Abstract
CD36 is a transmembrane glycoprotein of the class B scavenger receptor family. The CD36 gene is located on chromosome 7 q11.2 and is encoded by 15 exons. Defective CD36 is a likely candidate gene for impaired fatty acid metabolism, glucose intolerance, atherosclerosis, arterial hypertension, diabetes, cardiomyopathy, Alzheimer disease, and modification of the clinical course of malaria. Contradictory data concerning the effects of antiatherosclerotic drugs on CD36 expression indicate that further investigation of the role of CD36 in the development of atherosclerosis may be important for the prevention and treatment of this disease. This review summarizes current knowledge of CD36 gene structure, splicing, and mutations and the molecular, metabolic, and clinical consequences of these phenomena.
INTRODUCTION
CD36 is a type B scavenger receptor located on the surface of many types of cells. More than 20 mutations in the coding sequence of the CD36 gene that lead to type I receptor deficiency have been described, but the molecular basis of type II CD36 deficiency is still unclear. Different systems of nucleotide numbering in the CD36 gene are used by individual authors; thus descriptions of mutation locations are ambiguous. This review summarizes current knowledge of CD36 gene structure, splicing, and mutations and the molecular, metabolic, and clinical consequences of these phenomena.
STRUCTURE OF THE HUMAN CD36 GENE
The CD36 gene is located on chromosome 7 q11.2 (1). The structural organization of the human CD36 gene was described by Armesilla et al. (2,3). The CD36 gene is encoded by 15 exons (Table 1). Exons 1, 2, and 15 are non-coding. Exons 3 and 14 encode N-terminal and C-terminal domains of the CD36 protein, respectively (3). Interestingly, the 5′-untranslated region of CD36 mRNA is encoded by 3 exons. Exon 3 contains the last 89 nucleotides of the 5′-untranslated region and encodes the N-terminal cytoplasmic and transmembrane domains (2). The 3′-untranslated region is contained in exon 14 only or in exons 14 and 15 (3).
Table 1.
Exons, Introns, and Mutations in the Human CD36 Gene
| Exon number | Next intron length | mRNA nucleotidesa | Amino acids encoded | Change in nucleotide sequenceb | Change in amino acid sequence | Ref. |
|---|---|---|---|---|---|---|
| 1 | 7341 (43708)c | −289 to −184 (−356 to −184)c | None | }del exons 1–3 | no expression of CD36 protein | 59 |
| 2 | 470 | −183 to −90 | None | |||
| 3 | 9679 | −89 to +120 | 1–40 | |||
| 4 | 4362 | 121–281 | 41–94 | C268T | Pro90Ser | 48 |
| 5 | 1779 | 282–429 | 94–143 | 319–324delGCTGAG | inframe del AA 107–108 | 55 |
| 329–330del AC | frameshift at AA 110 | 55 | ||||
| G367A | Glu123Lys | 49 | ||||
| C380T | Ser127Leu | 50 | ||||
| T411C | Ala137Val | 51 | ||||
| 6 | 1236 | 430–609 | 144–203 | 560insT | frameshift at AA 187 | 56 |
| 7 | 1945 | 610–701 | 204–234 | 619–624del ACTGCA/ins AAAAC | frameshift at AA 207 | 57 |
| 691–696del AAAGGT | inframe del AA 231–232 | 53 | ||||
| 8 | 3463 | 702–748 | 234–250 | |||
| 9 | 954 | 749–818 | 250–273 | T760C | Phe254Leu | 52 |
| 10 | 757 | 819–1006 | 273–336 | 845–849del ACGTT | frameshift at AA 282 | 53 |
| 949insA | frameshift at AA 317 | 58 | ||||
| T975G | Tyr325Term | 53 | ||||
| 11 | 729 | 1007–1125 | 336–375 | T1079G | Leu360Term | 54 |
| 12 | 511 | 1126–1199 | 376–400 | Del tttagAT | skipping exon 12 | 56 |
| 1140–1146delTTTACAA/insCCAAA | frameshift at AA 380 | 53 | ||||
| G1150C + 1155delA | Ala384Pro + frameshift at AA 385 | 53 | ||||
| 13 | 573 | 1200–1254 | 400–418 | del tattacagAG | skipping exon 13 | 56 |
| dupl. 1204–1246 | frameshift at AA 416 | 56 | ||||
| 1218–1224delGAGGAAC | frameshift at AA 406 | 57 | ||||
| 1228-1239delATTGTGCCTATT | deletion of Ile-Val-Pro-Ile | 56,57 | ||||
| A1237C | Ile413Leu | 52 | ||||
| 14 | 2236d | 1255–1688 (1255–1419)d | 419–472 | |||
| 15d | — | 1420–2044d | None |
Genbank NM 000072; the first mRNA nucleotide encoding CD36 protein is +1.
Lowercase nucleotides are located in an intron.
Alternatively spliced exon 1 (Genbank NM 001001547).
At position 1709 within exon 14, there is an internal splicing donor site that can join nucleotide 1419 to the first nucleotide of exon 15, thus generating an alternative CD36 mRNA form (Genbank NM 001001548) containing exon 15 (3).
CD36 LOCATION
CD36 is present on the surface of platelets, endothelial cells, macrophages, dendrite cells, adipocytes, striated muscle cells, and hematopoietic cells (4–9).
CD36 GENE EXPRESSION REGULATION
CD36 gene expression is tissue dependent. In adipocytes, the nuclear peroxisome proliferation activator receptor γ (PPARγ) is the critical factor for feedback control of the CD36 gene (10–13). In monocytes, receptor expression can be upregulated by adhesion and by action of cytokines (such as M-CSF and GM-CSF), native and modified LDL, cellular cholesterol, interleukin-4, insulin, and glucose (14–17). Expression is inhibited by TGF-β, corticosteroids, HDL, and LPS (lipopolysaccharides) (14,17–19). CD36 expression in the heart and skeletal muscles is increased by triacylglycerides (TG) and fatty acids (FA) in plasma and regulated by energy requirements of the tissue (20).
