundle (34), were identified by sequence alignment from the protein 3D structure database. Homolog 1GHC was picked as the start point for modeling because it has a longer alignment, and its secondary structure fits better with the predicted secondary structure of the target sequence. Based on the 3D structure of the globular domain of histone H1, the globular domain of HILS1 was predicted to display a winged-helix structure similar to histone H1 (Fig. 2D).
Hils1 Gene Expression Is Confined to Elongating and Condensing Spermatids. As revealed in our in situ hybridization analysis, Hils1 mRNA displays a stage-specific expression pattern in the testis (see Fig. 8, which is published as supporting information on the PNAS web site). Higher levels of hybridization signals were observed in stages VI-IX, and lower levels of signals were found at stages X-XII and stages I-III. Hils1 mRNA initiated in step 4 spermatids, peaked in step 6-8 spermatids, decreased in step 9 spermatids, and was undetectable after step 15 spermatids.
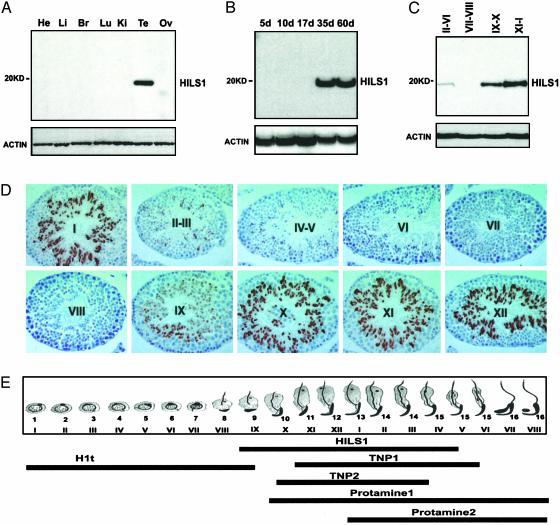
Western blot analysis demonstrated that the rabbit polyclonal anti-HILS1 antisera that we produced only recognized a 19-kDa protein band in the testis, indicating the high specificity of the antibody (Fig. 3A). Consistent with its spermatid-specific expression, HILS1 was not detected in testes before postnatal day 15 (Fig. 3B), when no spermatids are present in the testis. HILS1 protein levels are high at postnatal days 35 and 60, when elongating and condensing spermatids are abundant in the testis (Fig. 3B). By transillumination-assisted microdissection technique, we isolated seminiferous tubule segments from stages II-VI, VII-VIII, IX-X, and XI-I, and then performed Western blot analyses (Fig. 3C). HISL1 is highest at stages XI-I, and is undetectable at stages VII-VIII, indicating that HILS1 protein expression is highly stage specific. By immunohistochemistry, and consistent with our Western blot analyses, HILS1 is exclusively detected in the nuclei of spermatids (Fig. 3D), and is first expressed in step 9 spermatids (stage IX tubules). HILS1 persists at high levels in step 10-13 spermatids, decreases abruptly in step 14 spermatids (stage II-III tubules), and is barely detectable in step 15 spermatids. Thus, the stage-specific expression patterns of Hils1 mRNA and protein indicate that the gene is transcriptionally and translationally regulated.
Fig. 3.
Mouse HILS1 protein expression and localization. (A) Western blot analysis of HILS1 in mouse tissues. (B) HILS1 expression in developing testes. Proteins isolated from testes at different postnatal days (d) were analyzed by Western blot. (C) Stage-specific expression of HILS1 protein in the adult mouse testis. Seminiferous tubule segments as shown were dissected, and protein was prepared. ACTIN was used as a loading control (A-C). (D) Immunohistochemical detection of HILS1. Stages of adult mouse seminiferous tubules are marked with Roman numerals, and brown diaminobenzidine staining represents positive signals. HILS1 immunoreactivity is confined to step 9-15 spermatids. (E) Schematic summary of the expression profiles of HILS1, H1t, TNP1, TNP2, PRM1, and PRM 2 during spermiogenesis based on Figs. 3 and 9, and previously reported data on H1t expression (11, 12). Arabic numbers represent the steps of spermatids, and Roman numerals indicate the tubule stages. Bars represent the expression windows of these NPs in spermatids.
