Short abstract
Although, the S. cerevisiae genome is rightly considered as one of the most accurately sequenced and annotated eukaryotic genomes, it still benefits substantially from comparison to the completed sequence and syntenic gene map of Ashbya gossypii, an evolutionarily related fungus.
Abstract
Background
The recently sequenced genome of the filamentous fungus Ashbya gossypii revealed remarkable similarities to that of the budding yeast Saccharomyces cerevisiae both at the level of homology and synteny (conservation of gene order). Thus, it became possible to reinvestigate the S. cerevisiae genome in the syntenic regions leading to an improved annotation.
Results
We have identified 23 novel S. cerevisiae open reading frames (ORFs) as syntenic homologs of A. gossypii genes; for all but one, homologs are present in other eukaryotes including humans. Other comparisons identified 13 overlooked introns and suggested 69 potential sequence corrections resulting in ORF extensions or ORF fusions with improved homology to the syntenic A. gossypii homologs. Of the proposed corrections, 25 were tested and confirmed by resequencing. In addition, homologs of nearly 1,000 S. cerevisiae ORFs, presently annotated as hypothetical, were found in A. gossypii at syntenic positions and can therefore be considered as authentic genes. Finally, we suggest that over 400 S. cerevisiae ORFs that overlap other ORFs in S. cerevisiae and for which no homolog can be detected in A. gossypii should be regarded as spurious.
Conclusions
Although, the S. cerevisiae genome is rightly considered as one of the most accurately sequenced and annotated eukaryotic genomes, we have shown that it still benefits substantially from comparison to the completed sequence and syntenic gene map of A. gossypii, an evolutionarily related fungus. This type of approach will strongly support the annotation of more complex genomes such as the human and murine genomes.
Background
A major breakthrough in the field of genomics came with the publication of the 13 Mb genome of the budding yeast Saccharomyces cerevisiae [1], which was the first eukaryotic genome to be fully sequenced and annotated. Since then, DNA sequencing has developed with an increasing speed, and sequences of much larger genomes, such as those of Caenorhabditis elegans [2], Drosophila melanogaster [3], Arabidopsis thaliana [4]Homo sapiens [5,6], Anopheles gambiae [7] and Mus musculus [8] have been published. However, increased sequencing capacity was not matched by a corresponding development in annotation and the gene annotation process is now the rate-limiting step in whole-genome sequencing projects. Despite progress in gene prediction programs, comparisons to expressed sequence tag (EST) databases and to genomic sequences, preferably of related organisms, is still the most favored approach to the annotation of complex genomes.
The original annotation of the S. cerevisiae genome was especially challenging because, at the time of its completion [9], only limited genomic sequence information from other eukaryotes was available. Despite the functional characterization of a large number of orphan open reading frames (ORFs) and several efforts to re-evaluate the sequence at the gene level or for an entire chromosome [10,11], a significant number of uncertainties still remain. It is, for example, not known whether all protein-coding genes have been identified and which of the close to 2,000 genes annotated as hypothetical represent real genes. A careful comparison to a related genome should help clarify several of these issues.
The recently completed genome sequence of the filamentous ascomycete Ashbya gossypii revealed an unexpected high degree of gene homology and gene order conservation with S. cerevisiae (F.S.D., S.V., S.B., A.L., K.G., C. Mohr, S. Steiner, P. Luedi, T.G. and P.P., unpublished work). The two species diverged more than 100 million years ago, and both genomes differ substantially in their GC content (38.3% in S. cerevisiae and 51.9% in A. gossypii). However, 95% of the 4,700 A. gossypii protein-coding genes were found to have a homolog in S. cerevisiae and 90% of these homologous genes map at syntenic positions. Despite these striking genomic similarities, the average conservation at the DNA level is 55% in coding regions but drops to 33% in noncoding regions. Thus, significant sequence similarities are restricted to coding regions. Altogether, these findings open up the possibility of a whole-genome reinvestigation of the S. cerevisiae annotation.
We carried out an extensive search for homology at the amino-acid level between A. gossypii coding regions and S. cerevisiae 'annotation-free' regions: stretches of sequence bearing no annotated genomic features such as ORFs, RNA genes, or transposable elements. Focusing on syntenic regions, we identified a total of 95 inconsistencies, suggesting the following four types of changes in the S. cerevisiae annotation: novel genes, novel introns, potential ORF extensions, and neighboring ORF fusions. Furthermore, we provide evidence that information from the complete A. gossypii genome is also a major resource for recognizing real genes among the numerous S. cerevisiae hypothetical ORFs.
Results and discussion
We searched for homology at the amino-acid level between annotated A. gossypii coding regions and S. cerevisiae 'annotation-free regions'. As a result, we identified 95 regions in the S. cerevisiae genome, which had not been annotated as protein coding, that showed both homology and synteny to A. gossypii genomic sequences. In this context, synteny refers to a relaxed synteny (loose synteny), which results from several hundred genomic rearrangements in the A. gossypii and S. cerevisiae lineages and from frequent loss of one of the two gene copies (twin genes) in S. cerevisiae after the proposed doubling of the genome [12,13]. As a result, all remaining duplicated genes in S. cerevisiae have a single homolog in A. gossypii (F.S.D., S.V., S.B., A.L., K.G., C. Mohr, S. Steiner, P. Luedi, T.G. and P.P., unpublished work). On close inspection of these 95 S. cerevisiae syntenic loci, we found evidence for novel ORFs, and for substantial boundary changes of annotated ORFs. Figure 1 outlines the categories of changes suggested by this comparative genomics approach.
Figure 1.