ALTERNATIVE SPLICING
A marked feature of the CD36 gene is the presence of several (at least five) alternative and independent first exons and their promoters, which lack TATA-boxes and CpG islands (21). Another alternative promoter and first exon have recently been identified downstream of exon 2 (22). Alternative splicing of the first CD36 exon appears to be regulated differently in different tissues, indicating that the promoters are tissue specific. The alternative transcripts are all expressed in more than one human tissue, and their expression patterns are highly variable in skeletal muscle, heart, liver, adipose tissue, placenta, spinal cord, cerebrum, and monocytes (21).
Alternative splicing of 5′-untranslated and 3′-untranslated exons in CD36 pre-mRNA accounts for some of the heterogeneity in the molecular size of CD36 mRNA (23,24). All the splice acceptor and donor sequences conform to the consensus rule for splicing. Of the 11 splice junctions found around coding exons, type 0 occurs between codons, type 1 occurs after the first nucleotide, and type 2 occurs after the second nucleotide of a codon. Such diversity in the type of intron phase limits the number of mRNA forms that might arise by exon deletion while maintaining the same open reading frame (3). Alternatively spliced coding exons can be detected in CD36-expressing cells (24).
The functional diversity of CD36 may result from alternative splicing of CD36 pre-mRNA. Skipping of coding exons 4 and 5 gives rise to a CD36 isoform that lacks 103 amino acid residues, 41–143, and three potential N-glycosylation sites (25).
Andersen et al. have suggested that the molecular mechanisms regulating the CD36 gene expression are unusually complex, reflecting the multifunctional role of CD36 in different tissues and conditions (21).
PROTEIN STRUCTURE
CD36 is a transmembrane glycoprotein of the class B scavenger receptor family (26–28). Synonym names of CD36 are platelet glycoprotein, GPIV, glycoprotein IIIb, GPIIIB, leukocyte differentiation antigen CD36, CD36 antigen, SR-BI, PAS IV, PAS-4 protein, platelet collagen receptor, fatty acid translocase, FAT, and thrombospondin (TSP) receptor.
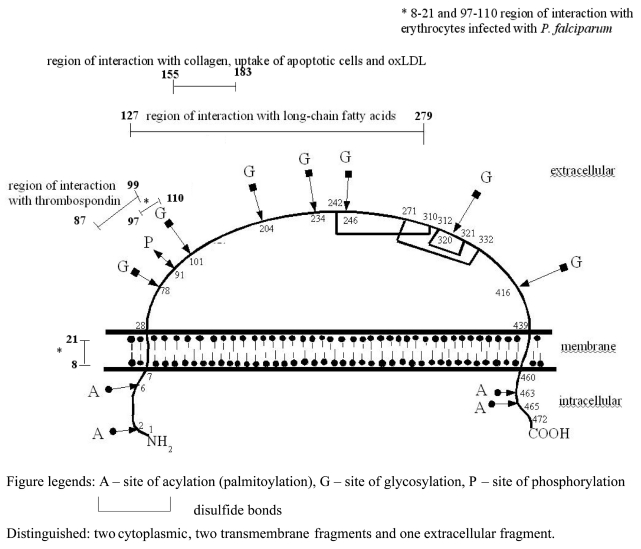
According to the nucleotide sequence, CD36 mRNA (Genbank: NM 000072) codes for 472 amino acids (AA) (SwissProt: P16671; Figure 1). The mature polypeptide is 471 AA long, beginning at the AA residue immediately following the initiator methionine (4). The molecular mass of the functional receptor is 78–88 kDa, depending on the cell type and on post-translational glycosylations (4,6,25). The receptor consists of 1 extracellular, 2 transmembrane, and two cytoplasmic fragments representing the C-terminal and N-terminal parts of the molecule (3). The cytoplasmic and transmembrane regions predicted at both terminal ends of the polypeptide chain are encoded by the single exons 3 and 14. The translation start codon is located 289 (or 356 with alternatively spliced exon 1) nucleotides downstream of the 5′-end of CD36 mRNA (25). The large number of exons encoding the highly glycosylated extracellular domain (exons 4–13 and part of exon 14) may be a consequence of the multiplicity of interactions in which CD36 seems to be involved (3).
Figure 1.
cDNA nucleotide and amino acid sequences of CD36 with mutations indicated as follows: substitutions (underline), deletions (line over and usually italic), insertions (bold) (based on: http://snpper.chip.org).
There are no free cysteines in CD36, and the six centrally clustered cysteines are linked by disulfide bonds: Cys242/Cys310, Cys271/Cys332, and Cys312/Cys321, resulting in a 1–3, 2–6, and 4–5 arrangement of the disulfide bridges (3). A diagram of the CD36 structure is presented in Figure 2.
Figure 2.
POSTTRANSLATIONAL MODIFICATIONS
The protein has two short intracellular segments (residues 1–6 and AA 461–472) that may undergo acylation (palmitoylation, Figure 2), and two transmembrane domains (residues 7–28 and AA 439–460) potentially acylated near the intracellular side of the membrane (29). The remaining part of CD36 is extracellular, comprising 7 glycosylation sites and 3 disulfide bridges (6,29,30).
CD36 FUNCTIONS
The CD36 functions presented in Table 2 include removal of oxidized LDL from plasma (in macrophages and monocytes) (10,31–34), apoptosis induction (together with other membrane proteins such as αVβ3 integrin and thrombospondin 1) (35–37), and enhancement of cytoadherence of abnormally shaped erythrocytes (infected with Plasmodium falciparum or containing hemoglobin S) (4,38). CD36 is a long-chain fatty acid receptor (39,40) with a potential binding site for the fatty acids in its extracellular segment between residues 127 and 279 (39). CD36 is also an adhesion molecule capable of interacting with collagen type I and IV and with thrombospondin (5,41–44). Leung et al. reported that the CD36-TSP interaction is a two-step process; the AA 139–155 region of CD36 binds first to TSP (thrombospondin), triggering a change in TSP structure, which then reveals a second site that binds the AA 93–110 region of CD36 with high affinity (45). On the other hand, Asch et al. (46) reported that three functional sequences exist; one of these mediates TSP binding (AA 87–99) and two other support malarial cytoadhesion (AA 8–21 and AA 97–110). The data reported by Asch et al. also suggest that CD36 ligand specificity is directed by the extracellular release or activation of threonine phosphatase in response to a platelet agonist and that CD36 dephosphorylation may affect signaling. Phosphorylation was observed to inhibit TSP binding to the peptide by 60% (46). Ren et al reported that transfection of the macrophage adhesion molecule CD36 into human Bowes melanoma cells specifically conferred greatly increased capacity, comparable to that exhibited by macrophages, to ingest apoptotic neutrophils, lymphocytes, and fibroblasts (35).