The onset and the peak of Hils1 mRNA expression are earlier (step 4) than HILS1 protein (step 9), a finding common to testis-expressed genes such as Prm1, Prm2 (35, 36), and Tnp1 (12), which have products needed at later stages of spermiogenesis. The mRNAs for these genes are produced at the round spermatid stages preceding the cessation of transcription, when spermatids start elongation and nuclear condensation (37, 38). To compare the protein expression profiles of HILS1, transition proteins, and PRMs, we analyzed their stage-specific expression in the mouse testis (see Fig. 9, which is published as supporting information on the PNAS web site). Immunohistochemical analyses demonstrated that TNP1 is expressed in step 11-15 spermatids, and TNP2 is expressed in step 10-15 spermatids (Fig. 9). PRM1 expression starts in step 10 spermatids, increases in step 11-12 spermatids, and peak in step 14-15 spermatids (Fig. 9). PRM2 expression initiates in step 13 spermatids and persists at high levels in step 15-16 spermatids (Fig. 9). Thus, the onset of HILS1 protein is earlier than in the TNPs, and the stage-specific expression pattern of HILS1 is different from TNPs and PRMs (Fig. 3 D and E and Fig. 9), suggesting that HILS1 may have a distinct role from the TNPs and PRMs.
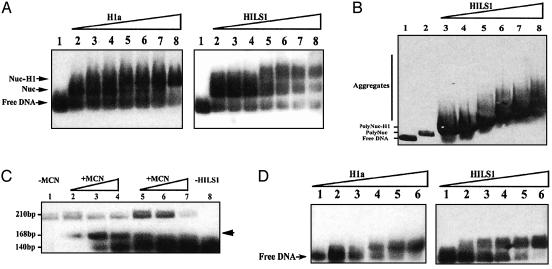
HILS1 Binds Nucleosome and DNA in Vitro as a Linker Histone. Members of the H1 histone family have a number of biochemical and physical features that distinguish them from other chromatin proteins in vitro (39). These characteristics include the following: (i) they bind to isolated nucleosomes and linker DNA; (ii) they produce a chromatosome stop during enzymatic digestion of native or reconstituted chromatin substrates; and (iii) they facilitate higher-order chromatin compaction under appropriate salt conditions. Our data reveal that HILS1 can bind reconstituted mononucleosomes with lower affinity, compared with linker histone H1a (Fig. 4A). HILS1 also aggregates polynucleosomes, suggesting HILS1 may have chromatin-compacting capacity (Fig. 4B). Micrococcal nuclease digestion of HILS1-bound reconstituted nucleosomes produced chromatosome stops (Fig. 4C), indicating that HILS1, as other members of linker histone H1 family, protects linker DNA from nuclease digestion. Moreover, at similar concentrations, HILS1 displays much higher binding capacity to naked DNA than histone H1a (Fig. 4D). Therefore, HILS1 is a DNA-binding protein and appears to act as a linker histone in vitro.
Fig. 4.
