Short abstract
Recent genomic and proteomic studies have identified many of the genes and gene products differentially expressed during bacterial biofilm formation, revealing the complexity of this developmental process.
Abstract
Bacterial communities that are attached to a surface, so-called biofilms, and their inherent resistance to antimicrobial agents are a cause of many persistent and chronic bacterial infections. Recent genomic and proteomic studies have identified many of the genes and gene products differentially expressed during biofilm formation, revealing the complexity of this developmental process.
In nature, the majority of bacteria live in close association with surfaces, as complex communities referred to as biofilms [1,2]. Biofilms (so called because macroscopically they do look like a thin layer of slime) have a distinct architecture, consisting of tower- and mushroom-shaped microcolonies encased in a hydrated matrix of exopolymeric substances, polysaccharides and proteins that are produced by the resident microorganisms. Compared with their planktonic (non-adherent) counterparts, the compact microbial consortia present in biofilms show extraordinary resistance to conventional biocides, antimicrobial treatments and the immune defense responses of the host. Formation of these sessile communities and their inherent resistance to antimicrobial agents are at the root of many persistent and chronic bacterial infections. Biofilms have been shown to colonize a wide variety of medical devices and to be associated with several human diseases, such as native valve endocarditis, burn wound infections, chronic otitis media with effusion and cystic fibrosis. Recent advances in our understanding of the genetic and molecular basis of bacterial community behavior point to therapeutic targets that may provide a means for the control of biofilm infections.
Rethinking biofilms
Looking back, research on biofilms has come a long way since the initial characterization of a biofilm by Antoni van Leeuwenhoek. The first descriptions of specific genes that are up- or down-regulated in biofilm bacteria were made using transcriptional lacZ reporter-gene fusions [3,4] and led to the belief that bacterial attachment initiates the expression of a set of genes that culminates in a biofilm phenotype (Figure 1) [2]. That significant fractions of the bacterial genome could be involved in, or affected during, biofilm formation was shown in Escherichia coli in a genome-wide screen using random chromosomal insertions of a lacZ reporter gene fusion construct [5]. Prigent-Combaret et al. [5] showed that bacteria within biofilms encounter higher-osmolarity conditions, greater oxygen limitation, and higher cell density than in the liquid phase. With so many genes involved, it is perhaps not surprising that biofilm formation is regarded as a developmental process (Figure 1), not unlike that observed in the formation of fruiting bodies containing spores by the soil bacterium Myxococcus xanthus and sporulation in Bacillus subtilis [6].
Figure 1.
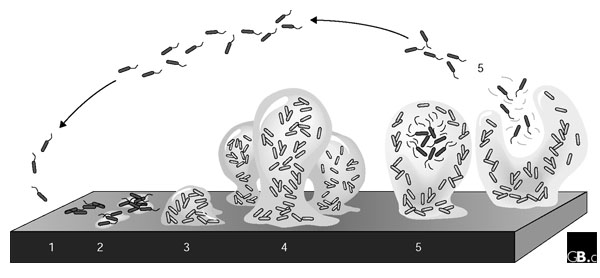
A model of the stages of bacterial biofilm development. At stage 1, the bacterial cells attach reversibly to the surface. Then, at stage 2, the cells attach irreversibly, a step mediated mainly by exopolymeric substances, and the cells lose their flagella-driven motility. At the next stage (3), the first maturation phase is reached, as indicated by early development of biofilm architecture. The second maturation phase is reached at stage 4 with fully mature biofilms, as indicated by the complex biofilm architecture. At the dispersion stage (5), single motile cells (dark cells on the figure) disperse from the microcolonies. Adapted from [27].
The availability of complete bacterial genome sequences, together with the development of microarrays with which the expression of the entire genome of an organism grown under two conditions can be assayed, has launched the post-genomic era of biofilm research and generated a wealth of new information. But a comparison of the differentially expressed gene sets identified in several recent DNA microarray studies [7-10] reveals that no common expression pattern for biofilms has yet emerged. Instead, in different studies different genes are found up- and down-regulated, in varying numbers ranging from 1% to 38% of the total genome. One explanation for these apparent discrepancies is that DNA microarrays provide a sensitive but transient snapshot of gene expression and that gene expression does not necessarily directly correlate with phenotype. This article will focus on the discrepancies that may arise from differences in experimental scenarios and between the species used; for example, Gram-negative bacteria differ from Gram-positive bacteria with respect to cell-wall composition, the molecules involved in quorum sensing (the ability of bacteria to communicate with each other in a population to coordinate population behavior in response to environmental cues), and some transcriptional regulators.
