Abstract
The F-box represents a protein motif originally identified as a conserved amino-terminal domain within the Neurospora crassa negative regulator sulfur controller-2. Recently, F-boxes have been found within a number of cell cycle regulatory proteins, where they mediate ubiquitin-driven proteolytic events required for major cell cycle transitions. F-box function, however, is not restricted solely to cell cycle pathways. Here we present evidence expanding F-box function to encompass gene regulatory processes independent of the cell cycle through in vivo analysis of an F-box acting within the N. crassa sulfur regulatory network. The Neurospora sulfur circuit features a set of regulatory genes acting to modulate gene expression based on environmental sulfur conditions. These sulfur regulatory genes include cys-3+, which encodes a basic region-leucine zipper transcriptional activator, as well as the negative regulatory gene scon-2+. Through site-directed mutagenesis of the SCON2 F-box, we have generated a sulfur auxotrophic phenotype previously unobserved in any scon-2 mutant. Using Northern analysis, we have traced this auxotrophy to a complete shutdown of cys-3+ gene expression. We have further analyzed F-box function by constructing a series of chimeric SCON2 proteins containing swapped F-box domains from the yeast transcriptional inhibitor Met30p and the Candida albicans cell cycle regulator Cdc4p. The ability of these chimeric proteins to restore partial wild-type sulfur regulation in vivo emphasizes the universal nature of this motif and confirms the functional importance of the F-box within noncell cycle regulatory pathways.
Keywords: sulfur controller-2, negative regulator, proteolysis, Cdc4p, eukaryotic cell cycle
Protein degradation stands as a key regulatory mechanism of the cell, used to maintain processes as fundamental as cell division itself. In fact, cells liberally use regulated proteolysis within the cell cycle; major cell cycle transitions require the precise destruction of a number of proteins, including cyclin-dependent kinase inhibitors, mitotic cyclins, and anaphase inhibitors (1). Recent studies have identified several cell cycle regulatory components containing a novel motif involved in ubiquitin-mediated proteolysis. This motif has been named the F-box, because of its characterization in the human cyclin-dependent kinase activator cyclin F (2). The F-box has been found to link cyclin F as well as the yeast cell cycle regulators Cdc4p and Skp2p to Skp1p, a potential component of an E3 ubiquitin-protein ligase complex (2). The F-box, however, does not appear to be confined to a limited proteolytic role within the cell cycle. We originally had discovered this same motif, initially termed the “N-terminal domain,” in the sulfur controller protein SCON2, a negative regulator operating within the sulfur regulatory circuit of the eukaryotic filamentous fungus Neurospora crassa (3).
In N. crassa, the uptake and assimilation of sulfur is accomplished through a complex multigene network encompassing a set of structural genes encoding a variety of sulfur-related enzymes such as arylsulfatase, sulfate permease I, and sulfate permease II (4, 5). These unlinked structural genes are subject to tight regulatory control, ensuring their coordinated expression only under conditions of sulfur limitation (derepressing conditions) (6). This transcriptional control is achieved through the concerted actions of a distinct set of trans-acting regulatory genes. The positive regulatory gene cys-3+ encodes a basic region-leucine zipper protein that functions as a DNA-binding transcriptional activator (7, 8). CYS3 protein activates expression of the aforementioned structural genes as well as its own expression only under low sulfur conditions (8). cys-3+ expression is governed by the negative regulatory sulfur controller genes scon-1+ and scon-2+ (9, 10). scon-1 and scon-2 mutants constitutively express the sulfur structural and cys-3+ genes (9, 10). We recently have cloned the scon-2+ gene. Surprisingly, it too is expressed only under low sulfur conditions, as shown by increases in mRNA content (10). scon-2+ expression is, in fact, stimulated by CYS3, indicating a novel control loop involving CYS3 activation of scon-2+ on sulfur limitation and SCON2 repression of cys-3+ expression under high sulfur conditions (3).