Biochemical characteristics of HILS1 in vitro.(A) Analysis of binding activity of histone H1a and HILS1 to mononucleosomes by gel shift assay. Nucleosomes reconstituted by using core histone particles prepared from mouse liver and digoxigenin-11-ddUTP end-labeled 210-bp mouse mammary tumor virus (MMTV) LTR DNA were incubated with 0, 0.375, 0.75, 1.5, 3, 6, and 12 ng of H1a or HILS1, corresponding to 0, 1, 2, 4, 8, 16, and 32 nM (lanes 2-8, respectively). Lane 1, free 210-bp DNA. Positions of free DNA, nucleosome (Nuc), and histone H1-nucleosome complexes (Nuc-H1) are indicated. (B) Chromatin aggregation properties of HILS1. Polynucleosomes reconstituted with digoxigenin-11-ddUTP end-labeled 1.3-kb MMTV LTR DNA were incubated with 0, 12, 24, 36, 48, 72, and 96 ng of HILS1, corresponding to 0, 45, 90, 135, 180, 270, and 360 nM (lanes 2-8, respectively). Lane 1, free 1.3-kb DNA. (C) Chromatosome stop assay. Nucleosomes reconstituted with digoxigenin-11-dUTP-labeled MMTV LTR promoter DNA were incubated with HILS1 [HILS1:DNA ratio (wt/wt) of 0.4 (lanes 2-4), and 0.1 (lanes 5-7)] and digested with 0.15, 0.3, or 0.6 units of micrococcal nuclease (MCN) for 6 min at 37°C. DNA fragments corresponding to nucleosome core particle (146 bp) and chromatosome (168 bp and arrow) are indicated. (D) Gel shift assay of binding activity of HILS1 and H1a to naked DNA. Digoxigenin-11-ddUTP end-labeled 210-bp MMTV LTR DNA was incubated with 0.375, 0.75, 1.5, 3, 6, and 12 ng of H1a and HILS1, corresponding to 1, 2, 4, 8, 16, and 32 nM (lanes 1-6, respectively).
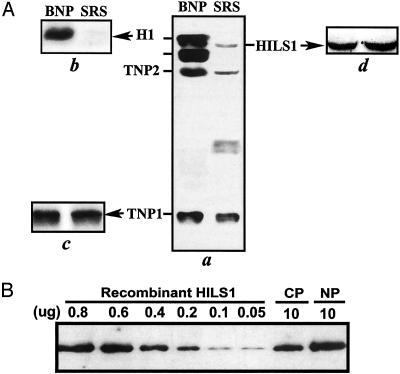
HILS1 Is a Chromatin-Associated Protein (CP). Coomassie blue staining of an acid-urea polyacrylamide gel separation of BNPs from total testis lysates and SRS (step 12-16 spermatids) revealed a faint band in the linker histone H1 region (Fig. 5Aa). Western blot analyses confirmed that this band is actually HILS1 (Fig. 5Ad). We can easily detect HILS1 in chromatin preparations from SRS, indicating that HILS1 is chromatin-associated (Fig. 5B). By using recombinant HILS1 as a standard, we estimated that HILS1 accounts for ≈10% of the total CPs, and ≈5% of the total nucleoproteins in SRS (Fig. 5B). These data are consistent with previous studies (20) showing that the basic nucleoproteins of SRS contains ≈5% histones, which was until now believed to be contamination from other cells.
Fig. 5.
Detection of HILS1 in total nuclear and chromatin protein preparations. (A) Coomassie blue-stained acid-urea 15% polyacrylamide gel (a) and Western blot analyses of HILS1 (d), H1 (b), and TNP1 (c). Total BNPs from unfractionated mouse testis nuclei were used as a marker (a). (B) Quantitative analysis of HILS1 in total CPs and total NPs. Ten grams of total NP or CP was fractionated through a 4-12% polyacrylamide gel. Serial dilution of recombinant mouse HILS1 protein was prepared and loaded in each lane as shown. After chemiluminescent detection, the film was scanned and quantified.
Subcellular Localization of HILS1, TNPs, and PRM1. Because the expression sites of HILS1, TNPs, and PRM1 overlap, we performed immunofluorescent microscopy to examine their subcellular localization in spermatid nuclei (see Fig. 10, which is published as supporting information on the PNAS web site). At stage XII, both HILS1 and TNP1 localize to step 12 spermatid nuclei, and the immunofluorescent patterns of these two proteins are similar, but not identical. HILS1 displays a pattern of patchy spots in the chromatin (Fig. 10A); and in the same plane, the pattern of TNP1 signals were not identical (Fig. 10B), suggesting that they are not localized to exactly the same portion of the chromatin. Interestingly, TNP2 and HILS1 display identical immunofluorescent patterns (Fig. 10 D-F), and PRM1 and HILS1 also show identical staining patterns in step 12 spermatid nuclei (Fig. 10 G-I).
