Abstract
Biosynthesis of sucrose-6-P catalyzed by sucrose-phosphate synthase (SPS), and the presence of sucrose-phosphate phosphatase (SPP) leading to the formation of sucrose, have both been ascertained in a prokaryotic organism: Anabaena 7119, a filamentous heterocystic cyanobacterium. Two SPS activities (SPS-I and SPS-II) were isolated by ion-exchange chromatography and partially purified. Four remarkable differences between SPSs from Anabaena and those from higher plants were shown: substrate specificity, effect of divalent cations, native molecular mass, and oligomeric composition. Both SPS-I and SPS-II accept Fru-6-P (Km for SPS-I = 0.8 ± 0.1 mM; Km for SPS-II = 0.7 ± 0.1 mM) and UDP-Glc as substrates (Km for SPS-I = 1.3 ± 0.4 mM; Km for SPS-II = 4.6 ± 0.4 mM), but unlike higher plant enzymes, they are not specific for UDP-Glc. GDP-Glc and TDP-Glc are also SPS-I substrates (Km for GDP-Glc = 1.2 ± 0.2 mM and Km for TDP-Glc = 4.0 ± 0.4 mM), and ADP-Glc is used by SPS-II (Km for ADP-Glc = 5.7 ± 0.7 mM). SPS-I has an absolute dependence toward divalent metal ions (Mg2+ or Mn2+) for catalytic activity, not found in plants. A strikingly smaller native molecular mass (between 45 and 47 kDa) was determined by gel filtration for both SPSs, which, when submitted to SDS/PAGE, showed a monomeric composition. Cyanobacteria are, as far as the authors know, the most primitive organisms that are able to biosynthesize sucrose as higher plants do.
Keywords: cyanobacterium sucrose synthesis
The enzymes that catalyze sucrose biosynthesis and cleavage in higher plants were first reported by Cardini et al. (1) and Leloir and Cardini (2) in 1955. Sucrose-phosphate synthase (SPS, UDP-glucose: d-fructose-6-phosphate glucosyltransferase, EC 2.4.1.14), its specific phosphatase (SPP, sucrose-6-phosphate phosphohydrolase, EC 3.1.3.00), and sucrose synthase (SS, UDP-glucose: d-fructose-2-glucosyltransferase, EC 2.4.2.13) were isolated and partially purified from wheat germ. Since then, the presence of sucrose enzymes has been reported as being ubiquitous in higher plants (3, 4). The regulation of SPS and SS activities has been thoroughly studied (5, 6, 7). The sequences of their genes have also been established in several plant species (8, 9, 10). On the other hand, there is little information on the specific phosphatase, SPP. It was purified from pea shoots and rice leaves (11, 12).
The knowledge of sucrose metabolism in unicellular organisms is limited. Buchanan et al. (13) determined the presence of UDP-Glc in Chlorella during their studies on photosynthesis in 1953, suggesting its participation in sucrose synthesis. Nevertheless, the existence of sucrose enzymes was demonstrated in unicellular eukaryotic organisms only two decades later by Durán and Pontis (14). They partially purified SPS and SS from Chlorella vulgaris and Scenedesmus obliquus and studied their biochemical characteristics. Later, Salerno investigated the properties of both enzymes in permeabilized C. vulgaris and Prototheca zopfii cells (15, 16). In both approaches (14, 15, 16), the biochemical properties of SS and SPS were shown to be similar to those of higher plants. An additional report by Müller and Wegmann (17) studied the properties of SPS in Dunaliella tertiolecta, a halotolerant phytoplankton alga.
Exogenous sucrose could be metabolized by bacteria through invertase and by a unique species of Pseudomonas (Pseudomonas saccharophila) through sucrose phosphorylase (18). However, the biosynthesis of sucrose was not elucidated in photosynthetic prokaryotic organisms even when the accumulation of sucrose was shown in cyanobacteria as a response to salt stress (19, 20, 21, 22) and SS activity was measured in extracts of Anabaena variabilis (23).