By conceptual translation, the scon-2+ gene encodes a polypeptide of 650 amino acids with a molecular mass of 72.2 kDa. Computational analysis of the SCON2 protein reveals the presence of several striking protein sequence motifs. The carboxy-terminal half of SCON2 is dominated by six β-transducin (or WD-40) repeats (11). These putative protein-interaction domains, which originally were found in the β-subunit of heterotrimeric G-proteins (12), are approximately 40 amino acids in length. Each repeat displays an Asp-His-Ser/Thr triad of residues responsible for hydrogen-bond stabilization of WD-protein structure (13). SCON2 additionally contains an F-box motif located roughly 180 residues N-terminal to the WD-repeats. We initially had identified this motif as an N-terminal domain conserved among a subset of the WD-protein family (3). The F-box subsequently has been identified outside of β-transducin homologs (2), suggesting that F-boxes operate independently of WD-repeats to fulfill single or multiple regulatory functions. To specifically address the question of F-box function within SCON2, we have undertaken a mutational analysis of this domain in vivo. Our study highlights a completely novel phenotype resulting from site-directed mutation of conserved SCON2 F-box residues. Additionally, our findings establish a significance for the F-box motif, with its putative role in targeting substrate proteins for proteolysis, beyond the bounds of cell cycle processes.
MATERIALS AND METHODS
Strains, Plasmids, and Culture Conditions.
740R23–1a was used as the wild-type strain for these studies. N. crassa Δcys-3(18–4) was constructed by gene replacement as detailed elsewhere (14). The scon-2 deletion strain Δscon-2 was constructed by gene replacement using a DNA segment containing scon-2+ noncoding sequence flanking a qa-2+ gene insert (replacing the scon-2+ coding sequence). An aro-9 qa-2 strain was transformed with the qa-2+/scon-2 fragment. The resulting qa-2+ transformants were made homokaryotic; transformants subsequently were screened for any defects in sulfur gene regulation. Only strains showing constitutive expression of the sulfur enzymes were observed. Proper chromosomal integration of the fragment was confirmed by Southern blot analysis. The N. crassa scon-2(PSD272) strain was described previously (10). The scon-2 his-3 strain was isolated from a cross of his-3(Y155M261) by scon-2(PSD272). The pDE1 vector was obtained from the Fungal Genetics Stock Center (Kansas City, KS). pJP11 carried the am+ gene on a 2.4-kb BamHI fragment in pBR322. N. crassa cultures were grown at 25°C on minimal Vogel medium (15) with supplements as required. Low and high sulfur conditions were achieved with Vogel-minus-sulfur medium supplemented with 0.25 mM methionine and 5.0 mM methionine, respectively.
Computational Methods.
The SCON2, cyclin F, Saccharomyces cerevisiae Cdc4p, Grr1p, and Pop1 F-box sequences were chosen as queries to initiate transitive searching of the National Center for Biotechnology Information nonredundant protein database (June 2, 1997 release date) using the blastp algorithm (16). In total, more than 50 blastp searches were performed as described using the pam240, blosum40, and blosum62 scoring matrices. Only sequences exhibiting high-scoring segment pairs in multiple independent searches were classified as F-box proteins. The local alignment program macaw (17) was used to further define the F-box motif in putative F-box proteins. Optimal sequence alignments were generated by using clustalw (18) (gap opening penalty of 2, gap extension penalty of 0.05, blosum series of protein weight matrices).
In Vitro Mutagenesis and Chimeric Gene Constructs.
Site-directed mutations were generated by the phosphorothioate method of Taylor and Eckstein (19). All site-directed constructs were sequenced by the dideoxy method (20).
Chimeric SCON2 constructs were generated through gene splicing by overlap extension (SOE) (21). SOE offers a restriction enzyme-independent approach to engineering hybrid genes involving the use of overlapping DNA fragments as template in a PCR. Briefly, scon2cdc was constructed as follows. The PCR primers 5′-CTTTGAGTGAGCTGATACCG-3′ and 5′-ACGTTTCAAGGCTTCGTTGACTTCTCT-3′ were used to amplify a 1.78-kb stretch of genomic scon-2+ sequence located immediately upstream of the scon-2+ F-box. Similarly, the PCR primers 5′-GAT ACTTGGGTCCGCATGTGCGAGCAA-3′ and 5′-TTACCCTTAGCATCTCCAGT-3′ were used to amplify a 2.25-kb region of genomic DNA immediately downstream of the scon-2+ F-box. These PCR products subsequently served as template, along with a synthetic DNA fragment encoding CDC4p amino acids 210–255, in a final SOE reaction with primers 5′-ATCGATGAATTCCTCGGATCGATGGCGA-3′ and 5′-GTGATCAAGCTTTGAAACCCGACATGCT-3′. The resulting PCR product was cloned into a 4.7-kb EcoRI–HindIII fragment from pDE1(22). scon2met was constructed in a similar manner by using the following primer pairs: 5′-CTTTGAGTGAGCTGATACCG-3′ and 5′-GATCTTGATGGCTTCGTTGACTTCTCT-3′; 5′-GGGTATGGGTCCGCATGTGCGAGCAAC-3′ and 5′-TTACCCTTAGCATCTCCAGT-3′. The resulting PCR products were combined with a double-stranded synthetic oligonucleotide, encoding Met30p amino acids 179–224, in a final SOE reaction using the same primer pair indicated above. Again, the resulting PCR product was cloned into pDE1 as before.