Biofilm formation in Gram-negative bacteria
Using DNA microarrays, gene expression in E. coli biofilms (grown in a silicone flow chamber for a total of 40 hours at varying flow rates) was compared with expression in planktonic cells in stationary phase [7]. The comparison revealed an overall change of more than 600 genes, with 9% of the whole genome being activated and 4.5% repressed in the biofilm cells. When the transcriptional profile of biofilm cells was compared with that of exponentially grown cells, a different expression pattern emerged: only 230 genes were found to be differentially expressed, with 4.8% up- and 0.5% down-regulated in biofilm cells [7]. Overall, the expression of only 79 genes, representing 1.84% of the E. coli genome, was significantly altered during biofilm growth compared with planktonic growth. Among the genes that showed increased expression in biofilms were three involved in adhesion and autoaggregation, several encoding structural proteins such as OmpC, OmpF and OmpT, lpxC (encoding a protein associated with lipid A biosynthesis), and slp (encoding an outer-membrane lipoprotein induced after carbon starvation). Some of these genes (slp and ompC) have recently been associated with the initial steps of E. coli biofilm formation on abiotic surfaces [5,11].
The microarray analysis of biofilms by Schembri et al. [7] also revealed differential expression of genes under oxygen- and nutrient-limiting conditions, and of genes associated with enhanced heavy-metal resistance. Interestingly, although quorum sensing has been shown to be important in biofilm formation in other species, such as Pseudomonas aeruginosa [12], no genes regulated in response to quorum sensing were found in the study of biofilm formation by Schembri et al. [7]. Furthermore, their E. coli transcriptome analysis did not reveal changes in the expression of components of the Cpx two-component signal transduction system, which senses changes in the environment and responds to general stress conditions in the extracytoplasmic compartment by activating genes that encode periplasmic protein-folding and protein-degradation factors. The Cpx transduction system in E. coli has previously been demonstrated to be involved in surface sensing and adhesion [13] and in the modulation of the expression of curli, thin bacterial appendices that are involved in adhesion and biofilm formation [14], so failure to find its genes regulated during biofilm formation is surprising.
A DNA microarray analysis of Pseudomonas aeruginosa detected only 1% of genes as differentially expressed in the biofilm growth mode, with 0.5% of the genes being activated and about 0.5% being repressed [8]. Whiteley et al. [8] assigned the differentially regulated genes to motility, attachment, translation, metabolism, transport and regulatory functions, and found that temperate phage genes were the most highly activated. This initial microarray analysis of P. aeruginosa biofilms [8] showed that, on average, gene expression in biofilm cells was remarkably similar to gene expression in planktonic cells maintained under similar environmental conditions, namely dense communities with high cell densities (1010 cells per milliliter). These conditions activate the bacterial communication system that would be expected to trigger quorum sensing and regulate between 353 [15] and 616 genes [16], but no quorum-sensing-regulated genes were identified in this study [8]. The latter is an unexpected result because the process of biofilm development has previously been shown in P. aeruginosa to involve quorum sensing [12]. Furthermore, bacterial communication via quorum sensing has been reported to be important in the production of virulence factors and antibiotic resistance. It has therefore been suggested that quorum sensing may contribute to the ability of P. aeruginosa to initiate infection and to persist in a host as a biofilm. Data from many models of both acute infection and chronic infection have supported the hypothesis that quorum sensing is important in P. aeruginosa pathogenesis and biofilm formation [16].
One protein known to play a key role in biofilm formation is RpoS, the σS subunit of RNA polymerase [17]. It governs the expression of many genes induced during the stationary phase of growth and is considered to be the master regulator of the general stress response in E. coli [18]. Schembri et al. [7] noted that 46% of the genes that were found to be differentially expressed during biofilm growth were under the control of RpoS, and deletion of rpoS rendered E. coli incapable of establishing sessile communities [7]. In P. aeruginosa, the role of a homolog of E. coli RpoS seems to be the opposite of the role of RpoS in E. coli: the P. aeruginosa rpoS gene was found to be repressed in biofilms, and rpoS-deficient mutants not only formed better biofilms than wild-type cells but were more resistant to antimicrobial treatment [8]. This finding is consistent with earlier reports that P. aeruginosa rpoS mutants were hypervirulent in a mouse model [19].