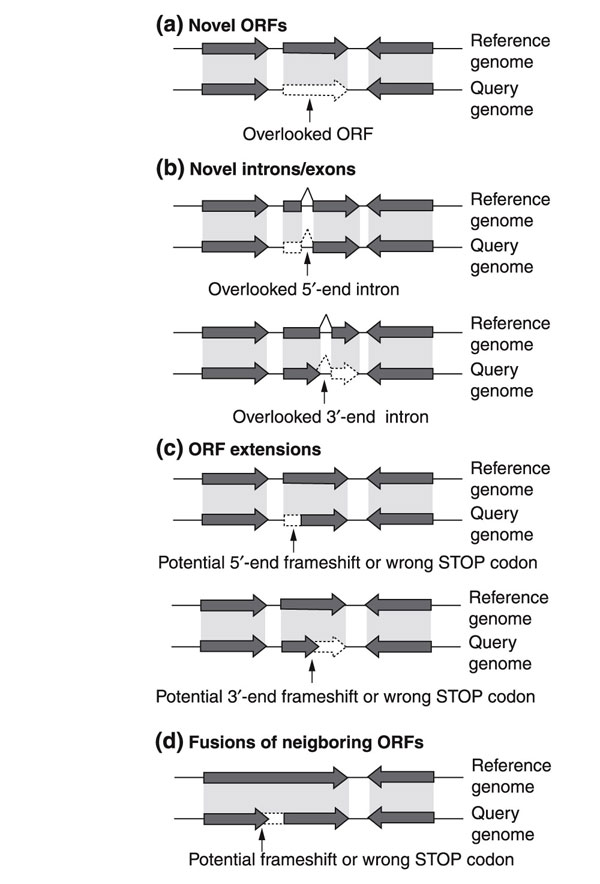
Genome reinvestigation using comparative genomics. Translated DNA comparison allows the detection of homology outside annotated features of a query genome. This can lead to the detection of (a) novel ORFs, (b) novel introns/exons, (c) ORF extensions, and (d) ORF fusions (fusion of adjacent ORFs or merging of overlapping ORFs with same transcription direction). Gray areas illustrate regions of homology at the protein level and dashed lines depict suggested modifications in the query genome. Cases of changed splice sites may also be detected but are not drawn here.
We first present data supporting novel protein-coding genes and provide detailed analysis of the different types of boundary changes of annotated ORFs due to novel exons, 5'- or 3'-end extension, or even fusion of adjacent ORFs. Second, we will focus on the validation of the approximately 2,000 hypothetical ORFs. We will present evidence that 50% of these hypothetical ORFs are real, and provide arguments to consider several hundred as probably spurious.
Novel ORFs
In 23 annotation-free regions, we discovered homology to syntenic small A. gossypii ORFs as outlined in Figure 1a and summarized in Table 1. These presumptive novel S. cerevisiae ORFs are 52 to 134 codons long. Twenty have a size below 100 codons, the arbitrary cut-off for small and nonhomologous yeast ORF annotation, several contain an intron, and one contains two introns. The short length and the presence of introns explain why these ORFs remained so far undiscovered. An additional example of a novel S. cerevisiae ORF identification by comparison to the A. gossypii genome was recently published [14].
Table 1.
Novel S. cerevisiae ORFs identified by homology and synteny to A. gossypii ORFs
| A. gossypii ORF | Novel S. cerevisiae ORF(s) | % Similarity | Homologs* | |||||
| Name | Size | Name(s) | Size(s) | Hemiascomycetes | C. albicans | S. pombe | N. crassa | |
| AGL322W | 57 | YBL039W-A | 59 | 53.70 | x | |||
| ADL343C | 99 | YBR111W-A | 98 | 65.28 | x | x | ||
| AAL005W | 72 | YCL005W-A | 73 | 79.17 | x | x | x | |
| AFR743C-A | 132 | YCL058W-A | 132 | 45.00 | x | |||
| AER271W | 81 | YCR075W-A/YNR034W-A† | 75/99 | 44.00/46.15 | x | |||
| ACL158W | 95 | YDL160C-A | 80 | 55.70 | x | |||
| AGR097W-A | 66 | YER180C-A | 85 | 83.70 | x | |||
| AFR298C | 73 | YIL156W-B | 73 | 71.23 | x | x | x | |
| AFL216C | 49 | YJL127C-B | 52 | 61.22 | x | x | ||
| AAL130W | 93 | YJR005C-A/YGR169C-A† | 92/93 | 74.44/50.82 | x | x | ||
| ABR148C-A | 117 | YJR112W-A | 109 | 56.44 | x | x | x | |
| ABR099W-A | 73 | YKL068W-A | 78 | 33.33 | x | |||
| AFR204W | 68 | YKL138C-A | 68 | 57.35 | x | |||
| AFR289W | 81 | YKR095W-A | 83 | 58.75 | x | x | x | |
| ADL036W-A | 75 | YLR307C-A | 87 | 50.00 | ||||
| AFL165W | 94 | YMR194C-B | 73 | 51.47 | x | x | x | x |
| ADL210W | 72 | YNL024C-A | 72 | 81.94 | x | x | x | x |
| AFR059W | 84 | YNL138W-A | 85 | 52.38 | x | |||
| AAR108W-A | 70 | YOR020W-A | 90 | 35.71 | x | x | ||
| ABR192C-A | 71 | YPL096C-A | 68 | 45.59 | x | |||
| AFL069C | 58 | YPL189C-A | 68 | 69.64 | x | x | ||
| ACR178C | 65 | YPR036W-A | 67 | 51.06 | x | x | ||
| AEL262C-A | 86 | YPR170W-B | 85 | 85.88 | x | x | x | |
We carried out homology searches for all novel ORFs against the available fungal databases. This analysis revealed that all but one of the novel ORFs are present in hemiascomycetes and for 15 ORFs, homologs were found in at least two of the following databases: hemiascomycetes, Candida albicans, Schizosaccharomyces pombe and Neurospora crassa (Table 1). This suggests that they represent conserved fungal proteins. For two genes, YMR194C-B and YNL024C-A, we identified homologs in higher eukaryotes, including mouse and human. The conservation in other species, and particularly their syntenic positions in A. gossypii, strongly support the authenticity of these novel genes.
We screened the 23 protein sequences for the presence of known domains but did not find any significant hits. These novel S. cerevisiae ORFs were not deleted by the yeast gene deletion consortium [15] but the A. gossypii homologs of YIL156W-B, YJL127C-B, YMR194C-B and YNL138W-A have been deleted (K.G. and T.G., unpublished data). One deletion is lethal; the others did not exhibit any apparent phenotype under normal growth conditions. Recently, two of the novel ORFs - YPL096C-A (ERI1, ER-associated Ras Inhibitor 1) and YKL138C-A (HSK3, Helper of Ask1) - were added to the Saccharomyces Genome Database (SGD) as reserved gene names, indicating unpublished functional data, and one novel ORF, YPR036W-A, was shown to be expressed in response to drug treatment [16].