Transformation and Homokaryon Isolation.
N. crassa was transformed by the Novozyme 234 spheroplasting technique (23). Homokaryons were isolated by filtration of iodoacetate-grown conidial cultures through 5 μm Millex filters (24).
Arylsulfatase and Chromate Resistance Assays.
Arylsulfatase specific activity was assayed by incubation of mycelial extracts with p-nitrophenyl sulfate according to standard methods (6). p-nitrophenyl liberation was monitored at 405 nm. Chromate resistance was determined as follows. Transformation mixes (spheroplasts and DNA) were overlaid on Vogel medium plates supplemented with 5 mM methionine. After a 14-hr incubation, 5 ml of sterile filtered 10 mM potassium chromate in 1.5% agar was added as an overlay. Transformants resistant to chromate were visible after 48 hr.
Northern Analysis.
Poly(A)+ mRNA was isolated by phenol extraction and subsequent oligo(dT)-cellulose chromatography as described previously (10). 32P-labeled probes were prepared by oligolabeling of DNA fragments (25). Northern blots were hybridized and washed as outlined elsewhere (10).
RESULTS
Sequence Analysis of the F-Box Motif.
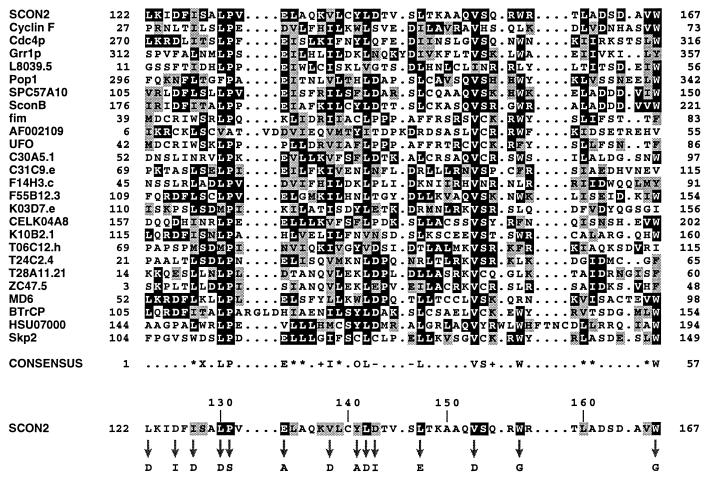
To characterize the F-box motif more rigorously, we adopted a computational approach entailing transitive database analysis in conjunction with additional local alignment methods. Initial database searches were undertaken with a small set of known F-box proteins. This preliminary analysis revealed a number of additional F-boxes, all of which were used as queries to initiate further database searches. This process was repeated to ensure exhaustive coverage of available database sequences. In total, transitive searching uncovered more than 45 potential F-box proteins in eukaryotic organisms ranging from yeast to humans. These proteins mediate a diverse array of cellular functions, implicating the F-box in a number of regulatory systems. A representative sample of F-boxes have been optimally aligned for presentation in Fig. 1. As shown, the F-box motif is typically 45 amino acids in length with relatively few insertions or deletions. The F-box is characterized by a strikingly conserved Leu-Pro dipeptide as well as numerous conserved hydrophobic, branched-chain amino acids. Specifically, 12 positions within the F-box consensus sequence in Fig. 1 are occupied by some combination of Val, Leu, and/or Ile, including nearly invariant Leu and Val residues at positions 9 and 37, respectively. The presence of two strongly conserved Trp residues toward the carboxy-terminus of the F-box is also noteworthy in light of the unusually large number of WD-proteins, which contain this motif.
Figure 1.