In this paper the presence of SPS and SPP was ascertained in a prokaryotic organism, Anabaena 7119, a filamentous heterocystic cyanobacterium. Two SPSs were isolated and partially purified. Four remarkable differences between SPSs from Anabaena and those from higher plants were shown: substrate specificity, effect of divalent cations, native molecular mass, and oligomeric composition.
MATERIALS AND METHODS
Culture Conditions.
Anabaena strain 7119 was provided by R. A. Wolosiuk (Instituto de Investigaciones Bioquímicas, Fundación Campomar, Buenos Aires, Argentina). Cells were grown in BG-11 liquid medium (24). They were bubbled with air supplemented with 5% CO2 and illuminated by fluorescent light. Cells from late exponential phase batch cultures were harvested by centrifugation at 2500 × g, for 15 min. The pellet was washed twice with 10 mM Hepes·NaOH buffer, pH 7.5, and stored at −80°C.
Sucrose Determination.
Cells (1 g fresh weight) were lyophilized and extracted with 80% (vol/vol) ethanol at 80°C three times. Pooled extracts were evaporated, and the residue was suspended in water. The solution was purified by passage through Dowex 50 and IR-4B(OH) columns (BDH). Sucrose was estimated by measuring Fru and Glc after hydrolysis with invertase, as described (25).
Purification of SPS.
All steps were carried out at 0–4°C, except when stated otherwise. Packed cells (8 g fresh weight) were resuspended in twice their volume with 100 mM Hepes·NaOH buffer, pH 7.5, containing 2 mM EDTA, 20 mM MgCl2, 2% ethyleneglycol, 50 mM phenylmethylsulfonyl fluoride, 20 mM 2-mercaptoethanol, and 20% glycerol. Cells were broken by either passage through a French press at 25,000 psi or by mortar disruption in the presence of glass powder. Cell debris were removed by centrifugation at 30,000 × g for 30 min. The supernatant was centrifuged at 100,000 × g for 1 h and the pellet was discarded. This supernatant, referred to as crude extract, was absorbed onto a column of DEAE–Sephacel (1 × 20 cm) that had been preequilibrated with 20 mM Hepes·NaOH buffer, pH 6.7, containing 1 mM EDTA, 10 mM MgCl2, 5 mM 2-mercaptoethanol, and 20% glycerol. After washing, the enzyme was eluted with a linear NaCl gradient (total volume, 100 ml) 0 to 0.5 M in the same equilibrium buffer. Fractions with SPS activities were pooled and concentrated in an Amicon ultrafiltration cell. Two pools with SPS activity (SPS-I and SPS-II) were obtained. Both were further purified by gel filtration in a Sephadex G-100 column (2 × 60 cm) eluted with 50 mM Hepes·NaOH buffer, pH 7.5, containing 5 mM 2-mercaptoethanol, 10 mM MgCl2, and 20% glycerol. Peak fractions were pooled and concentrated in Centriflo membrane cones, type CF 25 (Amicon). These partially purified enzymes, free from SS, SPP, unspecific phosphatases, and phosphoglucose isomerase (PGI) activities, were used for further studies.
Enzyme Assays.
Sucrose enzyme activities were first determined in permeabilized Anabaena cells as described (15). In routine assays, SPS and SS activities were measured by quantifying sucrose-6-P or sucrose by the thiobarbituric acid (TBA) method (26). SPS and SS assay mixtures (50 μl) contained 10 mM Fru-6-P or Fru, respectively, 10 mM UDP-Glc, 10 mM MgCl2, 100 mM buffer (Hepes·NaOH, pH 7.0), and variable amounts of enzyme. The radioactive method using UDP-[14C]Glc, as described previously, was used in crude extract enzyme assays or for product identification (27). SPP and PGI activities were assayed as described (12, 25).
Product Identification.
Sucrose and sucrose-6-P were identified in aliquots of the reaction mixture using either UDP-[14C]Glc or XDP-Glc (X = A, C, or T) as substrates, by descending paper chromatography (14). The sugar-P chromatographed at the position of sucrose-6-P was alkali resistant, and when treated with alkaline phosphatase, it liberated sucrose. UDP was determined according to Stitt et al. (28).
Effect of Divalent Cations.