Table 2.
Functions of the CD36 receptor
| Function | Type of cellsa | Domain location (Ref) |
|---|---|---|
| Thrombospondin receptor | Monocytes, platelets, some cancer cells | 87–99 AA (46) or 93–110 and 139–155 AA (45) |
| Receptor for erythrocytes infected with Plasmodium falciparum | Monocytes, endothelial cells, some cancer cells | 8–21 and 97–110 AA (4,38) |
| Collagen receptor | Platelets | (?)155–183 AA (3,43) |
| Uptake of apoptotic cells | Macrophages | 155–183 AA (35–37) |
| Receptor for oxidized LDL (oxLDL) | Macrophages, monocytes | 155–183 AA (10,31–34) |
| Long-chain fatty acid receptor | Endothelial cells, adipocytes, platelets | 127–279 AA (39,40) |
AA amino acids.
The list of cell types is incomplete and mentions only the most studied ones.
CD36 GENE MUTATIONS
Different systems of nucleotide numbering in the CD36 gene are used by individual authors. Therefore, to standardize the nomenclature, we consistently use the numbering system in which the position of the first protein-encoding mRNA nucleotide is +1 (Figure 1). This position corresponds to nucleotide +211 according to the nomenclature used by Kashiwagi et al. (47).
The following mutations in the CD36 gene (presented in Table 1) have been described so far: nucleotide substitutions (47–54), small deletions (53,55–57), small insertions (56,58), small indels (53,57), gross deletion (59), duplication (56), complex rearrangements (53), and repeat variation, a dinucleotide repeat (TG)n in intron 3 (50,60), where the repeat number n = 12 is involved in the nonproduction of the variant CD36 transcript that lacks exons 4 and 5 (50).
MOLECULAR CONSEQUENCES OF CD36 GENE MUTATIONS
CD36 deficiency is divided into two subgroups according to the phenotypes. In type I deficiency, neither platelets nor monocytes express CD36, whereas in type II deficiency (very rare in whites, 0.3% of the population, more frequent in Asians and Afro-Americans, 3%–4% of the population), CD36 is expressed in monocytes but not in platelets (7,44).
TYPE I DEFICIENCY
The type I CD36 deficiency is most commonly detected in subjects who are homozygous or compound heterozygous for the following three mutations: C268T, 949insA, and 329-330delAC (18,61). A survey of the CD36 mutations in type I CD36 deficiency has revealed that C268T is the most common mutation, and is responsible for more than 50% of the mutated allele frequency in Asians (57). This mutation is the most frequent factor responsible for the absence of CD36 receptor expression in platelets and monocytes/macrophages (47,48). Substitution leads directly to CD36 deficiency via posttranslational modification defects (probably due to defective glycosylation of the 81 kDa protein). In an expression assay using the C268 or T268 form of CD36 cDNA-transfected cells, the substitution markedly impaired maturation of the 81-kDa precursor to the 88-kDa mature form of CD36 (47,48). The mutated precursor form of CD36 was subsequently degraded in the cytoplasm.
Another mutation responsible for type I deficiency—deletion of 12 bp, ATTGTGCCTATT, at nt. 1228–1239 (exon 13)—is present with or without skipping of exon 9 (nt. 749–818). The mechanism of exon 9 skipping is unknown because there are no mutations at the junction sites of exon 9. The 12-bp deletion leads to an inframe 4-AA deletion, whereas exon 9 skipping leads to a frameshift and appearance of a premature stop codon. In an expression assay, both 1228–1239del and exon 9 skipping directly caused impairment of the surface expression of CD36 (57).
Individuals with type I CD36 deficiency may be at risk of developing an anti-CD36 isoantibody after receiving a transfusion or during pregnancy (44).
TYPE II DEFICIENCY
The genomic or molecular background of type II CD36 deficiency is not clearly understood and is probably heterogenous, leading to confounding interpretations of cause-and-effect relationships in human CD36 deficiency (18,51,61). Type II CD36 deficiency has been observed in individuals who lack a CD36 mutation but have a heterozygous mutation (most often C268T). The genotype-phenotype correlations indicated that the platelet CD36 protein expression level was regulated by other heritable platelet-restricted factors in addition to the CD36 coding region genotype (51). Kashiwagi et al. observed (47,55,57) that monocyte CD36 cDNA samples from two subjects with type II deficiency were heterozygous for the C268 and T268 forms, whereas their platelet CD36 cDNA consisted of only the T268 form.
As reported by Lipsky et al., in the Japanese population 6 different alleles (A1 to A6) of the CD36 gene were detected according to the size of a (TG)n repeat within intron 3 (60). To explain the tangled phenotype-genotype correlation in type II CD36 deficiency, Kashiwagi et al. hypothesized the “platelet-specific silent allele” and predicted its linkage to the A5 allele, which encodes (TG)12 (42,60). However, no DNA sequence corresponding to such a platelet-specific silent allele has yet been identified. Yanai et al. investigated the polymorphic site in the 5′-proximal flanking region and the 3′-untranslated region (61). According to these authors, an altered DNA sequence in or near the 3′-untranslated region may change the structure of mRNA and its stability in CD36 deficiency. Yanai et al. suggested that if the proposed platelet-specific silent allele exists, it may be remote to the CD36 gene, because the previously reported linkage with the A5 allele was not confirmed.
METABOLIC CONSEQUENCES OF CD36 GENE MUTATIONS
CD36 may play an important role in the pathogenesis of some metabolic diseases. The receptor is responsible for the removal of approximately 50% of oxidatively modified LDL (ox-LDL) from plasma (10,31). Binding takes place in the 155–183 amino acid region of the protein (34). Long-chain fatty acids (FA) are the main ligand for the receptor (26). Reduced expression of CD36 protects the vessels, according to some authors (62,63), and some have pointed out that CD36 gene mutations are associated with increased serum concentrations of total cholesterol and LDL (18,64). Other investigators have reported significant decreases in binding and uptake of ox-LDL in peritoneal macrophages of null mutation mice (65). Kashiwagi et al. demonstrated a marked reduction in the uptake of ox-LDL by CD36-deficient macrophages, a finding that suggests that differences in atherosclerosis may occur in type I or type II CD36-deficient individuals and CD36-positive individuals. In type II–deficient subjects, alleles having hypothetical platelet-specific mRNA transcription defects may be involved (42). Type I and type II deficiencies are believed to differ in their effects on the course of atherogenesis. Ox-LDL plays a role in macrophage transformation to foam cells, but the absence of expression of the macrophage CD36 can cause retention of ox-LDL in plasma (8,31,66).