A similar approach based on the so-called Génolevures project, a partial shotgun sequencing of 13 hemiascomycete genomes [17], suggested the presence of 50 overlooked ORFs in S. cerevisiae, distinct from the set of 23 described in this paper. These 50 ORFs were recently incorporated in the SGD. Arguing that the species under consideration were too closely related, Wood et al. [18] recommended further investigations before considering these 50 novel ORFs as real. Having the A. gossypii genome to hand allowed us to evaluate the authenticity of these proposed ORFs. Indeed, we found 20 of the suggested novel ORFs at syntenic positions (see Additional data files). Similarly, the comparison between the S. pombe and the S. cerevisiae genome annotations identified three additional novel S. cerevisiae ORFs [18,19], distinct from the 23 ORFs discussed above. All three correspond to syntenic homologs in A. gossypii, which confirms the assumption that they are real ORFs. More recently, 84 S. cerevisiae small ORFs, called smORFs, were identified on the basis of homology to a larger fungal database and experimental evidence for transcription products [20]. Upon re-evaluation, we found that five smORFs correspond to novel ORFs described here and five others match sequences of ORF extensions as discussed below. Several smORFs correspond to RNA genes or match the opposite strand of previously annotated ORFs in both S. cerevisiae and A. gossypii and thus do not represent protein-coding genes. For the remaining smORFs, there were no homologs found in A. gossypii.
Novel introns and exons
Splicing rules and intron positions are generally conserved in A. gossypii and S. cerevisiae. On this basis we were able to identify 13 cases of probably overlooked introns in S. cerevisiae, as schematically represented in Figure 1b. Splicing of the novel introns and fusion of the novel exon extend the S. cerevisiae ORFs up to 236 codons and lead in most cases to substantially increased similarity between homologs of the two species (data not shown). The ORFs under consideration, the overall size increases, and other supporting evidence are shown in Table 2, which summarizes all 72 ORF extensions. Perfect splice consensus sequences were found for only three genes, which explains the difficulty in recognizing these introns. Finally, for one gene, SEF1 (YBL066C), we propose a base-pair change in addition to an intron. We tested the authenticity of the proposed introns for YKR004C (ECM9), YML017W (PSP2) and YOL048C using 5' rapid amplification of cDNA ends (5' RACE). In all three cases, the intron could be confirmed by sequencing the cDNA obtained (AY245791, AY245792, and AY245793). cDNA and genomic sequence alignment confirms that the intron of YKR004C is spliced at perfect consensus splice sequences and that both YML017W and YOL048C bear non-consensus splice sites with the respective acceptor/donor sites GTATGT--CACTAAC--CAG and GTAAGT--GACTAAC--TAG. In three cases of novel introns - YBL091C-A, YHR079C-A and YOL048C - splicing has already been proposed by either Blandin et al. [17] or Wood et al. [18].
Table 2.
Summary of different types of ORF extensions proposed for annotated S. cerevisiae ORFs
| S. cerevisiae | Type* | Proposed reason† | Extension size (codons) | Supporting evidence‡ | ||||
| Hemiascomycetes | C. albicans | S. pombe | N. crassa | S. cerevisiae§ | ||||
| YAL013W | 3' extension | FS | 57 | ++ | ++ | ++ | Resequenced | |
| YAR044W/YAR042W | Fusion | FS | ++ | ++ | ++ | ++ | Resequenced; duplication | |
| YBL066C | 3' intron | Intron + FS | 3 | + | + | |||
| YBL091C-A | 5' intron | Intron | 76 | ++ | ++ | ++ | ||
| YBL104C | 5' and 3' extensions | FS | 128 | ++ | ++ | ++ | Resequenced | |
| YBR041W | 3' extension | FS | 46 | ++ | ++ | ++ | Resequenced | |
| YBR074W/YBR075W | Fusion | 2FS | + | + | + | + | Resequenced | |
| YBR098W/YBR100W | Fusion | FS | ++ | ++ | + | |||
| YBR157C | 3' extension | 1FS | 149 | ++ | Resequenced | |||
| YCL001W-A/YCL001W-B | Fusion | 2FS | Duplication | |||||
| YCL008C | 3' extension | 1FS | 89 | Resequenced [45] | ||||
| YCL025C | 3' extension | FS | 38 | ++ | ++ | + | ++ | gb: P25376; duplication |
| YCL069W | 5' extension | PS | 124 | ++ | ++ | ++ | ++ | |
| YDL115C | 5' intron | Intron | 113 | ++ | ++ | |||
| YDR179W-A | 5' extension | FS | 138 | ++ | ++ | gb: CAA86685 | ||
| YDR474C/YDR475C | Fusion | 2FS | ++ | ++ | Duplication | |||
| YDR494W | 3' extension | FS | 78 | ++ | ++ | |||
| YER039C/YER039C-A | Fusion | PS | ++ | ++ | ++ | ++ | Duplication | |
| YER066W | 5' extension | PS | 201 | ++ | ++ | ++ | ++ | |
| YFL007W/YFL006W | Fusion | FS | ++ | ++ | ++ | [46] | ||
| YFR038W | 3' extension | FS | 75 | ++ | ++ | ++ | ||
| YFR040W | 5' extension | FS | 97 | ++ | Duplication | |||
| YFR045W | 5' extension | FS + PS | 106 | ++ | ++ | ++ | ++ | [47] |
| YGL046W/YGL045W | Fusion | 3FS | ++ | ++ | + | |||
| YGL059W | 3' extension | FS | 46 | ++ | ++ | ++ | ||
| YGL183C | 5' intron | Intron | 45 | ++ | ++ | |||