EDTA to a final concentration of 100 mM was added to aliquots of the Sephadex G-100 pools of SPS-I and SPS-II. Enzyme fractions were agitated in a bath at 0°C for 30 min and were exhaustively dialyzed in 10 mM Hepes·NaOH buffer, pH 7.0, containing 5 mM 2-mercaptoethanol and 25% glycerol.
Molecular Mass Estimation.
A Sephadex G-100 (1 × 60 cm) column equilibrated with 100 mM Hepes·NaOH buffer, pH 7.5, containing 100 mM NaCl and 5 mM 2-mercaptoethanol was calibrated with standard molecular markers: blue dextran (2 × 103 kDa), alcohol dehydrogenase (150 kDa), bovine albumin (66 kDa), and carbonic anhydrase (29 kDa). The position of each protein in the eluate was ascertained by measuring the optical density at 230 nm. The relative molecular masses of SPS-I and SPS-II were estimated by chromatographying an aliquot of the concentrate enzyme from the DEAE–Sephacel step. Elution was performed with the equilibrating buffer at a flow rate of 0.5 ml/h. Fractions of 0.25 ml were collected and sucrose enzymes were localized by their activity.
Polyacrylamide Gel Electrophoresis.
Proteins were separated by SDS/PAGE on 10% polyacrylamide (29) and stained with Coomassie blue. To ascertain the SPS polypeptide position, a gel was made in which half was stained with Coomassie blue and the other half was used for determining SPS activity after removal of SDS, according to Spanos and Hübscher (30). The activity was measured in gel sections (2-mm thick), as described (31), by both the TBA method (26) and the radioactive method (27).
Protein was measured by Bradford’s dye-binding assay using bovine serum albumin as a standard (32).
RESULTS
Presence of Sucrose and Sucrose Enzymes in Anabaena.
After determining the presence of sucrose in Anabaena cells harvested during late exponential growth phase (2 μmol/g fresh weight), a search for the enzymes involved in sucrose metabolism as reported in plants was carried out in toluene-permeabilized cells (15). SPS as well as SS activities were determined using UDP-[14C]Glc and fructose-6-P or fructose. The products [14C]sucrose-6-P and [14C]sucrose were identified by paper chromatography.
Isolation of SPS from Anabaena.
We focused our interest on the isolation and characterization of SPS, which, together with the specific phosphatase SPP, are considered responsible for sucrose synthesis in plants. Cell-free extracts were prepared for that purpose and chromatographed through a DEAE–Sephacel column. Fractions were analyzed for SPS and SPP activities. Two SPS activity peaks were eluted at 0.25 and 0.40 M NaCl (SPS-I and SPS-II, respectively) (Fig. 1). Each peak fraction was pooled and concentrated. Products of enzyme activities (UDP and sucrose-6-P) were identified as described in Materials and Methods in both SPS pools. SPS-I and SPS-II were further purified by gel filtration through a Sephadex G-100 column (Fig. 2). None of the two Sephadex SPS pools showed SS, SPP, or PGI activity. Later purification steps were not possible due to high lability of enzyme activity. That is why all the following studies were done using the enzymes from Sephadex pools. A summary of the purification procedure of both SPSs corresponding to an average preparation is shown in Table 1.
Figure 1.
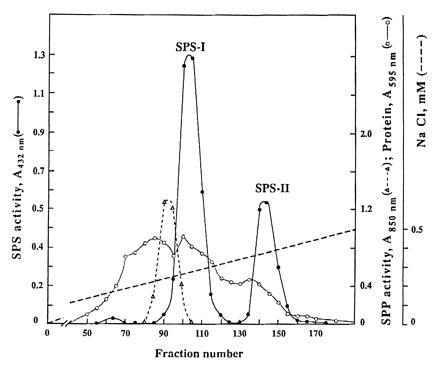
DEAE–Sephacel chromatography of crude extracts from Anabaena 7119. SPS (•) and SPP (▴) activities. Protein (○). The broken line represents the NaCl gradient.
Figure 2.