Other authors have observed abnormalities of glucose and lipid metabolism in type I CD36 deficiency. For example, increased plasma triglycerides, decreased HDL-cholesterol, impaired glucose tolerance, and delayed response of insulin secretion (57,67). Kadlecova et al. proposed that defective CD36 is probably a candidate gene for disordered fatty-acid metabolism, glucose intolerance, and insulin resistance in spontaneously hypertensive rats (SHR), but other genes may also play a role in the pathogenesis of the metabolic syndrome in Prague hereditary hypertriglyceridemic rats (68). As shown by Glazier, chromosomal deletion at the CD36 locus in SHR was mediated by unequal recombination between the CD36 gene and the second pseudogene CD36-ps2, and was induced by extensive homology between these sequences. This recombination resulted in the creation of a single chimeric gene in SHR. The trait of insulin resistance and defective fatty acid metabolism is probably caused by CD36 deficiency (69). Recently, CD36 has been reported to play an important role in atherogenicity. To understand the role of CD36, it is important to clarify the condition of CD36-deficient subjects. Some authors suggest that upregulation of class B scavenger receptors could have therapeutic potential for the treatment of atherosclerosis (70,71). CD36 expression in tissues with very active fatty acid metabolism (skeletal muscle, heart, mammary epithelium, and adipose tissue), and its involvement in foam cell formation (macrophages), suggest that lipoprotein binding to CD36 might contribute to the regulation of lipid metabolism and to the pathogenesis of atherosclerosis (72).
Long-chain fatty acids (LCFA) are the major energy substrate for the heart, and their oxidation is important to ensure maximum cardiac performance. CD36 acts as a major myocardial-specific LCFA transporter in humans (73,74). FA translocase/CD36 is probably involved in regulating FA oxidation along with the well-known regulator carnitine palmitoyltransferase I (74). Some authors suggest that polymorphisms at the CD36 gene modulate lipid metabolism and cardiovascular risk in whites (75,76). Homozygous or compound heterozygous mutations of the CD36 gene in humans result in severe defects of the uptake of long-chain fatty acids (LCFAs) in the heart (56,77). Type I CD36 deficiency is closely related to the absence of LCFA accumulation and metabolism in the myocardium (19,78). CD36 deficiency leads also to decreased myocardial accumulation of 123I-BMIPP (iodine-123-(R,S)-15-(p-iodophenyl)-3-methylpentadecanoic acid) (79). Tanaka et al. hypothesized that this deficiency is a possible etiology of hereditary hypertrophic cardiomyopathy (HCM) (80). Some patients with HCM demonstrate abnormal myocardial LCFA metabolism. Data reported by Okamoto et al. suggest that abnormal myocardial LCFA metabolism seen in HCM patients may be related to abnormality of the CD36 molecule, and that abnormalities of this molecule may be the cause of some HCM types. On the other hand, patients in the Okamoto et al. study with apical HCM showed normal CD36 molecule expression and no severe impairment of myocardial BMIPP accumulation. Thus, abnormalities of the CD36 molecules may be related to the pathogenesis of some HCM types (81). Moreover, some CD36 mutations found in persons in Thailand are associated with reduced risk of cerebral malaria (50,60). In Africans, such mutations may be maintained by unidentified selection pressures and probably protect against some infections other than malaria (53).
PERSPECTIVES
Since the CD36 receptor was first isolated in 1973 (82), its function has been associated with an increased risk of or protection against many pathological conditions, including metabolic diseases. There are indications that abnormalities of the CD36 gene are implicated in the pathogenesis of hypercholesterolemia, peripheral vessel atherosclerosis, arterial hypertension, diabetes, cardiomyopathy, Alzheimer disease, and malaria (9,10,16,25,83–86). An animal model revealed that lack of CD36 expression restrained atherosclerosis (87). Increased expression of CD36 was shown in atheroslerotic plaques and in damaged vascular tissue (88). Contradictory data (89–91) concerning the effects of antiatherosclerotic drugs on CD36 expression indicate the necessity for further investigation of the role of CD36 in the development of atherosclerosis. The results may be important for the prevention and treatment of this disease. The association between cardiovascular risk and CD36 deficiency should be analyzed in extensive prospective studies in various populations.
The spectrum of CD36 mutations differs in various populations. The influence on CD36 expression of polymorphisms in noncoding regions of the CD36 gene is currently under investigation. Further research is needed to characterize the roles of numerous alternative CD36 transcripts and tissue-specific promoters. The results of these studies may elucidate the still unclear molecular background of type II CD36 deficiency.