| YGL211W | 3' extension | 3FS | 164 | ++ | ++ | ++ | ++ | |
| YGR006W | 3' extension | FS | 32 | ++ | ++ | ++ | [48] | |
| YGR225W | 3' extension | FS | 31 | ++ | ++ | ++ | ++ | [49] |
| YGR272C/YGR271C-A | Fusion | FS | ++ | ++ | ||||
| YHR056C | 5' extension | FS | 51 | gb: P38781; duplication | ||||
| YHR079C-A | 3' intron | Intron | 45 | ++ | ++ | ++ | ++ | |
| YHR176W | 3' extension | FS | 67 | ++ | ++ | ++ | ||
| YJL012C/YJL012C-A | Fusion | FS | ++ | ++ | ++ | Resequenced | ||
| YJL017W/YJL016W | Fusion | FS | ++ | Resequenced | ||||
| YJL019W/YJL018W | Fusion | FS | Resequenced | |||||
| YJL020C/YJL021C | Fusion | FS | Resequenced | |||||
| YJL031C | 5' intron | Intron | 37 | ++ | ++ | ++ | ++ | |
| YJL108C/YJL107C | Fusion | FS | ++ | ++ | ++ | ++ | Resequenced | |
| YJL159W | 3' extension | FS | 4 | ++ | ++ | Resequenced; duplication | ||
| YJL160C | 3' extension | FS | 107 | ++ | ++ | Resequenced; duplication | ||
| YJL178C | 5' extension | FS | 75 | ++ | ++ | Resequenced | ||
| YJR013W | 5' extension | FS | 98 | ++ | ++ | ++ | ++ | Resequenced |
| YKL033W-A | 3' extension | FS | 176 | ++ | ++ | ++ | Resequenced | |
| YKL199C/YKL198C | Fusion | FS | ++ | ++ | ++ | ++ | Resequenced; duplication | |
| YKL207W | 5' extension | FS | 22 | ++ | ++ | ++ | Resequenced | |
| YKR004C | 5' intron | Intron | 85 | 5' RACE verified | ||||
| YKR056W | 5' extension | FS | 21 | ++ | ++ | ++ | ++ | Resequenced |
| YKR058W | 5' extension | 2FS | 138 | ++ | ++ | Resequenced [50]; duplication | ||
| YKR100C | 3' extension | FS | 114 | ++ | Duplication | |||
| YKR103W/YKR104W | Fusion | PS | ++ | ++ | ++ | ++ | ||
| YLL017W/YLL016W | Fusion | FS | + | Duplication | ||||
| YLL052C/YLL053C | Fusion | FS | ++ | ++ | ++ | |||
| YLR054C | 5' intron | Intron | 212 | ++ | ++ | |||
| YLR205C | 5' extension | 2FS | 44 | ++ | ++ | Resequenced | ||
| YLR389C | 3' extension | FS | 40 | ++ | ++ | Resequenced | ||
| YLR401C | 3' extension | FS | 59 | ++ | ++ | ++ | ++ | |
| YLR445W | 3' intron | Intron | 54 | |||||
| YML002W/YML003W | Fusion | FS | ++ | ++ | ++ | |||
| YML017W | 5' intron | Intron | 15 | 5' RACE verified | ||||
| YMR084W/YMR085W | Fusion | FS | ++ | ++ | ++ | ++ | Duplication | |
| YMR207C | 5' extension | FS | 59 | ++ | ++ | Duplication | ||
| YMR269W | 5' extension | FS | 69 | ++ | ++ | Resequenced | ||
| YNL083W | 3' extension | FS | 51 | ++ | ++ | ++ | ++ | [47] |
| YNL130C | Other intron | Intron | 21 | ++ | ++ | ++ | [21]; duplication | |
| YOL048C | 5' intron | Intron | 236 | ++ | ++ | 5' RACE verified; duplication | ||
| YOL163W/YOL162W | Fusion | PS+FS | ++ | ++ | ++ | ++ | ||
| YOR069W | 5' extension | Annotation | 364 | ++ | ++ | ++ | gb: Q92331; duplication | |
| YOR298C-A | 5' extension | FS | 92 | ++ | ++ | ++ | ++ | Resequenced; gb: BAA33217 |
| YPL109C | 3' intron | Intron | 47 | ++ | ++ | ++ | ++ | |
| YPR090W/YPR089W | Fusion | FS | ++ | ++ | ++ | Resequenced | ||
| YPR098C | 5' intron | Intron | 53 | ++ | ++ | |||
*Extension, ORF extension. †FS, frameshift; PS, premature STOP. ‡Supporting evidence from other fungal sequences: ++ indicates that homology supports the corrected S. cerevisiae ORF; + indicates that weak homology supports the corrected S. cerevisiae ORF. §Supporting evidence from other S. cerevisiae sequences: from published work (reference given); from sequence databases (GenBank accession number); from this study (resequenced or 5'RACE); from duplicated S. cerevisiae sequences matching the proposed sequence (duplication).
In addition to overlooked introns, we identified one case of a potentially wrongly assigned 5' splice site in CPT1 (YNL130C), which codes for the sn-1,2-diacyglycerol cholinephosphotransferase [21]. The current annotation proposes that CPT1 would be spliced at a mismatched splice acceptor sequence. However, comparison with the A. gossypii homolog strongly suggests an intron of 92 base-pairs (bp), instead of 441 bp, with perfect consensus splice sequences. This would result in a protein of 407 amino acids with increased similarity to its A. gossypii homolog. This suggestion is supported by comparison with other fungal species, for example C. albicans and S. pombe (Table 2). Finally, a size of 407 amino acids for this enzyme was already proposed in the first publication describing it [21].
A special case, not listed in Table 2, is the intron in STO1 (YMR125W), a gene that encodes the large subunit of the nuclear cap-binding protein complex, a transcriptional activator of glycolytic genes. The comparison with A. gossypii cannot distinguish between two alternatives: presence or absence of an intron, as shown in Figure 2. The S. cerevisiae sequence currently available at SGD is annotated with an intron. Although we noticed the presence of an equivalent intron in A. gossypii, homology is conserved between the two non-spliced forms of these genes in the two organisms as well. Therefore, it may be possible that the STO1 locus in both organisms encodes two proteins with differently charged amino ends.