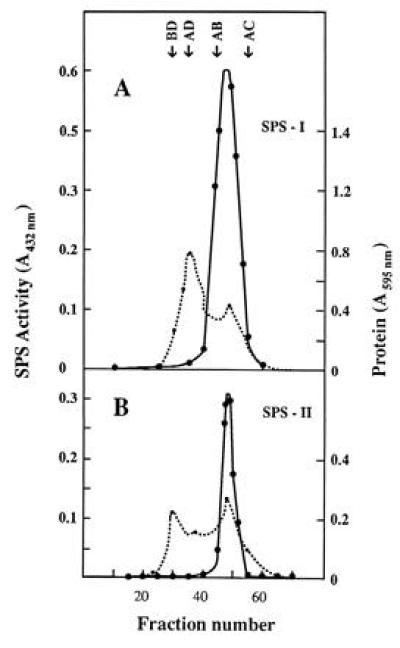
Sephadex G-100 chromatography of SPS-I (A) and SPS-II (B) pools. SPS activity (•——•) and protein (• - - - •). The arrows indicate the relative position of standard markers: BD, blue dextran (2 × 103 kDa); AD, alcohol dehydrogenase (150 kDa); AB, bovine albumin (66 kDa); AC, carbonic anhydrase (29 kDa).
Table 1.
Partial purification of sucrose-phosphate synthase from Anabaena 7119
| Steps | Total protein, mg | Total activity, nkat* | Recovery, % | Specific activity, nkat/mg | Purification, -fold |
|---|---|---|---|---|---|
| Crude extract | 92.0 | 2.80 | 100 | 0.03 | 1 |
| DEAE–Sephacel (SPS-I) | 6.5 | 2.10 | 75 | 0.32 | 10 |
| Sephadex G-100 (SPS-I) | 0.9 | 0.58 | 20 (27) | 0.64 | 21 |
| DEAE–Sephacel (SPS-II) | 1.83 | 0.74 | 26 | 0.40 | 13 |
| Sephadex G-100 (SPS-II) | 0.12 | 0.50 | 17 (67) | 4.17 | 139 |
Percentages in parentheses indicate recovery from previous step.
Amount of enzyme that converts 1 nmol of substrate to product in 1 sec.
SPS activity present in the homogenate was recovered in the two SPS peaks of the DEAE–Sephacel column. Total SPS-I activity at this step was three times higher than in the case of SPS-II. Conversely, the recovery and specific activity of SPS-II after Sephadex G-100 filtration were higher than in SPS-I.
Substrate Specificity and Kinetic Constants.
Specificity for the glucosyl donor was investigated for both SPSs. Unlike higher plant enzymes, SPSs from Anabaena were not specific for UDP-Glc. They also used GDP-Glc and TDP-Glc (SPS-I substrates) and ADP-Glc (SPS-II substrate). The identification of the product sucrose-6-P was ascertained in each case. The apparent Km and Vmax values from Wolf’s plots are summarized in Table 2. SPS-I has 3- to 4-fold more affinity for UDP-Glc than SPS-II.
Table 2.
Kinetic constants calculated for Anabaena sucrose-phosphate synthases
| Substrate | SPS-I
|
SPS-II
|
||
|---|---|---|---|---|
| Kmapp, mM | Vmax, nkat·ml−1 | Kmapp, mM | Vmax, nkat·ml−1 | |
| Fru-6-P* | 0.8 ± 0.1 | 4.87 ± 0.90 | 1.1 ± 0.4 | 8.0 ± 0.9 |
| UDP-Glc | 1.3 ± 0.4 | 5.30 ± 0.52 | 4.3 ± 0.4 | 8.5 ± 1.5 |
| ADP-Glc | ND | ND | 4.9 ± 0.9 | 3.2 ± 0.4 |
| GDP-Glc | 1.2 ± 0.2 | 4.53 ± 0.80 | ND | ND |
| TDP-Glc | 4.0 ± 0.4 | 0.25 ± 0.04 | ND | ND |
ND, not detected.
(+UDP-Glc).
Effect of Divalent Cations.