The mechanisms of nuclear condensation of the developing male gamete have attracted considerable attention over the last 40 years, and most of these efforts have been directed toward understanding chromatin structure. During the initial steps of spermiogenesis, the haploid spermatids display the typical nucleosomal chromatin pattern, and are engaged in considerable nonribosomal RNA transcription. As spermiogenesis advances, the beaded chromatin configuration is replaced by a smooth chromatin fiber that is transcriptionally silenced. Packing of DNA in fixed, dehydrated mammalian spermatozoa approaches the physical limits of molecular compaction, making mammalian sperm chromatin the most condensed eukaryotic DNA. The fundamental packaging unit of sperm chromatin is a toroid with an ≈900-Å outer diameter, 200-Å thickness, and a 150-Å diameter hole. Each toroid contains 60 kb of DNA and is linked to other toroids by uncoiled DNA stretches. Whereas the roles of PRMs in sperm chromatin condensation and nuclear shaping are evident, the roles of H1t (16-18), TNP1 (20), and TNP2 (21) in the intermediate steps before the appearance of PRMs remain uncertain. Because HILS1 has diverged significantly from other linker histone family members and is expressed in elongating and elongated spermatids where core histones are absent, HILS1 is unlikely to function as a typical linker histone. HILS1 may alternatively interact with TNP1 and TNP2 in the transition to the transcriptional inert toroid structure. In bacteria through humans, the winged helix is a highly conserved DNA-binding peptide motif. In mammals, an array of DNA/CPs belonging to the winged-helix/Forkhead box (Fox) proteins are involved not only in the control of gene transcription (e.g., FoxA-FoxM, HNF-3, ILF, RFX5, RAP30, etc.) but also in DNA replication (e.g., 1BM9, FoxM1b), DNA repair (HFH-11B, CHES1), and other chromosome processes (40). Likewise, HILS1 could belong to this large winged-helix superfamily, and may function in one or more of these processes. Future biochemical and genetic studies including generation of Hils1 knockout mice will help to define the physiological roles of this testis-specific histone-like protein.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Kistler for the TNP1 and TNP2 rabbit polyclonal antibodies; Dr. Rod Balhorn for PRM1 and PRM2 antibodies; Dr. Marvin Meistrich for helpful discussions; Dr. Michael Mancini for assistance with microscopic imaging; and Drs. Heribert Talasz and Jiemin Wong for help in nucleosome reconstitution and nucleoprotein binding assays. This work was supported in part by National Institutes of Health Specialized Cooperative Centers Program in Reproductive Research Grant HD07495. W.Y. is supported by a postdoctoral fellowship from the Ernst Schering Research Foundation (Berlin). K.H.B. is a student in the Medical Scientist Training Program at Baylor College of Medicine and is supported in part by National Institutes of Health Training Grant GM07330.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HILS1, a spermatid-specific linker histone H1-like protein; NP, nuclear protein; TNP, transition NP; BNP, basic NP; PRM, protamine; SRS, sonication-resistant spermatids.
Data deposition: The Hils1 cDNA sequences reported in this paper have been deposited in the GenBank database [accession nos. AY286318 (mouse) and AY286319 (human)].