Figure 2.
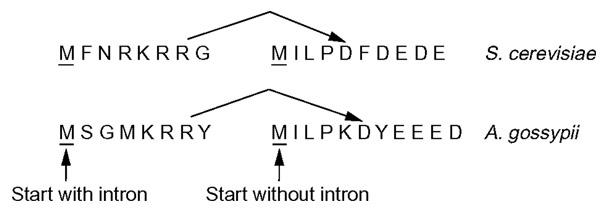
S. cerevisiae STO1 (YMR125W) and its A. gossypii syntenic homolog show two possible amino termini. The STO1 mRNA was proposed to be spliced in S. cerevisiae. However, both the spliced and non-spliced versions show homology to the A. gossypii genome, suggesting two alternative variants.
5' and 3'ORF extensions
In 35 cases, it was possible to extend the boundaries of ORFs into annotation-free regions by artificially introducing single base-pair changes in the S. cerevisiae genomic sequence. These changes eliminated presumptive frameshifts or premature stop codons, as outlined in Figure 1c. The ORFs affected, the increase in ORF size (more than 70 amino acids for 50% of the ORFs), and other supporting evidence are listed as part of Table 2. In 15 cases, we resequenced the region of the proposed change and confirmed the sequencing error. Finally, homology searches also supported the proposed sequence corrections. All regions of suggested change were inspected using BLAST searches against the Génolevures, C. albicans, S. pombe and N. crassa sequence data. In more than 70% of cases, we found homologous sequences in two or more databases that matched the A. gossypii annotation (Table 2).
A special case of ORF extension concerns VPS5 (YOR069W), for which we propose an annotation rather than a sequence correction. The A. gossypii homolog is much longer and consideration of a further upstream start codon for VPS5 would result in an 5' extension 364 codons long with strongly enhanced homology to the A. gossypii homolog.
ORF fusions
Another 22 proposed modifications resulted in the fusion of two previously distinct S. cerevisiae ORFs, as outlined in Figure 1d. A compilation of the A. gossypii ORFs, the fused S. cerevisiae ORFs, and their sizes is given in Table 2 and Table 3. As for the ORF extensions, we obtained supporting evidence for the validity of these fusions from database searches. For 17 of the proposed fusions, we found homologs of similar sizes in two or more fungal databases. Moreover, 10 of the ORF fusions had already been reported but not yet been included in databases, and seven are supported by a much better alignment to a duplicated copy in S. cerevisiae.
Table 3.
Size comparison of proposed fused ORF in S. cerevisiae with A. gossypii homolog
| S. cerevisiae ORF1 | S. cerevisiae ORF2 | Fused ORF | A. gossypii | Supporting evidence | ||||||
| Systematic name | Common name | Size (amino acids) | Systematic name | Common name | Size (amino acids) | Size (amino acids) | Reason* | Name | Size (amino acids) | |
| YAR044W | 859 | YAR042W | 256 | 1,188 | FS | AER225W | 1,300 | Resequenced | ||
| YBR074W | 103 | YBR075W | 460 | 976 | 2FS | AGL209W | 1,012 | Resequenced | ||
| YBR098W | MMS4 | 108 | YBR100W | 112 | 691 | FS | ADL318C | 715 | ||
| YCL001W-A† | 84 | YCL001W-B | 153 | 313 | 2FS | AAL001W | 386 | |||
| YDR474C† | 155 | YDR475C | JIP4 | 555 | 876 | 2FS | AEL233C | 794 | ||
| YER039C† | HVG1 | 249 | YER039C-A | 72 | 341 | PS | AFR236C | 330 | Pseudogene [22] | |
| YFL007W† | BLM3 | 1,804 | YFL006W | 254 | 2,143 | FS | AGL022W | 2,146 | [46] | |
| YGL046W | 262 | YGL045W | 229 | 540 | 3FS | AFR108W | 549 | |||
| YGR272C | 152 | YGR271C-A‡ | 63 | 233 | FS | ABR143C | 214 | |||
| YJL012C | VTC4 | 648 | YJL012C-A‡ | 65 | 721 | FS | AGR316C | 714 | Resequenced | |
| YJL017W | 325 | YJL016W | 171 | 561 | FS | AGR313W | 471 | Resequenced | ||
| YJL019W | MPS3 | 620 | YJL018W | 102 | 682 | FS | AGR312W | 617 | Resequenced | |
| YJL020C | BBC1 | 446 | YJL021C | 710 | 1,156 | FS | AGR306C | 924 | Resequenced | |
| YJL108C† | PRM10 | 383 | YJL107C | 387 | 770 | FS | AFR075C | 741 | Resequenced | |
| YKL199C† | YKT9 | 279 | YKL198C | PTK1 | 399 | 649 | FS | AFR372W | 775 | Resequenced |
| YKR103W | 1,218 | YKR104W | 306 | 1,558 | PS | NB§ | NB§ | |||
| YLL017W† | SDC25 | YLL016W | SDC25 | 1,252 | FS | ADL038W | 1,510 | Pseudogene (SGD) | ||
| YLL052C† | AQY2 | 149 | YLL053C | 152 | 286 | FS | AGL266C | 451 | ||
| YML002W | 737 | YML003W | 290 | 1,090 | FS | ACR006C | 1,072 | |||
| YMR084W† | 262 | YMR085W | 432 | 719 | FS | ABL036C | 707 | |||
| YOL163W† | 169 | YOL162W | 215 | 553 | PS | ACL203C | 538 | Pseudogene [22] | ||
| YPR090W | 736 | YPR089W | 155 | 888 | FS | ABR111C | 790 | Resequenced | ||
*PS, premature STOP codon; FS, frameshift. †Agreement with proposal of Blandin et al. [17] and Wood et al. [18]. ‡Novel ORFs identified by Blandin et al. [17]. §No homolog was found in A. gossypii but YKR103W/YKR104W is a member of a gene family including YLL048C and YHL035C for which A. gossypii homologs were found.