Another significant difference between Anabaena and eukaryotic SPS was that SPS catalytic activity was highly dependent on the presence of a divalent metal ion (Mg2+ or Mn2+) in the incubation mixture. After EDTA treatment, the prokaryotic enzyme was practically inactive. The addition of 10 mM of divalent ions (Mn2+ and Mg2+) restored the activity to control values. Furthermore, at equivalent concentration of cations (10 mM), SPS activity was 1.5- to 2-fold higher in the presence of Mn2+ than in Mg2+. The calculated activation constants for Mn2+ as well as Mg2+ for both SPSs ranged from 3 to 5 mM.
Molecular Mass of Anabaena SPSs.
Relative molecular masses of ≈46,000 ± 1000 Da were determined by gel filtration chromatography for both SPS-I and SPS-II. An analysis of polypeptide composition of SPSs was performed by submitting aliquots of the Sephadex pools to denaturation conditions, and the polypeptides were separated by SDS/PAGE. Stained polypeptides are shown in Fig. 3A. Sephadex SPS-I pool presented several polypeptides (Fig. 3A, lane 3). A major band of ≈47 kDa and a minor one of smaller molecular mass are shown in the Sephadex SPS-II pool (Fig. 3A, lane 4). SPS activity measured in the parallel lanes of the second half of the gel was detected in the position corresponding to 45–47 kDa. Fig. 3B shows SPS-II activity. A similar pattern was obtained with the SPS-I pool (data not shown).
Figure 3.
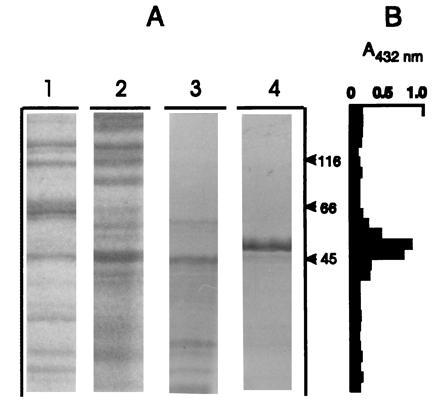
SDS/PAGE of Anabaena SPS purification steps. (A) Coomassie blue staining. Lanes: 1, DEAE–Sephacel SPS-I pool; 2, DEAE–Sephacel SPS-II pool; 3, Sephadex G-100 SPS-I pool; 4, Sephadex G-100 SPS-II pool. (B) SPS activity determined by using the TBA method.
DISCUSSION
The outcome of the present study is the first evidence of the presence of SPS and SPP activities in a prokaryotic organism. The data presented herein indicate that these enzymes may be considered responsible for sucrose biosynthesis in Anabaena 7119 as well as in higher plants. In an earlier report, Schilling and Ehrnsperger (23), investigating the biosynthesis and degradation of sucrose in cell- free extracts from A. variabilis, were only able to detect SS activity mainly in vegetative cells. According to their report, SS might be responsible for sucrose synthesis. Additionally, a sucrose-cleaving activity was attributed to an alkaline invertase (23).
However, unlike the previously cited report, we were able to demonstrate the presence of SPS and SPP in Anabaena 7119. As a consequence, we might conclude that these could be the enzymes that synthesize sucrose in cyanobacteria.
The central role of SPS in the regulation of sucrose biosynthesis led us to focus our studies on the determination of the biochemical characteristics of SPS in Anabaena. Attempts to purify SPSs to homogeneity have been unsuccessful. Instability in low protein concentration solutions could be the major impediment to this goal. This is a general feature observed in the purification of SPS from leaves of various plant species (33, 34). SPS-I was not extensively purified because it was accompanied by the bulk of the proteins; however, SPS-II eluted in a NaCl concentration where there was a small amount of protein (Figs. 1 and 3A, lanes 1–4). After Sephadex G-100 filtration, SPS-I was purified 21-fold and SPS-II, 139-fold (Table 1). These partially purified enzymes, free from SS, SPP, unspecific phosphatases, and PGI activities, were used to study their biochemichal properties. The two SPSs isolated from ion exchange chromatography (SPS-I and SPS-II) clearly show four differences from the enzymes of algae and higher plants: sugar nucleotide specificity, high dependence on divalent ions for activity, relative native molecular mass, and oligomeric composition.