References
- 1.Wolffe, A. P. (1997) Int. J. Biochem. Cell Biol. 29, 1463-1466. [DOI] [PubMed] [Google Scholar]
- 2.Hartzog, G. A. & Winston, F. (1997) Curr. Opin. Genet. Dev. 7, 192-198. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe, A. P. (2001) Essays Biochem. 37, 45-57. [DOI] [PubMed] [Google Scholar]
- 4.Lennox, R. W. & Cohen, L. H. (1983) J. Biol. Chem. 258, 262-268. [PubMed] [Google Scholar]
- 5.Lennox, R. W. & Cohen, L. H. (1984) Dev. Biol. 103, 80-84. [DOI] [PubMed] [Google Scholar]
- 6.Lennox, R. W. & Cohen, L. H. (1988) Biochem. Cell Biol. 66, 636-649. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka, M., Hennebold, J. D., Macfarlane, J. & Adashi, E. Y. (2001) Development (Cambridge, U.K.) 128, 655-664. [DOI] [PubMed] [Google Scholar]
- 8.Wang, Z. F., Tisovec, R., Debry, R. W., Frey, M. R., Matera, A. G. & Marzluff, W. F. (1996) Genome Res. 6, 702-714. [DOI] [PubMed] [Google Scholar]
- 9.Wang, Z. F., Sirotkin, A. M., Buchold, G. M., Skoultchi, A. I. & Marzluff, W. F. (1997) J. Mol. Biol. 271, 124-138. [DOI] [PubMed] [Google Scholar]
- 10.Meistrich, M. L. (1993) Nuclear Morphogenesis During Spermiogenesis (Academic, San Diego).
- 11.Doenecke, D., Albig, W., Bode, C., Drabent, B., Franke, K., Gavenis, K. & Witt, O. (1997) Histochem. Cell Biol. 107, 1-10. [DOI] [PubMed] [Google Scholar]
- 12.Doenecke, D., Drabent, B., Bode, C., Bramlage, B., Franke, K., Gavenis, K., Kosciessa, U. & Witt, O. (1997) Adv. Exp. Med. Biol. 424, 37-48. [DOI] [PubMed] [Google Scholar]
- 13.Cole, K. D., Kandala, J. C. & Kistler, W. S. (1986) J. Biol. Chem. 261, 7178-7183. [PubMed] [Google Scholar]
- 14.Kistler, W. S., Heidaran, M. A., Cole, K. D., Kandala, J. C. & Showman, R. M. (1987) Ann. N.Y. Acad. Sci. 513, 102-111. [DOI] [PubMed] [Google Scholar]
- 15.Meistrich, M. L., Bucci, L. R., Trostle-Weige, P. K. & Brock, W. A. (1985) Dev. Biol. 112, 230-240. [DOI] [PubMed] [Google Scholar]
- 16.Fantz, D. A., Hatfield, W. R., Horvath, G., Kistler, M. K. & Kistler, W. S. (2001) Biol. Reprod. 64, 425-431. [DOI] [PubMed] [Google Scholar]
- 17.Drabent, B., Saftig, P., Bode, C. & Doenecke, D. (2000) Histochem. Cell Biol. 113, 433-442. [DOI] [PubMed] [Google Scholar]
- 18.Lin, Q., Sirotkin, A. & Skoultchi, A. I. (2000) Mol. Cell. Biol. 20, 2122-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho, C., Willis, W. D., Goulding, E. H., Jung-Ha, H., Choi, Y. C., Hecht, N. B. & Eddy, E. M. (2001) Nat. Genet. 28, 82-86. [DOI] [PubMed] [Google Scholar]
- 20.Yu, Y. E., Zhang, Y., Unni, E., Shirley, C. R., Deng, J. M., Russell, L. D., Weil, M. M., Behringer, R. R. & Meistrich, M. L. (2000) Proc. Natl. Acad. Sci. USA 97, 4683-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao, M., Shirley, C. R., Yu, Y. E., Mohapatra, B., Zhang, Y., Unni, E., Deng, J. M., Arango, N. A., Terry, N. H., Weil, M. M., et al. (2001) Mol. Cell. Biol. 21, 7243-7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajkovic, A., Yan, C., Yan, W., Klysik, M. & Matzuk, M. M. (2002) Genomics 79, 711-717. [DOI] [PubMed] [Google Scholar]