It should be pointed out that S. cerevisiae carries pseudogenes [22] and that confirmed pseudogenes may have homology over their entire length to single A. gossypii ORFs; see examples in Table 3 for three pseudogenes annotated as YER039C/YER039C-A, YLL017W/YLL016W, and YOL163W/YOL162W. Consequently, discrepancies observed between ORFs of the two species may either result from sequencing errors or may represent real pseudogenes. Therefore, we experimentally investigated nine of the proposed ORF fusions by resequencing the respective genomic regions in S. cerevisiae strain S288C, the reference strain of the yeast genome sequencing project [1]. In eight cases, a sequencing error was found, confirming the fusion of eight pairs of neighboring genes (Table 2 and Table 3). On the other hand, resequencing also revealed that YJL107W/YJL108W is a novel pseudogene that bears a single point mutation in S288C.
In addition, programmed ribosomal frameshifting has been demonstrated in S. cerevisiae [23,24] and this might explain some of the observed differences between S. cerevisiae and A. gossypii genomic sequences. Therefore, resequencing all the questioned regions in S. cerevisiae would be needed to be able to discriminate between sequencing errors, pseudogenes and functional frameshifts.
Gene extensions revealing additional functional domains
We analyzed the presence of known functional domains in S. cerevisiae proteins with or without the proposed changes. Most of these extensions did not generate additional domains in the proteins. YKL033W-A, however, could be extended at the 3' end by 176 codons, adding a HAD (haloacid dehalogenase) domain (InterPro: PF00702) and suggesting that this protein of previously unknown function may have a role in the assimilation of halogenated compounds. Similarly, YMR269W was described as a hypothetical protein of 164 amino acids. We propose an amino-terminal extension of 69 amino acids, which would generate a protein of 211 amino acids with 11% greater similarity to its A. gossypii homolog (Figure 3a,b). This proposal was confirmed by resequencing. Domain analysis revealed the presence of a putative RNA-binding domain (D111: PS50174) in both the extended S. cerevisiae and the annotated A. gossypii proteins (Figure 3c). YMR269W might therefore have a role in RNA-mediated cellular processes such as splicing, transcription, or translation. Indeed, YMR269W was recently found to interact with the translation initiation factor GCN3 (YKR026C) in a whole-genome two-hybrid screen [25], which also points to a role in translation. Finally, evaluation of expression data from over 100 genome-scale experiments showed that YMR269W is regulated in a very similar manner as genes involved in protein synthesis [26]. It thus appears very likely that the 'extended' version of YMR269W is involved in protein synthesis.
Figure 3.
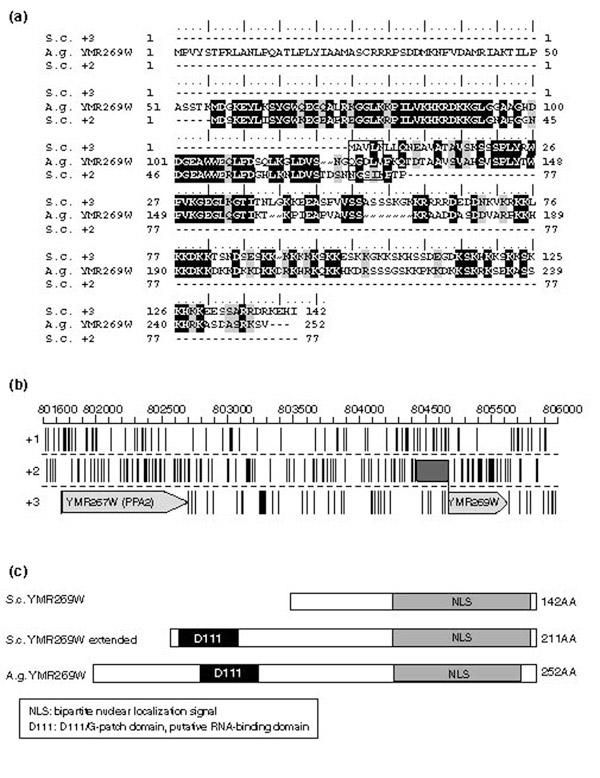
Proposed changes in the hypothetical protein YMR269W. (a) Multiple alignment of the translated +2 and +3 frames of the S. cerevisiae YMR269W region and the syntenic A. gossypii protein. The boxed sequence indicates the region of the potential shift from frame +3 to +2 as suggested by the multiple alignment. (b) S. cerevisiae YMR269W region. Light gray depicts the current annotation as available at SGD. Dark gray shows the proposed elongation of the frame +2 translation on the 5' end of the yeast YMR269W gene. (c) Domain organization of YMR269W proteins in S. cerevisiae and A. gossypii. While the current YMR269W protein sequence of S. cerevisiae carries only a nuclear localization signal, both the A. gossypii homolog and the proposed extended YMR269W protein have an additional G-patch domain, which has been described as a putative RNA-binding domain.
Confirmation of hypothetical ORFs as real ORFs
In 1996 the yeast genome sequencing consortium faced the difficulty of annotating the first eukaryotic genome. Many potential ORFs in the newly sequenced genome did not have homology with entries in the existing databases, and discrimination between 'real' and 'chance' ORFs was often not possible. Novel S. cerevisiae genes lacking homology to any database were annotated if they were at least 100 codons long. The use of this arbitrary cut-off permitted the annotation of most of the 'real' genes but also led to the annotation of many questionable ORFs. Since then, a substantial number of these ORFs have been functionally characterized. However, many are still annotated as hypothetical ORFs because of a lack of functional data. The identification of homologs of these ORFs in related organisms can be taken as strong evidence for their biological significance. Because A. gossypii shares as much as 95% of its genes with S. cerevisiae (90% being in synteny) it is an excellent organism to evaluate the authenticity of yeast hypothetical ORFs.