Anabaena SPSs are not specific for UDP-Glc. GDP-Glc and TDP-Glc are also SPS-I substrates. ADP-Glc is also used by SPS-II (Table 2). SS is the only known enzyme among the many that catalyze sugar nucleotide glucosyl transfer that can use various sugar nucleotides as substrates (3, 4). In the case of SS from plants and algae, the affinity for each sugar nucleotide is variable, and in general, the Km for UDP-Glc is smaller (3, 4). SPS-I from Anabaena showed similar higher affinities for UDP-Glc and GDP-Glc than for TDP-Glc. SPS-II evidenced no difference in affinities for UDP-Glc and ADP-Glc (Table 2). Little is known about the carbohydrate metabolism of cyanobacteria. The idea that each sugar nucleotide may be used under different physiological situations or is part of different pathways is apt to be under speculation.
SPS-I showed an absolute requirement for the presence of Mn2+ or Mg2+ for catalytic activity. On the contrary, higher plant SPSs are activated at different degrees by those metal ions but enzyme activities do not depend on their presence (3, 4). Similar behavior was found for SPS-II.
The difference in native molecular mass between Anabaena and eukaryotic SPSs is quite remarkable. While SPSs from higher plants and algae were reported to have molecular masses ranging from 260,000 to 520,000 Da (33, 34), the apparent molecular masses determined by gel filtration for both Anabaena SPSs were 5 to 10 times smaller (≈47,000 Da) (Fig. 3).
The analysis of Anabaena SPS polypeptide composition by SDS/PAGE showed SPS activity in the position of 45–47 kDa (Fig. 3B). Thus, SPS is composed by a unique polypeptide chain. It is the first SPS described having a monomeric composition. In cyanobacteria exposed to salt stress, several small organic molecules accumulate as osmotic solutes, including sucrose (20). As mentioned, sucrose is also present in nonstressed culture conditions. It should be noted that cyanobacteria are, as far as the authors know, the most primitive organisms in the chain of evolution in which sucrose biosynthesis takes place as in higher plants. This paper demonstrates that these prokaryotic cells contain ancestral sucrose enzymes. The important unanswered question that we have been asking ourselves for several years is why and when sucrose metabolism appeared in nature. This crucial metabolism could have originated in cyanobacteria 3 billion years ago, due to the wide spectrum of environmental stresses to which cyanobacteria had to adapt, and/or to the appearance of new photosynthetic events as known in higher plants, and/or to the need of a carbon carrier molecule from vegetative cells and heterocysts (23). Further studies on cyanobacteria SPS proteins will give a better insight into the origin and evolution of this key enzyme.
Acknowledgments
We are grateful to Prof. H. G. Pontis for stimulating discussions and to Ms. Clara Fernández and Carmen Rodríguez for their able technical assistance. G.L.S. is a Career Investigator of the Consejo Nacional de Investigaciones Científicas y Tecnológicas (Argentina). This work is part of the Ph.D. thesis of A.C.P. (Universidad Nacional de Mar del Plata). This work was supported in part by grants from Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina and Fundación para Investigaciones Biológicas Aplicadas.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviations: SPP, sucrose-phosphate phosphatase; SPS, sucrose-phosphate synthase; SS sucrose synthase; TBA, thiobarbituric acid; PGI, phosphoglucose isomerase.