Footnotes
Online address: http://www.molmed.org
REFERENCES
- 1.Fernandez-Ruiz E, Armesilla AL, Sanchez-Madrid F, Vega MA. Gene encoding the collagen type I and thrombospondin receptor CD36 is located on chromosome 7q11.2. Genomics. 1993;17:759–61. doi: 10.1006/geno.1993.1401. [DOI] [PubMed] [Google Scholar]
- 2.Armesilla AL, Calvo D, Vega MA. Structural and functional characterization of the human CD36 gene promoter: identification of a proximal PEBP2/CBF site. J Biol Chem. 1996;271:7781–7. doi: 10.1074/jbc.271.13.7781. [DOI] [PubMed] [Google Scholar]
- 3.Armesilla AL, Vega MA. Structural organization of the gene for human CD36 glycoprotein. J Biol Chem. 1994;269:18985–91. [PubMed] [Google Scholar]
- 4.Oquendo P, Hundt E, Lawler J, Seed B. CD36 directly mediates cytoadherence of Plasmodium falciparum parasitized erythrocytes. Cell. 1989;58:95–101. doi: 10.1016/0092-8674(89)90406-6. [DOI] [PubMed] [Google Scholar]
- 5.McGregor JL, Catimel B, Parmentier S, Clezardin P, Dechavanne M, Leung LL. Rapid purification and partial characterization of human platelet glycoprotein IIIb. Interaction with thrombospondin and its role in platelet aggregation. J Biol Chem. 1989;264:501–6. [PubMed] [Google Scholar]
- 6.Greenwalt DE, Lipsky RH, Ockenhouse CF, Ikeda H, Tandon NN, Jamieson GA. Membrane glycoprotein CD36: a review of its roles in adherence, signal transduction, and transfusion medicine. Blood. 1992;80:1105–15. [PubMed] [Google Scholar]
- 7.Take H, Kashiwagi H, Tomiyama Y, et al. Expression of GPIV and N(aka) antigen on monocytes in N(aka)-negative subjects whose platelets lack GPIV. Br J Haematol. 1993;84:387–91. doi: 10.1111/j.1365-2141.1993.tb03091.x. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson AC, Han J, Febbraio M, Silversterin RL, Hajjar DP. Role of CD36, the macrophage class B scavenger receptor, in atherosclerosis. Ann N Y Acad Sci. 2001;947:224–8. doi: 10.1111/j.1749-6632.2001.tb03944.x. [DOI] [PubMed] [Google Scholar]
- 9.Rusinol AE, Yang L, Thewke D, Panini SR, Kramer MF, Sinensky MS. Isolation of a somatic cell mutant resistant to the induction of apoptosis by oxidized low density lipoprotein. J Biol Chem. 2000;275:7296–303. doi: 10.1074/jbc.275.10.7296. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto K, Hirano K, Nozaki S, et al. Expression of macrophage (Mphi) scavenger receptor, CD36, in cultured human aortic smooth muscle cells in association with expression of peroxisome proliferator activated receptor-gamma, which regulates gain of Mphi-like phenotype in vitro, and its implication in atherogenesis. Arterioscler Thromb Vasc Biol. 2000;20:1027–32. doi: 10.1161/01.atv.20.4.1027. [DOI] [PubMed] [Google Scholar]
- 11.Nicholson AC, Hajjar DP. CD36, oxidized LDL and PPAR gamma: pathological interactions in macrophages and atherosclerosis. Vascul Pharmacol 2004. 2004;41:139–46. doi: 10.1016/j.vph.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson AC. Expression of CD36 in macrophages and atherosclerosis: the role of lipid regulation of PPARgamma signaling. Trends Cardiovasc Med. 2004;14:8–12. doi: 10.1016/j.tcm.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Sato O, Kuriki C, Fukui Y, Motojima K. Dual promoter structure of mouse and human fatty acid translocase/CD36 genes and unique transcriptional activation by peroxisome proliferator-activated receptor alpha and gamma ligands. J Biol Chem. 2002;277:15703–11. doi: 10.1074/jbc.M110158200. [DOI] [PubMed] [Google Scholar]
- 14.Feng J, Han J, Pearce SF, Silverstein RL, Gotto AM, Jr, Hajjar DP, Nicholson AC. Induction of CD36 expression by oxidized LDL and IL-4 by a common signaling pathway dependent on protein kinase C and PPAR-gamma. J Lipid Res. 2000;41:688–96. [PubMed] [Google Scholar]
- 15.Griffin E, Re A, Hamel N, Fu C, Bush H, McCaffrey T, Asch AS. A link between diabetes and atherosclerosis: Glucose regulates expression of CD36 at the level of translation. Nat Med. 2001;7:840–6. doi: 10.1038/89969. [DOI] [PubMed] [Google Scholar]
- 16.Liang CP, Han S, Okamoto H, Carnemolla R, Tabas I, Accili D, Tall AR. Increased CD36 protein as a response to defective insulin signaling in macrophages. J Clin Invest. 2004;113:764–73. doi: 10.1172/JCI19528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yesner LM, Huh HY, Pearce SF, Silverstein RL. Regulation of monocyte CD36 and thrombospondin-1 expression by soluble mediators. Arterioscler Thromb Vasc Biol. 1996;16:1019–25. doi: 10.1161/01.atv.16.8.1019. [DOI] [PubMed] [Google Scholar]
- 18.Yanai H, Chiba H, Morimoto M, et al. Human CD36 deficiency is associated with elevation in low-density lipoprotein-cholesterol. Am J Med Genet. 2000;93:299–304. doi: 10.1002/1096-8628(20000814)93:4<299::aid-ajmg9>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe K, Ohta Y, Toba K, et al. Myocardial CD36 expression and fatty acid accumulation in patients with type I and II CD36 deficiency. Ann Nucl Med. 1998;12:261–6. doi: 10.1007/BF03164911. [DOI] [PubMed] [Google Scholar]
- 20.Bastie CC, Hajri T, Drover VA, Grimaldi PA, Abumrad NA. CD36 in myocytes channels fatty acids to a lipase-accessible triglyceride pool that is related to cell lipid and insulin responsiveness. Diabetes. 2004;53:2209–16. doi: 10.2337/diabetes.53.9.2209. [DOI] [PubMed] [Google Scholar]