Sequence analysis of the F-box motif. A representative sample of F-box sequences were globally aligned as described. The F-box proteins included are as follows (GenBank accession numbers are indicated in parentheses): N. crassa SCON2 (U17251); Homo sapiens Cyclin F (Z36714); S. cerevisiae proteins Cdc4p (X05625), Grr1p (M592407), L8039.5 gene product (U19103); Schizosaccharomyces pombe proteins Pop1(Y08391) and unknown protein (Z94864); Emericella nidulans SconB (U21220); Antirrhinum majus fim (S71192); Arabidopsis thaliana hypothetical protein (AF002109) and unusual floral organs (X89224); Caenorhabditis elegans proteins C30A5.1 (L10990), C31C9.e (Z83219), F14H3.c (Z83105), F55B12.3 (Z79757), K03D7.e (Z81562), hypothetical protein (U64849), K10B2.1 (U28730), T06C12.h (Z81116), T24C2.4 (Z68120), T28A11.21 (U80027), ZC47.5 (Z81141); Mus musculus MD6 (X54352); Xenopus laevis β-TrCP (M98268); H. sapiens unknown protein (U07000) and Skp2 (U33761). The numbers bracketing each line of sequence represent the location of each F-box within its parent protein. The most prevalent residue at each position within the alignment is shown as white on black, and similar residues are represented as black on gray. Alignment positions exhibiting less than 20% overall similarity have been left unshaded. The consensus sequence highlights any position within the alignment exhibiting 14 or more occurrences of a single amino acid or class of amino acid (∗, aliphatic; X, hydroxl- or sulfur-containing amino acids and derivatives; +, basic; –, acidic; O, aromatic). The SCON2 F-box sequence has been isolated at the bottom of this alignment to facilitate comparison with the indicated consensus sequence. For this purpose, residues within SCON2 that are conserved with respect to the consensus sequence are represented as black on gray. SCON2 residues that are identical to indicated amino acids in the consensus are shown as white on black. Arrows designate residues within the SCON2 F-box analyzed by site-directed mutagenesis, with corresponding amino acid substitutions indicated below.
Mutational Analysis of the SCON2 F-Box.
To assess the functional significance of the F-box motif within SCON2, we have undertaken a site-directed mutagenesis study targeting a series of strongly conserved F-box residues. Nonconservative amino acid substitutions were generated at 14 potentially critical positions spanning the SCON2 F-box (Fig. 1). Site-directed mutants subsequently were transformed into a mutant scon-2 background for analysis of F-box function under both low and high sulfur conditions. As shown in Table 1, we have characterized the sulfur regulatory phenotype of each mutant strain through a series of in vivo assays. Specifically, transformants were categorized as sulfur prototrophs based on their ability to grow under permissive conditions on minimal media in the absence of any supplemental sulfur sources. We additionally have analyzed the sulfur regulatory state of F-box mutants through functional assays measuring arylsulfatase as well as sulfate permease activity. The sulfate analog chromate served as a useful indicator of sulfate permease activity. When expressed, sulfate permease will transport the toxic compound chromate into N. crassa, resulting in cell death. The ability of strains to grow in the presence of chromate, therefore, indicates a lack of sulfate permease resulting from sulfur system repression (10). Determination of arylsulfatase activity served to further characterize the sulfur regulatory state of given mutants, with elevated arylsulfatase levels indicating sulfur system activation as discussed previously (6).
Table 1.
In vivo characterization of SCON2 F-box site-directed mutants
| Strain | Sulfur prototroph | Chromate resistance*
|
ARS specific activity†
|
||
|---|---|---|---|---|---|
| High S | Low S | High S | Low S | ||
| WT | + | + | − | − | + |
| Δcys-3 | − | + | + | − | − |
| Δscon-2 | + | − | − | + | + |
| L122D | − | + | + | − | − |
| D1251 | + | + | − | − | + |
| I127D | − | + | + | − | − |
| L130D | − | + | + | − | − |
| P131S | + | + | − | − | + |
| E133A | − | + | + | − | − |
| V138D | − | + | + | − | − |
| Y141A | + | + | − | − | + |
| L142D | − | + | + | − | − |
| D143I | − | + | + | − | − |
| L147E | − | + | + | − | − |
| V153D | − | + | + | − | − |
| W157G | − | + | + | − | − |
| W167G | − | + | + | − | − |
All functional analyses of site-directed mutants were performed on a minimum of three individual homokaryotic transformants per strain. Functional analysis of the wild-type (WT), Δcys-3(18-4), and Δscon-2 strains were performed in triplicate.
*Resistance to chromate has been indicated with a +.
Mutant strains exhibited two distinct levels of arylsulfatase activity: −, mean values <0.35 nm p-nitrophenyl/min per mg with an average SD of 0.10; +, mean values >50.00 nm p-nitrophenyl/min per mg with an average SD of 17.42.