- 23.Rajkovic, A., Lee, J. H., Yan, C. & Matzuk, M. M. (2002) Mech. Dev. 112, 173-177. [DOI] [PubMed] [Google Scholar]
- 24.Yan, W., Rajkovic, A., Viveiros, M. M., Burns, K. H., Eppig, J. J. & Matzuk, M. M. (2002) Mol. Endocrinol. 16, 1168-1184. [DOI] [PubMed] [Google Scholar]
- 25.Yan, W., Burns, K. H., Ma, L. & Matzuk, M. M. (2002) Mech. Dev. 118, 233-239. [DOI] [PubMed] [Google Scholar]
- 26.Suzumori, N., Yan, C., Matzuk, M. M. & Rajkovic, A. (2002) Mech. Dev. 111, 137-141. [DOI] [PubMed] [Google Scholar]
- 27.Sirotkin, A. M., Edelmann, W., Cheng, G., Klein-Szanto, A., Kucherlapati, R. & Skoultchi, A. I. (1995) Proc. Natl. Acad. Sci. USA 92, 6434-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talasz, H., Sapojnikova, N., Helliger, W., Lindner, H. & Puschendorf, B. (1998) J. Biol. Chem. 273, 32236-32243. [DOI] [PubMed] [Google Scholar]
- 29.Zheng, H., Yan, W., Toppari, J. & Harkonen, P. (2000) BioTechniques 28, 832-834. [DOI] [PubMed] [Google Scholar]
- 30.Roberds, S. L., Leturcq, F., Allamand, V., Piccolo, F., Jeanpierre, M., Anderson, R. D., Lim, L. E., Lee, J. C., Tome, F. M., Romero, N. B., et al. (1994) Cell 78, 625-633. [DOI] [PubMed] [Google Scholar]
- 31.McNally, E. M., Yoshida, M., Mizuno, Y., Ozawa, E. & Kunkel, L. M. (1994) Proc. Natl. Acad. Sci. USA 91, 9690-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piccolo, F., Roberds, S. L., Jeanpierre, M., Leturcq, F., Azibi, K., Beldjord, C., Carrie, A., Recan, D., Chaouch, M., Reghis, A., et al. (1995) Nat. Genet. 10, 243-245. [DOI] [PubMed] [Google Scholar]
- 33.Kasinsky, H. E., Lewis, J. D., Dacks, J. B. & Ausio, J. (2001) FASEB J. 15, 34-42. [DOI] [PubMed] [Google Scholar]
- 34.Ramakrishnan, V., Finch, J. T., Graziano, V., Lee, P. L. & Sweet, R. M. (1993) Nature 362, 219-223. [DOI] [PubMed] [Google Scholar]
- 35.Mali, P., Kaipia, A., Kangasniemi, M., Toppari, J., Sandberg, M., Hecht, N. B. & Parvinen, M. (1989) Reprod. Fertil. Dev. 1, 369-382. [DOI] [PubMed] [Google Scholar]
- 36.Lee, K., Haugen, H. S., Clegg, C. H. & Braun, R. E. (1995) Proc. Natl. Acad. Sci. USA 92, 12451-12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steger, K. (1999) Anat. Embryol. 199, 471-487. [DOI] [PubMed] [Google Scholar]
- 38.Steger, K. (2001) Anat. Embryol. 203, 323-334. [DOI] [PubMed] [Google Scholar]
- 39.Wolffe, A. P., Khochbin, S. & Dimitrov, S. (1997) BioEssays 19, 249-255. [DOI] [PubMed] [Google Scholar]
- 40.Gajiwala, K. S. & Burley, S. K. (2000) Curr. Opin. Struct. Biol. 10, 110-116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.