- 21.Andersen M, Lenhard B, Whatling C, Eriksson P, Abumrad NA. Alternative promoter usage of the membrane glycoprotein CD36. BMC Mol Biol. 2006;7:8. doi: 10.1186/1471-2199-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato O, Takanashi N, Motojima K. Third promoter and differential regulation of mouse and human fatty acid translocase/CD36 genes. Mol Cell Biochem. 2007;299:37–43. doi: 10.1007/s11010-005-9035-0. [DOI] [PubMed] [Google Scholar]
- 23.Noguchi K, Naito M, Tezuka K, et al. cDNA expression cloning of the 85-kDa protein overexpressed in adriamycin-resistant cells. Biochem Biophys Res Commun. 1993;192:88–95. doi: 10.1006/bbrc.1993.1385. [DOI] [PubMed] [Google Scholar]
- 24.Taylor KT, Tang Y, Sobieski DA, Lipsky RH. Characterization of two alternatively spliced 5′-untranslated exons of the human CD36 gene in different cell types. Gene. 1993;133:205–12. doi: 10.1016/0378-1119(93)90639-k. [DOI] [PubMed] [Google Scholar]
- 25.Tang Y, Taylor KT, Sobieski DA, Medved ES, Lipsky RH. Identification of a human CD36 isoform produced by exon skipping. Conservation of exon organization and pre-mRNA splicing patterns with a CD36 gene family member, CLA-1. J Biol Chem. 1994;269:6011–5. [PubMed] [Google Scholar]
- 26.Abumrad NA, el-Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation: homology with human CD36. J Biol Chem. 1993;268:17665–8. [PubMed] [Google Scholar]
- 27.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–20. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 28.Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. 2001;108:785–91. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen JT, Berglund L, Rasmussen MS, Petersen TE. Assignment of disulfide bridges in bovine CD36. Eur J Biochem. 1998;257:488–94. doi: 10.1046/j.1432-1327.1998.2570488.x. [DOI] [PubMed] [Google Scholar]
- 30.Gruarin P, Thorne RF, Dorahy DJ, Burns GF, Sitia R, Alessio M. CD36 is a ditopic glycoprotein with the N-terminal domain implicated in intracellular transport. Biochem Biophys Res Commun. 2000;275:446–54. doi: 10.1006/bbrc.2000.3333. [DOI] [PubMed] [Google Scholar]
- 31.Endemann G, Stanton LW, Madden KS, Bryant CM, White RT, Protter AA. CD36 is a receptor for oxidized low density lipoprotein. J Biol Chem. 1993;268:11811–6. [PubMed] [Google Scholar]
- 32.Nicholson AC, Hajjar DP. Herpesvirus in atherosclerosis and thrombosis: etiologic agents or ubiquitous bystanders? Arterioscler Thromb Vasc Biol. 1998;18:339–48. doi: 10.1161/01.atv.18.3.339. [DOI] [PubMed] [Google Scholar]
- 33.Nozaki S, Kashiwagi H, Yamashita S, et al. Reduced uptake of oxidized low density lipoproteins in monocyte-derived macrophages from CD36-deficient subjects. J Clin Invest 1995. 1995;96:1859–65. doi: 10.1172/JCI118231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puente-Navazo MD, Daviet L, Ninio E, McGregor JL. Identification on human CD36 of a domain (155-183) implicated in binding oxidized low-density lipoproteins (Ox-LDL) Arterioscler Thromb Vasc Biol. 1996;16:1033–9. doi: 10.1161/01.atv.16.8.1033. [DOI] [PubMed] [Google Scholar]
- 35.Ren Y, Silverstein RL, Allen J, Savill J. CD36 gene transfer confers capacity for phagocytosis of cells undergoing apoptosis. J Exp Med. 1995;181:1857–62. doi: 10.1084/jem.181.5.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savill J, Hogg N, Ren Y, Haslett C. Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J Clin Invest. 1992;90:1513–22. doi: 10.1172/JCI116019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silverstein RL, Asch AS, Nachman RL. Glycoprotein IV mediates thrombospondin-dependent platelet-monocyte and platelet-U937 cell adhesion. J Clin Invest. 1989;84:546–52. doi: 10.1172/JCI114197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alessio M, Greco NJ, Primo L, et al. Platelet activation and inhibition of malarial cytoadherence by the anti-CD36 IgM monoclonal antibody NL07. Blood. 1993;82:3637–47. [PubMed] [Google Scholar]
- 39.Baillie AG, Coburn CT, Abumrad NA. Reversible binding of long-chain fatty acids to purified FAT, the adipose CD36 homolog. J Membr Biol. 1996;153:75–81. doi: 10.1007/s002329900111. [DOI] [PubMed] [Google Scholar]
- 40.Hwang EH, Taki J, Yasue S, et al. Absent myocardial iodine-123-BMIPP uptake and platelet/monocyte CD36 deficiency. J Nucl Med. 1998;39:1681–4. [PubMed] [Google Scholar]
- 41.Frieda S, Pearce A, Wu J, Silverstein RL. Recombinant GST/CD36 fusion proteins define a thrombospondin binding domain. Evidence for a single calcium-dependent binding site on CD36. J Biol Chem. 1995;270:2981–6. doi: 10.1074/jbc.270.7.2981. [DOI] [PubMed] [Google Scholar]
- 42.Kashiwagi H, Tomiyama Y, Kosugi S, et al. Family studies of type II CD36 deficient subjects: linkage of a CD36 allele to a platelet-specific mRNA expression defect(s) causing type II CD36 deficiency. Thromb Haemost. 1995;74:758–63. [PubMed] [Google Scholar]
- 43.Tandon NN, Kralisz U, Jamieson GA. Identification of glycoprotein IV (CD36) as a primary receptor for platelet-collagen adhesion. J Biol Chem. 1989;264:7576–83. [PubMed] [Google Scholar]
- 44.Yamamoto N, Akamatsu N, Sakuraba H, Yamazaki H, Tanoue K. Platelet glycoprotein IV (CD36) deficiency is associated with the absence (type I) or the presence (type II) of glycoprotein IV on monocytes. Blood. 1994;83:392–7. [PubMed] [Google Scholar]
- 45.Leung LL, Li WX, McGregor JL, Albrecht G, Howard RJ. CD36 peptides enhance or inhibit CD36-thrombospondin binding. A two-step process of ligand-receptor interaction. J Biol Chem. 1992;267:18244–50. [PubMed] [Google Scholar]