Applying the assays described above, wild-type Neurospora is a sulfur prototroph, exhibiting chromate resistance and arylsulfatase deficiency under high sulfur growth conditions. The D125I, P131S, and Y141A mutants each exhibited this pattern of low sulfur induction/high sulfur repression characteristic of wild-type sulfur regulation. The restoration of wild-type function in the P131S strain was particularly surprising because Pro131 is so strongly conserved throughout the F-box protein family. In an even more surprising twist, the remaining 11 site-directed mutants all exhibited a completely novel phenotype distinct from both scon-2 null and wild-type strains. Each mutant was a sulfur auxotroph, unable to survive under normal growth conditions in the absence of a supplemental sulfur source such as 5 mM methionine. These auxotrophic F-box mutants were chromate resistant and arylsulfatase deficient under both low and high sulfur conditions, consistent with constitutive sulfur system repression. In contrast, the scon-2 deletion strain Δscon-2 is a sulfur prototroph with constitutively elevated levels of arylsulfatase enzyme activity. The site-directed F-box mutants most closely resemble the cys-3 deletion strain Δcys-3(18–4), a sulfur auxotroph with constitutively depressed levels of arylsulfatase activity (14).
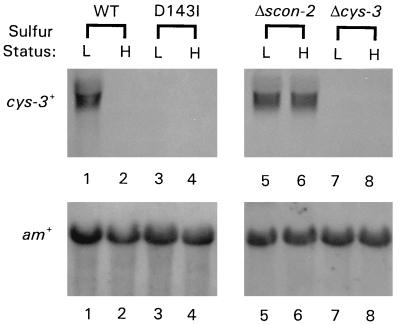
The clear phenotypic similarity between Δcys-3 and the F-box mutants may reflect an underlying commonality of cause. Accordingly, we have investigated cys-3+ expression in the representative scon-2 site-directed mutant strain D143I. Northern blots of poly(A)+ mRNA isolated from wild-type, D143I, Δscon-2, and Δcys-3 strains were probed with the cloned cys-3+ gene (Fig. 2). cys-3+ expression could not be detected under low or high sulfur conditions in either the D143I or Δcys-3 strains. As expected, the cys-3+ gene was constitutively expressed in the Δscon-2 strain and was induced under conditions of sulfur limitation in the wild-type strain.
Figure 2.
Northern hybridization analysis of cys-3+ mRNA levels in the scon-2 mutant strain D143I. Poly(A)+ mRNA was isolated from N. crassa wild-type, D143I, Δscon-2, and Δcys-3(18–4) strains grown under low and high sulfur conditions (L has been used to designate low sulfur growth conditions; H designates high sulfur growth conditions). Northern blots were prepared and probed with 32P-labeled cys-3+ and am+ DNA. The am+ gene (10) served as a control to ensure comparability between samples. Lanes 1–2, mRNA from wild-type N. crassa; lanes 3–4, mRNA from the site-directed F-box mutant strain D143I; lanes 5–6, mRNA from the scon-2 deletion strain Δscon-2; lanes 7–8, mRNA from the cys-3 deletion strain Δcys-3(18–4). As indicated in lanes 3–4, cys-3+ gene expression could not be detected in the D143I mutant strain under either low or high sulfur growth conditions.
In Vivo Analysis of Chimeric SCON2 F-box Constructs.
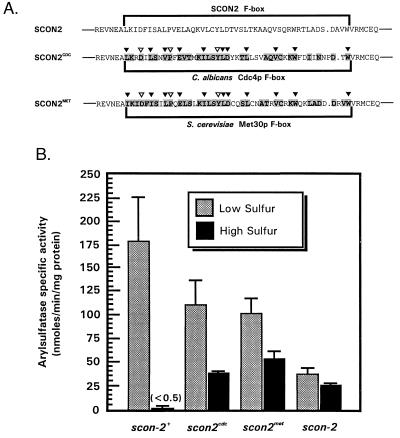
The findings presented here establish a role for the F-box motif in SCON2 inhibition of cys-3+ expression. Given the diverse nature of F-box proteins, we wondered whether other F-box sequences actually fulfilled analogous regulatory roles in their respective proteins. To address this question, we have used splice-overlap extension PCR to construct a series of chimeric SCON2 proteins containing foreign F-box sequences. These constructs are shown in Fig. 3A. The C. albicans CDC4 gene encodes a cell cycle regulator whose yeast homolog is required for proteolysis of the kinase inhibitor Sic1p and subsequent entry into S phase (2, 27). Met30p is a transcriptional inhibitor responsible for regulating the biosynthesis of sulfur amino acids in S. cerevisiae (26). Both proteins share strong sequence similarity with SCON2 within their respective F-box regions. Specifically, the Cdc4p and Met30p F-boxes exhibit 63% and 72% similarity to SCON2, respectively. Additionally, each F-box sequence contains conserved or identical residues at sites resulting in sulfur auxotrophy within SCON2.