Biofilm formation in Gram-positive bacteria
To analyze global gene expression in B. subtilis using microarrays, biofilms were grown under stagnant growth conditions in a beaker, and after 8, 12, and 24 h the total content of the growth vessel, which contained both biofilm and planktonic cells, was harvested for RNA isolation [9]. For comparison, the RNA of a purer population of planktonic cells was isolated from cells grown with shaking to late exponential phase. A total of 519 genes were identified as differentially expressed during the time-course of biofilm formation. More than 55% of these were expressed at only one of the three time points, indicating temporal control of gene expression during biofilm formation. Most of the differentially expressed genes were involved in phage-related functions, membrane bioenergetics, glycolysis, and the tricarboxylic acid cycle, and in addition there were many genes involved in motility and chemotaxis.
Although the role of motility in biofilm formation has been shown in other experimental models, such as E. coli and P. aeruginosa biofilms [5,20], it is noteworthy that motility and chemotaxis seem to be associated with only the very initial steps of biofilm formation, namely the transition to the sessile mode of growth (Figure 1). It should be noted that batch cultures grown in beakers and microtiter plates are models in which it is difficult to produce steps past the initiation of biofilm formation, so the findings should be considered as somewhat limited as fully mature biofilms cannot be studied under these experimental conditions [21].
Stanley et al. [9] used expression profiling to identify transcriptional regulators that were affected during biofilm formation in B. subtilis, by extrapolating from the expressed genes expressed to their regulators. Using this indirect approach, several transcription factors were identified, including SpooA and the starvation-activated transcription factor σH. SpooA was previously shown to be required for biofilm formation [22] and for directing the development of endospores. Furthermore, 40 genes responsive to glucose concentration were found in the study by Stanley et al. [9], who concluded that glucose inhibits biofilm formation through the catabolite control protein CcpA. A role for glucose in biofilm formation has also been proposed for Streptococcus mutans [10] and E. coli [23] when grown under stagnant batch growth conditions. In the case of E. coli, the availability of glucose affected biofilm formation through the carbon storage regulator CsrA; disruption of csrA significantly decreased biofilm formation [23].
Alternative approaches to studying biofilms
In addition to microarray analyses, in vivo expression technology has recently been used to study global gene expression in P. aeruginosa biofilms [24]. This technology was originally developed to study virulence genes that are repressed during growth on laboratory media but expressed in pathogenic bacteria during an infectious process in a host. Finelli et al. [24] have used gene-fusion constructs that complement an attenuating adenine auxotrophic mutation to identify promoters that are turned on in vivo in P. aeruginosa biofilms. P. aeruginosa derivatives under flowing conditions were allowed to establish biofilms on the interior surface of silicone tubing, and after five days the biofilm was harvested and cells plated on adenine-containing medium to isolate single positive clones for further analysis.
Using in vivo expression technology, only five genes essential for P. aeruginosa biofilms were identified and subsequently confirmed by insertional mutation [24]. Mutation in PA5065, a homolog of E. coli ubiB (involved in ubiquinone biosynthesis), was lethal, and mutation in PA2247 (encoding 2-oxoisovalerate dehydrogenase) has not been characterized in detail [24]. Mutations in three other genes, PA0240 (encoding a putative porin), PA3710 (encoding aliphatic alcohol dehydrogenase), and PA3782 (encoding an AraC-like transcriptional regulator), had no effect on planktonic growth but caused defects in biofilm formation in static and flowing systems. Interestingly, in competition experiments, these three P. aeruginosa mutants had reduced fitness compared with the parent strain: they comprised less than 0.0001% of total biofilm cells after five days growth in culture, indicating a role for the mutated genes in the establishment of sessile communities. Finelli et al. [24] concluded that they had identified novel genes that did not affect planktonic growth but were important for biofilm formation, development, and fitness. None of the genes identified in this study was detected in previous DNA microarray analyses of E. coli [7] or P. aeruginosa [8].