- 46.Asch AS, Liu I, Briccetti FM, et al. Analysis of CD36 binding domains: ligand specificity controlled by dephosphorylation of an ectodomain. Science. 1993;262:1436–40. doi: 10.1126/science.7504322. [DOI] [PubMed] [Google Scholar]
- 47.Kashiwagi H, Tomiyama Y, Honda S, et al. Molecular basis of CD36 deficiency. Evidence that a 478C→ T substitution (proline90→ serine) in CD36 cDNA accounts for CD36 deficiency. J Clin Invest. 1995;95:1040–6. doi: 10.1172/JCI117749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashiwagi H, Honda S, Tomiyama Y, et al. A novel polymorphism in glycoprotein IV (replacement of proline-90 by serine) predominates in subjects with platelet GPIV deficiency. Thromb Haemost 1993. 1993;69:481–4. [PubMed] [Google Scholar]
- 49.Gelhaus A, Scheding A, Browne E, Burchard GD, Horstmann RD. Variability of the CD36 gene in West Africa. Hum Mutat. 2001;18:444–50. doi: 10.1002/humu.1215. [DOI] [PubMed] [Google Scholar]
- 50.Omi K, Ohashi J, Patarapotikul J, Hananantachai H, Naka I, Looareesuwan S, Tokunaga K. CD36 polymorphism is associated with protection from cerebral malaria. Am J Hum Genet. 2003;72:364–74. doi: 10.1086/346091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imai M, Tanaka T, Kintaka T, Ikemoto T, Shimizu A, Kitaura Y. Genomic heterogeneity of type II CD36 deficiency. Clin Chim Acta. 2002;321:97–106. doi: 10.1016/s0009-8981(02)00102-x. [DOI] [PubMed] [Google Scholar]
- 52.Hanawa H, Watanabe K, Nakamura T, et al. Identification of cryptic splice site, exon skipping, and novel point mutations in type I CD36 deficiency. J Med Genet. 2002;39:286–91. doi: 10.1136/jmg.39.4.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aitman TJ, Cooper LD, Norsworthy PJ, et al. Malaria susceptibility and CD36 mutation. Nature. 2000;405:1015–6. doi: 10.1038/35016636. [DOI] [PubMed] [Google Scholar]
- 54.Lepretre F, Vasseur F, Vaxillaire M, et al. A CD36 nonsense mutation associated with insulin resistance and familial type 2 diabetes. Hum Mutat. 2004;24:104–10. doi: 10.1002/humu.9256. [DOI] [PubMed] [Google Scholar]
- 55.Kashiwagi H, Tomiyama Y, Kosugi S, et al. Identification of molecular defects in a subject with type I CD36 deficiency. Blood. 1994;83:3545–52. [PubMed] [Google Scholar]
- 56.Tanaka T, Nakata T, Oka T, et al. Defect in human myocardial long-chain fatty acid uptake is caused by FAT/CD36 mutations. J Lipid Res. 2001;42:751–9. [PubMed] [Google Scholar]
- 57.Kashiwagi H, Tomiyama Y, Nozaki S, et al. Analyses of genetic abnormalities in type I CD36 deficiency in Japan: identification and cell biological characterization of two novel mutations that cause CD36 deficiency in man. Hum Genet. 2001;108:459–66. doi: 10.1007/s004390100525. [DOI] [PubMed] [Google Scholar]
- 58.Kashiwagi H, Tomiyama Y, Nozaki S, et al. A single nucleotide insertion in codon 317 of the CD36 gene leads to CD36 deficiency. Arterioscler Thromb Vasc Biol. 1996;16:1026–32. doi: 10.1161/01.atv.16.8.1026. [DOI] [PubMed] [Google Scholar]
- 59.Curtis BR, Ali S, Glazier AM, Ebert DD, Aitman TJ, Aster RH. Isoimmunization against CD36 (glycoprotein IV): description of four cases of neonatal isoimmune thrombocytopenia and brief review of the literature. Transfusion. 2002;42:1173–9. doi: 10.1046/j.1537-2995.2002.00176.x. [DOI] [PubMed] [Google Scholar]
- 60.Lipsky RH, Ikeda H, Medved ES. A dinucleotide repeat in the third intron of CD36. Hum Mol Genet. 1994;3:217. doi: 10.1093/hmg/3.1.217. [DOI] [PubMed] [Google Scholar]
- 61.Yanai H, Chiba H, Fujiwara H, et al. Phenotype-genotype correlation in CD36 deficiency types I and II. Thromb Haemost. 2000;84:436–41. [PubMed] [Google Scholar]
- 62.Kincer JF, Uittenbogaard A, Dressman J, Guerin TM, Febbraio M, Guo L, Smart EJ. Hyper-cholesterolemia promotes a CD36-dependent and endothelial nitric-oxide synthase-mediated vascular dysfunction. J Biol Chem. 2002;277:23525–33. doi: 10.1074/jbc.M202465200. [DOI] [PubMed] [Google Scholar]
- 63.Steinberg HO, Baron AD. Vascular function, insulin resistance and fatty acids. Diabetologia. 2002;45:623–34. doi: 10.1007/s00125-002-0800-2. [DOI] [PubMed] [Google Scholar]
- 64.Hayek T, Aviram M, Heinrich R, Sakhnini E, Keidar S. Losartan inhibits cellular uptake of oxidized LDL by monocyte-macrophages from hypercholesterolemic patients. Biochem Biophys Res Commun. 2000;273:417–20. doi: 10.1006/bbrc.2000.2963. [DOI] [PubMed] [Google Scholar]
- 65.Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, Pearce SF, Silverstein RL. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J Biol Chem. 1999;274:19055–62. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 66.Rać ME. CD36 receptor mutations and atherosclerosis. Prog Med Res. 2005;3:9. (on-line) [Google Scholar]
- 67.Miyaoka K, Kuwasako T, Hirano K, Nozaki S, Yamashita S, Matsuzawa Y. CD36 deficiency associated with insulin resistance. Lancet. 2001;357:686–7. doi: 10.1016/s0140-6736(00)04138-6. [DOI] [PubMed] [Google Scholar]
- 68.Kadlecova M, Cejka J, Zicha J, Kunes J. Does CD36 gene play a key role in disturbed glucose and fatty acid metabolism in Prague hypertensive hypertriglyceridemic rats? Physiol Res. 2004;53:265–71. [PubMed] [Google Scholar]
- 69.Glazier AM, Scott J, Aitman TJ. Molecular basis of the Cd36 chromosomal deletion underlying SHR defects in insulin action and fatty acid metabolism. Mamm Genome. 2002;13:108–113. doi: 10.1007/s00335-001-2132-9. [DOI] [PubMed] [Google Scholar]