Figure 3.
In vivo functional analysis of chimeric SCON2 F-box mutants. (A) Sequence comparison of SCON2CDC and SCON2MET. SCON2CDC contains a precise replacement of the native SCON2 F-box sequence (encoded within residues 122–167) with the corresponding F-box region from the C. albicans cell cycle regulator Cdc4p (2); SCON2MET features a replacement of this same SCON2 region with the F-box sequence from the S. cerevisiae transcriptional inhibitor Met30p (26). Both chimeric proteins are presented below wild-type SCON2, with F-box domains bracketed within each sequence. F-box sequences were optimally aligned by using clustalw (multiple alignment parameters as previously reported). Residues within SCON2CDC and SCON2MET that are similar or identical to residues within the SCON2 F-box have been printed in bold on a gray background. The C. albicans Cdc4p F-box exhibits 63% similarity and 39% identity with SCON2; the Met30p F-box exhibits 72% similarity and 57% identity to the native SCON2 F-box. Inverted triangles highlight positions within SCON2CDC and SCON2MET corresponding to sites within scon-2+ that already have been studied through site-directed mutagenesis. Specifically, black triangles indicate sites that resulted in sulfur auxotrophy on mutagenesis in scon-2+. (B) Arylsulfatase specific activity in SCON2CDC and SCON2MET. scon2cdc and scon2met strains were assayed for recovery of wild-type sulfur regulation by measuring arylsulfatase enzyme specific activity under low and high sulfur conditions. Full-length scon-2+ sequence as well as the scon-2 mutant strain (10) served as controls. In all cases, reported values indicate the mean arylsulfatase specific activity (in nmols/min per mg of total protein) ± SD from a minimum of five trials of independent homokaryotic transformants. Under high sulfur conditions, scon-2+ transformants displayed an average arylsulfatase activity of 0.37 ± 0.01 nmols p-nitrophenyl/min per mg.
To ensure accurate analysis of scon2cdc and scon2met in vivo, we have targeted integration of these chimeric genes to the his-3 locus, a transcriptionally active site under both low and high sulfur conditions. Briefly, scon2cdc and scon2met coding sequences were fused to a truncated form of the wild-type his-3 gene known to confer His prototrophy upon recombination with a mutant allele of his-3 (28). These gene fusions subsequently were transformed into an scon-2 his-3 double-mutant strain. Restoration of His prototrophy in scon-2 his-3 after transformation with scon2cdc and scon2met would, therefore, indicate homologous integration of these plasmids at the his-3 locus. Resulting homokaryotic transformants were assayed for arylsulfatase enzyme activity under low and high sulfur conditions (Fig. 3B). In wild-type Neurospora, sulfur system activity is strongly repressed in response to high levels of sulfur, with arylsulfatase activity barely detectable under such repressing conditions. The scon2cdc strain exhibited a somewhat similar pattern of repression under high sulfur conditions, with arylsulfatase activity depressed to 33% of its corresponding low sulfur level. The scon2met strain also functioned similarly, exhibiting 54% arylsulfatase specific activity under high sulfur repressing conditions. Although these chimeric strains did not inhibit sulfur system activity to the degree observed within wild-type Neurospora, both scon2cdc and scon2met displayed appreciably decreased levels of arylsulfatase activity under repressing conditions. In contrast, scon-2 null mutant strains exhibit nearly equivalent levels of arylsulfatase activity under both low and high sulfur conditions. This result suggests at least partial function of foreign Cdc4p and Met30p F-boxes within the context of SCON2 and the Neurospora sulfur system in vivo. Surprisingly, the C. albicans Cdc4p F-box seemed to function slightly more effectively in SCON2 than did the Met30p F-box, despite the stronger sequence similarity between SCON2 and Met30p. This finding may indicate fundamental differences between the N. crassa and S. cerevisiae sulfur regulatory systems that render Met30p somewhat ineffective in Neurospora.
DISCUSSION
At present, the F-box stands as a novel protein motif whose full functional significance is only beginning to be understood. Previous studies already have established a role for the F-box motif in ubiquitin-mediated proteolysis at the G1-S transition in budding yeast (2). Given the diverse nature of F-box proteins, however, it is unlikely that the F-box functions exclusively at cell cycle transitions. In fact, the majority of F-box proteins highlighted within Fig. 1 are thought to operate independently of cell cycle events. The S. cerevisiae gene GRR1, for example, encodes an F-box protein required for catabolite repression (29). Furthermore, grr1 mutants proceed rapidly into S phase despite lacking Grr1p-associated proteolytic capabilities (30). It is possible that F-boxes do strictly mediate proteolytic events, although not solely within cell cycle pathways. Alternatively, the F-box may represent a more general protein-interaction domain, initially implicated in cell cycle processes because of its preliminary characterization in a limited number of proteins.