References
- 1.Cardini C E, Leloir L F, Chiriboga J. J Biol Chem. 1955;214:149–155. [PubMed] [Google Scholar]
- 2.Leloir L F, Cardini C E. J Biol Chem. 1955;214:157–165. [PubMed] [Google Scholar]
- 3.Pontis H G. In: Plant Biochemistry II. Northcote D H, editor. Vol. 13. Baltimore: University Press; 1977. pp. 79–117. [Google Scholar]
- 4.Avigad G. In: Encyclopedia of Plant Physiology. Loewus F A, Tanner W, editors. 13A. Berlin: Springer; 1982. pp. 217–347. [Google Scholar]
- 5.Wolosiuk R A, Pontis H G. Mol Cell Biochem. 1974;4:115–123. doi: 10.1007/BF01770292. [DOI] [PubMed] [Google Scholar]
- 6.Doehlert D C, Huber S C. Plant Physiol. 1983;73:989–994. doi: 10.1104/pp.73.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stitt M, Huber S C, Kerr P. In: The Biochemistry of Plants. Preiss J, editor. Vol. 10. New York: Academic; 1987. pp. 327–409. [Google Scholar]
- 8.Geiser M, Doring H P, Wostemeyer J, Behrens U, Tillman E, Starlinger P. Nucleic Acids Res. 1980;8:6175–6188. doi: 10.1093/nar/8.24.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worrell A C, Bruneau J-M, Summerfelt K, Boersig M, Voelker T A. Plant Cell. 1991;3:1121–1130. doi: 10.1105/tpc.3.10.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein R R, Crafts-Brandner S J, Salvucci M E. Planta. 1993;190:498–510. doi: 10.1007/BF00224789. [DOI] [PubMed] [Google Scholar]
- 11.Whitaker D P. Phytochemistry. 1984;23:2429–2430. [Google Scholar]
- 12.Echeverría E, Salerno G. Plant Sci. 1994;96:15–19. [Google Scholar]
- 13.Buchanan J G, Lynch V H, Benson A A, Bradley D F, Calvin M. J Biol Chem. 1953;203:935–945. doi: 10.2172/915412. [DOI] [PubMed] [Google Scholar]
- 14.Durán W R, Pontis H G. Mol Cell Biochem. 1977;16:149–152. doi: 10.1007/BF01732056. [DOI] [PubMed] [Google Scholar]
- 15.Salerno G L. Physiol Plant. 1985;64:259–264. [Google Scholar]
- 16.Salerno G L. Plant Sci. 1985;42:5–8. [Google Scholar]
- 17.Müller W, Wegmann K. Planta. 1978;141:159–163. doi: 10.1007/BF00387883. [DOI] [PubMed] [Google Scholar]
- 18.Doudoroff M, Kaplan N O, Hassid W Z. J Biol Chem. 1943;148:67–75. [Google Scholar]
- 19.Warr S R C, Reed R H, Stewart W D P. New Phytol. 1985;100:285–292. [Google Scholar]
- 20.Reed R H, Borowitzka L J, Mackay M A, Chudek J A, Foster R, Warr S R C. FEMS Microbiol Rev. 1986;39:51–56. [Google Scholar]
- 21.Stal L, Reed R H. FEMS Microbiol Ecol. 1987;45:305–312. [Google Scholar]
- 22.Hershkovitz N, Oren A, Cohen Y. Appl Environ Microbiol. 1991;57:645–648. doi: 10.1128/aem.57.3.645-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schilling N, Ehrnsperger K. Z Naturforsch C. 1985;40:776–779. [Google Scholar]
- 24.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 25.Pontis H G, Babio J R, Salerno G L. Proc Natl Acad Sci USA. 1981;78:6667–6669. doi: 10.1073/pnas.78.11.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Percheron F. C R Acad Sci Paris. 1962;255:2521–2522. [Google Scholar]
- 27.Salerno G L, Gamundi S S, Pontis H G. Anal Biochem. 1979;93:196–200. [PubMed] [Google Scholar]
- 28.Stitt M, Wilke I, Feil R, Heldt H W. Planta. 1988;174:217–230. doi: 10.1007/BF00394774. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Spanos A, Hübscher U. Methods Enzymol. 1983;91:263–277. doi: 10.1016/s0076-6879(83)91024-8. [DOI] [PubMed] [Google Scholar]
- 31.Salerno G L, Crespi M D, Zabaleta E J, Pontis H G. Physiol Plant. 1991;81:541–547. doi: 10.1104/pp.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 33.Salvucci M E, Drake R R, Haley B E. Arch Biochem Biophys. 1990;281:212–218. doi: 10.1016/0003-9861(90)90434-z. [DOI] [PubMed] [Google Scholar]
- 34.Bruneau J-M, Worrell A C, Cambou B, Lando D, Volker T A. Plant Physiol. 1991;96:473–478. doi: 10.1104/pp.96.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]