Proteomics refers to the comparative identification of all proteins expressed under various conditions, as found for example, by two-dimensional gel electrophoresis. Although low resolution and detection limits are common pitfalls, this technique is an essential complement to transcriptome analysis because it allows the detection of proteins, the functional entities of a cell, and of post-translational protein modifications, which cannot be predicted by mRNA expression analysis. Unfortunately, to date only limited information about biofilm proteomics is available, making a thorough comparison between transcriptomes and proteomes difficult. My colleagues and I [25] showed by two-dimensional gel electrophoresis combined with reporter-gene analysis and microscopy that biofilm communities of P. aeruginosa displayed at least five distinct physiologies during biofilm development (Figure 1) [25]. The five stages were visible when biofilms were grown on the interior surface of silicone tubing under flowing conditions over a 12-day period.
A large number of proteins were found to be differentially produced during the different stages of biofilm development; several proteins were differentially expressed after one day of biofilm growth and the protein-expression pattern showed maximum change compared with the expression pattern in planktonic cells in mature biofilm cells [25]. At each of the five stages, the majority of differently produced protein spots on gels were found to be overexpressed; 23 of the overproduced proteins were involved in oxidative damage, production of exopolymeric substances, aerobic and anaerobic metabolism, and membrane transport. After maturation, biofilm dispersion and reversion to the planktonic mode of growth occurred (Figure 1) and most of the differentially produced proteins were repressed. Similar observations were made for Bacillus cereus, in which distinct and reproducible protein patterns were observed between biofilms of different ages [26].
In conclusion, although it is apparent that biofilms have gene-expression patterns that differ from those of planktonic bacteria, it is also clear that we still have to decipher the genetic basis of biofilm formation. Much more work is also still needed if we are to completely describe the physiological changes that occur during biofilm formation. The detection of stage-specific physiologies and the display of multiple phenotypes during biofilm development may hold clues to the differences among the various DNA microarray analyses described so far.
References
- Costerton JW, Cheng K-J, Geesey GG, Ladd T, Nickel JC, Dasgupta M, Marrie JT. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.micro.41.1.435. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell D, Korber D, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.micro.49.1.711. [DOI] [PubMed] [Google Scholar]
- Dagostino L, Goodman AE, Marshall KC. Physiological responses induced in bacteria adhering to surfaces. Biofouling. 1991;4:113–119. [Google Scholar]
- Davies DG, Charabarty AM, Geesey GG. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Schembri MA, Kjaergaard K, Klemm P. Global gene expression in Escherichia coli biofilms. Mol Microbiol. 2003;48:253–627. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Stanley NR, Britton RA, Grossman AD, Lazazzera BA. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J Bacteriol. 2003;185:1951–1957. doi: 10.1128/JB.185.6.1951-1957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen ZT, Burne RA. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl Environ Microbiol. 2002;68:1196–1203. doi: 10.1128/AEM.68.3.1196-1203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto K, Norbeck J, Larsson T, Karlsson KA, Hermansson M. Adhesion of type 1-fimbriated Escherichia coli to abiotic surfaces leads to altered composition of outer membrane proteins. J Bacteriol. 2001;183:2445–2453. doi: 10.1128/JB.183.8.2445-2453.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Otto K, Silhavy TJ. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc Natl Acad Sci USA. 2002;99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorel C, Vidal O, Prigent-Combaret C, Vallet I, Lejeune P. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol Lett. 1999;178:169–175. doi: 10.1016/S0378-1097(99)00347-X. [DOI] [PubMed] [Google Scholar]
- Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JL, McLean RJ. Impact of rpoS deletion on Escherichia coli biofilms. Appl Environ Microbiol. 1999;65:4285–4287. doi: 10.1128/aem.65.9.4285-4287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh SJ, Silo-Suh L, Woods DE, Hassett DJ, West SE, Ohman DE. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Ghigo JM. Are there biofilm-specific physiological pathways beyond a reasonable doubt? Res Microbiol. 2003;154:1–8. doi: 10.1016/S0923-2508(02)00012-8. [DOI] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Jackson DW, Simecka JW, Romeo T. Catabolite repression of Escherichia coli biofilm formation. J Bacteriol. 2002;184:3406–3410. doi: 10.1128/JB.184.12.3406-3410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli A, Gallant CV, Jarvi K, Burrows LL. Use of in-biofilm expression technology to identify genes involved in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2003;185:2700–2710. doi: 10.1128/JB.185.9.2700-2710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosthuizen MC, Steyn B, Theron J, Cosette P, Lindsay D, Von Holy A, Brozel VS. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl Environ Microbiol. 2002;68:2770–2780. doi: 10.1128/AEM.68.6.2770-2780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]