To advance understanding of the F-box motif, we have directly analyzed F-box function in the negative regulator SCON2 through extensive mutational analysis in vivo. Site-directed mutagenesis of conserved residues within the SCON2 F-box resulted in sulfur auxotrophy, with mutants unable to grow under standard conditions in the absence of supplemental levels of methionine. As indicated in Fig. 2, this auxotrophy stems from transcriptional inhibition of the positive regulatory gene cys-3+. The SCON2 F-box, therefore, is functionally significant, mediating gene regulation within the Neurospora sulfur regulatory system. To examine the mechanism by which the F-box inhibits cys-3+ expression, we constructed chimeric SCON2 proteins containing foreign F-box sequences from the regulatory proteins Cdc4p and Met30p. The ability of these swapped domains to at least partially function within SCON2 suggests a common underlying mechanism of action, possibly involving F-box-mediated proteolysis.
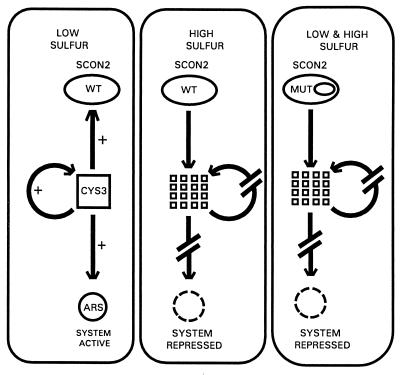
The possibility that SCON2 may function via a proteolytic process analogous to the pathway followed by Cdc4p holds significant implications for our understanding of the intricate N. crassa sulfur control system. Fig. 4 provides an updated model of this system, synthesizing current as well as previous data. As indicated in this diagram, CYS3 stands as a prime regulatory target for SCON2. When present, the CYS3 protein is capable of activating expression of the entire family of sulfur-related structural genes. CYS3 can additionally activate its own expression as well as initiate a negative feedback loop by activating expression of scon-2+. Loss of CYS3, therefore, would eliminate sulfur structural gene expression, allowing for shutdown of the sulfur system. The absence of CYS3 would necessarily interrupt its own autoregulatory loop, explaining the lack of cys-3+ mRNA observed on sulfur system repression. Decreased CYS3 levels eventually would result in decreased levels of SCON2, offering a mechanism for sulfur system reactivation under appropriate conditions. Our mutational analysis clearly defines a role for the F-box motif in regulating expression of cys-3+. As suggested in Fig. 4, site-directed mutations within the SCON2 F-box may result in the deregulated proteolysis of CYS3 under both low and high sulfur conditions. These mutations may prevent formation of a complete protein complex at the SCON2 F-box; the altered SCON2 protein instead may become part of an incomplete complex unresponsive to repression and derepression signals. As a consequence, the SCON2 F-box mutants still may be partially functional and capable of sequestering CYS3 protein for subsequent proteolysis irrespective of sulfur levels. Along similar lines, mutations within the SCON2 F-box may mimic some form of posttranslational modification normally responsible for activating SCON2 function specifically under high sulfur conditions. These prematurely active F-box mutants might effect CYS3 proteolysis under both repressing and derepressing conditions, explaining the sulfur auxotrophy evident in strains such as D143I. In either case, our results indicate that a single site-directed mutation restricted solely to the SCON2 F-box can result in CYS3 repression under conditions of sulfur limitation; therefore, any additional components necessary for CYS3 proteolysis also must be present under low sulfur conditions. Accordingly, work currently is underway to identify other proteins involved in sulfur regulation within N. crassa.
Figure 4.
Diagrammatic model of SCON2 function within the N. crassa sulfur regulatory network. To emphasize the different functional states exhibited by SCON2, this model has been partitioned according to sulfur status as well as the mutational state of scon-2+. Under low sulfur conditions, wild-type scon-2+ does not effect repression of the transcriptional activator cys-3+, allowing CYS3-mediated activation of itself, scon-2+, and the entire set of sulfur-regulated structural genes (represented by arylsulfatase). Under high sulfur conditions, however, wild-type scon-2+ functions to repress cys-3+, possibly through a pathway ultimately resulting in CYS3 proteolysis. CYS3 destruction would eliminate the activation pathways mentioned previously, thereby resulting in sulfur-system repression. (Right) Site-directed mutagenesis of conserved residues within the SCON2 F-box results in cys-3+ repression (again, possibly through CYS3 proteolysis) under both low and high sulfur conditions. The SCON2 F-box, therefore, stands as a key regulatory target mediating the functional state of SCON2 within the N. crassa sulfur control system.