- 70.Hsu LA, Ko YL, Wu S, et al. Association between a novel 11-base pair deletion mutation in the promoter region of the scavenger receptor class B type I gene and plasma HDL cholesterol levels in Taiwanese Chinese. Arterioscler Thromb Vasc Biol. 2003;23:1869–74. doi: 10.1161/01.ATV.0000082525.84814.A9. [DOI] [PubMed] [Google Scholar]
- 71.Huszar D, Varban ML, Rinninger F, et al. Increased LDL cholesterol and atherosclerosis in LDL receptor-deficient mice with attenuated expression of scavenger receptor B1. Arterioscler Thromb Vasc Biol. 2000;20:1068–73. doi: 10.1161/01.atv.20.4.1068. [DOI] [PubMed] [Google Scholar]
- 72.Calvo D, Gomez-Coronado D, Suarez Y, Lasuncion MA, Vega MA. Human CD36 is a high affinity receptor for the native lipoproteins HDL, LDL, and VLDL. J Lipid Res. 1998;39:777–88. [PubMed] [Google Scholar]
- 73.Nozaki S, Tanaka T, Yamashita S, et al. CD36 mediates long-chain fatty acid transport in human myocardium: complete myocardial accumulation defect of radiolabeled long-chain fatty acid analog in subjects with CD36 deficiency. Mol Cell Biochem. 1999;192:129–135. [PubMed] [Google Scholar]
- 74.Bonen A, Campbell SE, Benton CR, et al. Regulation of fatty acid transport by fatty acid translocase/CD36. Proc Nutr Soc. 2004;63:245–9. doi: 10.1079/PNS2004331. [DOI] [PubMed] [Google Scholar]
- 75.Ma X, Bacci S, Mlynarski W, et al. A common haplotype at the CD36 locus is associated with high free fatty acid levels and increased cardiovascular risk in Caucasians. Hum Mol Genet. 2004;13:2197–2205. doi: 10.1093/hmg/ddh233. [DOI] [PubMed] [Google Scholar]
- 76.Ibrahimi A, Abumrad NA. Role of CD36 in membrane transport of long-chain fatty acids. Curr Opin Clin Nutr Metab Care. 2002;5:139–45. doi: 10.1097/00075197-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 77.Kintaka T, Tanaka T, Imai M, Adachi I, Narabayashi I, Kitaura Y. CD36 genotype and long-chain fatty acid uptake in the heart. Circ J. 2002;66:819–25. doi: 10.1253/circj.66.819. [DOI] [PubMed] [Google Scholar]
- 78.Nozaki S, Tanaka T, Yamashita S, et al. CD36 mediates long-chain fatty acid transport in human myocardium: complete myocardial accumulation defect of radiolabeled long-chain fatty acid analog in subjects with CD36 deficiency. Mol Cell Biochem. 1999;192:129–35. [PubMed] [Google Scholar]
- 79.Teraguchi M, Ohkohchi H, Ikemoto Y, Higashino H, Kobayashi Y. CD36 deficiency and absent myocardial iodine-123-(R,S)-15-(p-iodophenyl)-3-methylpentadecanoic acid uptake in a girl with cardiomyopathy. Eur J Pediatr. 2003;162:264–6. doi: 10.1007/s00431-002-1118-2. [DOI] [PubMed] [Google Scholar]
- 80.Tanaka T, Sohmiya K, Kawamura K. Is CD36 deficiency an etiology of hereditary hypertrophic cardiomyopathy? J Mol Cell Cardiol. 1997;29:121–7. doi: 10.1006/jmcc.1996.0257. [DOI] [PubMed] [Google Scholar]
- 81.Okamoto F, Tanaka T, Sohmiya K, Kawamura K. CD36 abnormality and impaired myocardial long-chain fatty acid uptake in patients with hypertrophic cardiomyopathy. Jpn Circ J. 1998;62:499–504. doi: 10.1253/jcj.62.499. [DOI] [PubMed] [Google Scholar]
- 82.Kobylka D, Carraway KL. Proteolytic digestion of proteins of the milk fat globule membrane. Biochim Biophys Acta. 1973;307:133–40. doi: 10.1016/0005-2736(73)90031-x. [DOI] [PubMed] [Google Scholar]
- 83.Ge Y, Elghetany MT. CD36: a multiligand molecule. Lab Hematol. 2005;11:31–7. [PubMed] [Google Scholar]
- 84.Hajri T, Han XX, Bonen A, Abumrad NA. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. J Clin Invest. 2002;109:1381–9. doi: 10.1172/JCI14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tedgui A, Mallat Z. Platelets in atherosclerosis: a new role for beta-amyloid peptide beyond Alzheimer’s disease. Circ Res. 2002;90:1145–6. doi: 10.1161/01.res.0000023048.87638.92. [DOI] [PubMed] [Google Scholar]
- 86.Uittenbogaard A, Shaul PW, Yuhanna IS, Blair A, Smart EJ. High density lipoprotein prevents oxidized low density lipoprotein-induced inhibition of endothelial nitric-oxide synthase localization and activation in caveolae. J Biol Chem. 2000;275:11278–83. doi: 10.1074/jbc.275.15.11278. [DOI] [PubMed] [Google Scholar]
- 87.Febbraio M, Podrez EA, Smith JD, et al. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J Clin Invest. 2000;105:1049–56. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakata A, Nakagawa Y, Nishida M, et al. CD36, a novel receptor for oxidized low-density lipoproteins, is highly expressed on lipid-laden macrophages in human atherosclerotic aorta. Arterioscler Thromb Vasc Biol. 1999;19:1333–9. doi: 10.1161/01.atv.19.5.1333. [DOI] [PubMed] [Google Scholar]
- 89.Nakamura T, Saito Y, Ohyama Y, Uchiyama T, Sumino H, Kurabayashi M. Effect of cerivastatin on endothelial dysfunction and aortic CD36 expression in diabetic hyperlipidemic rats. Hypertens Res. 2004;27:589–98. doi: 10.1291/hypres.27.589. [DOI] [PubMed] [Google Scholar]
- 90.Ruiz-Velasco N, Dominguez A, Vega MA. Statins upregulate CD36 expression in human monocytes, an effect strengthened when combined with PPAR-gamma ligands Putative contribution of Rho GTPases in statin-induced CD36 expression. Biochem Pharmacol. 2004;67:303–13. doi: 10.1016/j.bcp.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 91.Puccetti L, Sawamura T, Pasqui AL, Pastorelli M, Auteri A, Bruni F. Atorvastatin reduces platelet-oxidized-LDL receptor expression in hypercholesterolaemic patients. Eur J Clin Invest. 2005;35:47–51. doi: 10.1111/j.1365-2362.2005.01446.x. [DOI] [PubMed] [Google Scholar]