Currently, 1,885 S. cerevisiae ORFs are classified as hypothetical ORFs in the Saccharomyces Genome Database (SGD). We compared these genes to the A. gossypii genome annotation and identified a homolog in A. gossypii for 1,041 of them. Most important, 999 of these (96%) share both homology and synteny and can therefore be considered to be orthologs. The full list of hypothetical ORFs that should be regarded as real ORFs because of their homology and synteny with A. gossypii is available as an Additional data file. The Munich Information Center for Protein Sequences (MIPS), the other publicly accessible S. cerevisiae genome database, lists 988 of the S. cerevisiae ORFs classified as hypothetical or questionable (with questionable referring to hypothetical ORFs overlapping functionally characterized ORFs). We found homologs for 279 of these ORFs at syntenic positions in the genome of A. gossypii and all belong to the group of ORFs identified above as real among the 1,885 hypothetical ORFs at SGD. This comparison therefore provides strong evidence for the authenticity of a substantial part of the ORFs annotated as hypothetical in both SGD and MIPS.
Spurious ORFs among S. cerevisiae hypothetical ORFs
We assume it unlikely that all the remaining 844 hypothetical S. cerevisiae ORFs in SGD encode proteins, as only 10% of the known functional S. cerevisiae genes have no homolog in A. gossypii (F.S.D., S.V., S.B., A.L., K.G., C. Mohr, S. Steiner, P. Luedi, T.G. and P.P., unpublished work). They cannot, however, be directly investigated using this comparative approach owing to the absence of homologous genes in A. gossypii. Nevertheless, indirect evidence of the dubiousness of a subgroup of the ORFs absent in A. gossypii can be obtained by taking into consideration that they overlap other ORFs. The inspection of all overlapping pairs among annotated S. cerevisiae ORFs (based on our revised version of the S. cerevisiae ORF boundaries) reveals that, in the vast majority of the cases, one of the two ORFs belongs to the group of hypothetical ORFs lacking a syntenic homolog in A. gossypii. Furthermore, although there is some experimental evidence that, in rare cases, functional fungal genes overlap at the 3' ends of their ORFs; in other words, the opposite strand of one ORF acts as transcription terminator for the other ORF and vice versa [27], there is no experimental demonstration that a sequence within a functional ORF can act as promoter for another gene. We used these rules, in addition to the absence or presence of homologs in A. gossypii, to validate S. cerevisiae overlapping ORFs. We found three different categories among the 419 pairs of overlaps as schematically shown in Figure 4. For only seven pairs, A. gossypii carries homologs of both ORFs. Two pairs of homologs overlap in A. gossypii; the other five do not. Two cases of 5' end overlapping ORF pairs are probably explained by the assignment of the wrong start codon, and the remaining cases relate to overlapping 3' ends, which supports the hypothesis that ORF overlaps are rare in S. cerevisiae and that they involve 3' ends of ORFs (see Figure 4 legend).
Figure 4.
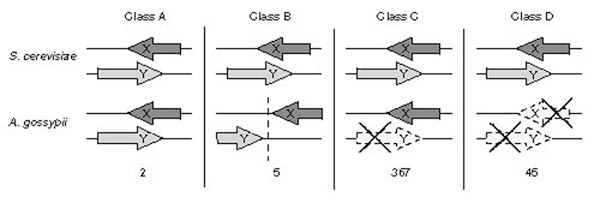
Classes of overlapping annotated ORFs in S. cerevisiae derived from comparison with the A. gossypii genome. Class A, homologs for both overlapping S. cerevisiae ORFs are found at syntenic positions in A. gossypii and also overlap. Class B, homologs for both overlapping S. cerevisiae ORFs are found in A. gossypii but do not overlap (see comments at end of legend). Class C, gene X but not gene Y has a syntenic homolog in A. gossypii. Class D, both overlapping S. cerevisiae ORFs lack a homolog in A. gossypii. Numbers refer to the frequency of the four types of overlapping pairs. Although all three possible directions of overlaps were observed, for convenience only 3'/3'-end overlaps are depicted here. YPL166W/YPL165C and YLR360W/YLR361C are the only two cases for which overlap was observed both in S. cerevisiae and A. gossypii (class A). The two overlaps are short (24 and 35 nucleotides, respectively, in S. cerevisiae and 24 and 14 nucleotides in A. gossypii, and involve only terminator-ORF sequence overlap. Class B overlaps comprise three syntenic ORF pairs (YJR012C/YJR013W, YML095C/YML096W, and YGR074W/YGR075C) and two non-syntenic (YPL018W/YPL017C and YBR262C/YBR263W). The lack of synteny reflects chromosomal rearrangements in one or the other species which resulted in either the separation of the two ORFs in A. gossypii or in their joining in S. cerevisiae. Two of the five class B ORF pairs refer to YML096W/YML095C and YGR074W/YGR075C, both with 6-nucleotide 3-end overlaps. The syntenic A. gossypii homologs are separated at their 3' ends by 2 and 51 nucleotides respectively, implying some overlap of terminator and ORF sequences very similar to their syntenic S. cerevisiae homologs. For two pairs of ORFs overlapping at their 5' ends in S. cerevisiae (YBR262C/YBR263W and YJR012C/YJR013W) both ORFs have a homolog in A. gossypii. The alignments of YBR263W and YJR012C with their respective A. gossypii homologs strongly suggest an error in selection of their start codons. Both S. cerevisiae ORFs are very likely to be 75 codons shorter, thus eliminating the presumptive promoter-ORF overlaps. Directions are provided for classes C and D in the additional data files.
For 367 pairs, one ORF homolog was found in A. gossypii, the other not and we propose that the later ones are very likely to be spurious (see Figure 4, class B). These ORFs are listed in the additional data files with information about their present functional annotation, their sizes, and the type of overlap. For 66% of the pairs in this class, one or both presumptive promoter regions overlap an ORF sequence. In the remaining 34%, both terminator regions overlap ORF sequences. These latter cases should be viewed with caution as a very small percentage of them might turn out to be real. Indeed, in two cases marked in the additional data files, published data confirm the authenticity of a suggested spurious ORF. It should be noted that some of the proposed spurious ORFs are reported in very close relative of S. cerevisiae such as S. bayanus. However, the similarity is often restricted to the overlapping regions and is likely to result from the transfer of conservation from the real coding region to the other frames or strand. As A. gossypii is more distantly related, such homology can be found, though seldom, but does not match ORFs owing to the presence of STOP codons. Finally, in the remaining 45 pairs, for two ORFs we could find no homolog in A. gossypii and the criteria applied above cannot be used here. However, 36 ORFs could be considered as likely to be spurious as they overlap ORFs with described function or with a size of at least 500 codons (Figure 4, class D, and see Additional data files).