As an alternative to this model, it is possible that the CYS3 protein is not in fact proteolyzed through a pathway featuring the SCON2 F-box. Rather, SCON2 itself might be targeted for destruction by its own F-box. Presumably such F-box-mediated proteolysis would be induced only in response to sulfur limitation, allowing for SCON2 accumulation and subsequent CYS3 inactivation under repressing conditions. This alternative, however, seems somewhat unlikely because no other F-box protein, as of yet, has been itself identified as an actual target of proteolysis; in all known cases, F-box proteins instead have mediated proteolytic processes targeting substrate molecules for destruction.
Although some aspects of SCON2 function still remain uncertain, our findings leave little uncertainty concerning the importance of the F-box within SCON2: the F-box motif does indeed function in SCON2-mediated gene regulation within the N. crassa sulfur control system. The F-box, therefore, mediates processes governing the regulation of gene expression outside of cell cycle pathways. Considering the diverse variety of eukaryotic regulatory systems containing F-box proteins, this finding suggests a potential role for the F-box as a regulatory motif of widespread significance to eukaryotes.
Acknowledgments
A.K. was supported by a fellowship from the Biomedical Sciences Ph.D. Program at Wright State University.
References
- 1.King R W, Deshaies R J, Peters J-M, Kirschner M W. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 2.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 3.Kumar A, Paietta J V. Proc Natl Acad Sci USA. 1995;92:3343–3347. doi: 10.1073/pnas.92.8.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metzenberg R L. Microbiol Rev. 1979;43:361–383. doi: 10.1128/mr.43.3.361-383.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marzluf G A. Annu Rev Microbiol. 1993;47:31–55. doi: 10.1146/annurev.mi.47.100193.000335. [DOI] [PubMed] [Google Scholar]
- 6.Paietta J V. Mol Cell Biol. 1989;9:3630–3637. doi: 10.1128/mcb.9.9.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y-H, Paietta J V, Mannix D G, Marzluf G A. Mol Cell Biol. 1989;9:1120–1127. doi: 10.1128/mcb.9.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paietta J V, Akins R A, Lambowitz A M, Marzluf G A. Mol Cell Biol. 1987;7:2506–2511. doi: 10.1128/mcb.7.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton E G, Metzenberg R L. J Bacteriol. 1972;109:140–151. doi: 10.1128/jb.109.1.140-151.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paietta J V. Mol Cell Biol. 1990;10:5207–5214. doi: 10.1128/mcb.10.10.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neer E J, Schmidt C J, Nambudripad R, Smith T F. Nature (London) 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 12.Fong H K W, Hurley J B, Hopkins R S, Miake-Lye R, Johnson M S, Doolittle R F, Simon M I. Proc Natl Acad Sci USA. 1986;83:2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Nature (London) 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 14.Paietta J V. Mol Cell Biol. 1992;12:1568–1577. doi: 10.1128/mcb.12.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis R H, DeSerres E F. Methods Enzymol. 1970;17:79–143. [Google Scholar]
- 16.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Schuler G D, Altschul S F, Lipman D J. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor J W, Ott J, Eckstein R. Nucleic Acids Res. 1985;13:8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 22.Sachs M S, Ebbole D. Fungal Genet Newsl. 1990;37:35–36. [Google Scholar]
- 23.Vollmer S J, Yanofsky C. Proc Natl Acad Sci USA. 1986;83:4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebbole D, Sachs M S. Fungal Genet Newsl. 1990;37:17–18. [Google Scholar]
- 25.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 26.Thomas D, Kuras L, Barbey R, Cherest H, Blaiseau P-L, Surdin-Kerjan Y. Mol Cell Biol. 1995;15:6526–6534. doi: 10.1128/mcb.15.12.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwob E, Boehm T, Mendenhall M D, Nasmyth K. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 28.Overton L K, Dubins J S, DeSerres F J. Mutat Res. 1989;214:267–283. doi: 10.1016/0027-5107(89)90171-1. [DOI] [PubMed] [Google Scholar]
- 29.Flick J S, Johnston M. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barral Y, Jentsch S, Mann C. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]