In summary, comparison of pairs of overlapping S. cerevisiae ORFs with the A. gossypii genome suggests that probably 403 of the remaining 844 hypothetical ORFs should be considered with care as they are likely to be spurious. Wood et al. [18] used ORF overlaps as one criterion to disregard 371 S. cerevisiae genes annotated as hypothetical, 289 of which are spurious according to the criteria applied above. Our analysis leaves about 450 hypothetical ORFs for which no information is currently available to categorize them as likely to be real or spurious. Additional evidence from similar analyses with other yeast species will be necessary to resolve these problems. Therefore, projects such as the current Saccharomyces Genome Project [28] will hopefully allow for a final S. cerevisiae genome annotation, seven years after completion of the genome sequence.
Conclusions
We have demonstrated the power of comparative genomics for the annotation of two completely sequenced fungal genomes. Whole-genome comparison guided the identification of novel ORFs, improved gene annotations, revealed sequencing errors, and helped to distinguish between real and spurious ORFs, thereby enhancing our view of the S. cerevisiae genome. As a consequence, these results will also contribute to the validity of genome-scale experiments, such as gene-expression profiling, an area where accurate gene annotation is crucial. The forthcoming availability of more yeast species genomes will, in an analogous way, drastically improve speed and accuracy of genome annotation of S. cerevisiae and the newly sequenced species. The method described here is straightforward for lower eukaryotes and should be applicable to any two closely related genomes of any complexity.
Materials and methods
Yeast strain
S. cerevisiae AB972 (S288C) strain was used for resequencing and 5' RACE-based intron verification.
5' RACE-based verification of proposed introns in S. cerevisiae
Reverse transcription and 5' RACE was done using SMART RACE cDNA amplification system (BD Bioscience Clontech). Gene-specific primer (GSP) sequences were selected approximately 200 bp downstream of the putative introns for YKR004C, YML017W, and YOL048C. The amplified cDNA fragments were purified from the gel, cloned in the TOPO-TA cloning vector (Invitrogen) and sequenced on an ABI Prism 310 sequencer (Applied Biosystems). The sequences are available in GenBank (accession numbers AY245791, AY245792, and AY245793).
Resequencing of S. cerevisiae genomic regions
The following 25 S. cerevisiae genomic regions were resequenced: YAL013W (AY260888), YAR044W/YAR042W (AY260892), YBL104C (AY260889), YBR074W/YBR075W (AY260891), YBR157W (AY260879), YCL008C (AY260880), YJL012C/YJL012C-A (AY227894), YJL016W/YJL017W (AY260898), YJL019W/YJL018W, YJL020C/YJL021C, YJL108C/YJL107C (AY227895), YJL159W (AY260881), YJL160C (AY260893), YJL178C (AY260894), YJR013W (AY260895), YKL033W-A (AY260896), YKL199C/YKL198C, YKL207W (AY260882), YKR056W (AY260897), YKR100C (AY260883), YLR205C (AY260884), YLR389C (AY260885), YMR269W (AY22789), YOR298C-A (AY260886), and YPR089W/YPR090W (AY260887). Primers flanking the region of the putative frameshift or premature stop mutation were selected to be unique within the yeast genome, and to have similar melting points. PCR was carried out using standard protocols on the AB972 (S288C) genomic DNA. PCR products were confirmed by agarose gel electrophoresis and sequencing was carried out on an ABI 310 sequencer using big dye chemistry and protocols from ABI. Sequence assembly was performed using the phred/phrap/consed analysis package [29-31]. Sequence corrections for YJL019W/YJL018W, YJL020C/YJL021C and YKL199C/YKL198C were recently corrected in SGD and were, therefore, not submitted to GenBank.
Sequence databases and sequence analysis
S. cerevisiae, S. pombe and hemiascomycete genomic sequence information was retrieved from GenBank at the National Center for Biotechnology Information (NCBI) [32,33]. The S. cerevisiae genome was used as available at NCBI on 27 October, 2002. This release was submitted by the SGD [34,35]. Sequence data from C. albicans and N. crassa were obtained from the Stanford Genome Technology Center [36] and from the Neurospora Sequencing Project, Whitehead Institute/MIT Center for Genome Research [37], respectively. Sequence analysis was carried out using the GCG Wisconsin Package (Accelrys), BLAST [38] and FASTA tools [39,40]. Domain analysis was carried out using the InterProScan.pl algorithm from the European Institute of Bioinformatics [41,42]. Functional classifications of S. cerevisiae ORFs were taken from the SGD [34] and MIPS [43].
Annotation of A. gossypii chromosomes
The 9 Mb genome was sequenced by combining three strategies: end-sequencing of chromosome-sorted plasmid and BAC clones, shotgun sequencing of sheared genomic DNA fragments, and extensive gap filling by primer walking, which resulted in an average accuracy of 99.8% (F.S.D., S.V., S.B., A.L., K.G., C. Mohr, S. Steiner, P. Luedi, T.G. and P.P., unpublished work). In the first round of annotation, all ORFs longer than 50 codons were searched using BLAST against the set of S. cerevisiae ORF translations available from SGD [34]. A. gossypii ORFs with a hit lower or equal to E = 1e-2 were automatically annotated as S. cerevisiae homologs. This first draft of the A. gossypii genome annotation together with the BLAST results were re-evaluated case by case in a non-automatic procedure. A. gossypii ORFs sharing low or high homology with syntenic S. cerevisiae ORFs were kept. The synteny was independently assessed by two people. Inter-ORF regions were then compared with the six translation frames of the S. cerevisiae genome sequence, leading to the identification of potentially overlooked ORFs in S. cerevisiae and A. gossypii. In a final step, the remaining A. gossypii potential ORFs were searched against other databases and led to the annotation of A. gossypii genes with no homolog in S. cerevisiae.
Homology screening of all S. cerevisiae inter-ORF regions
A. gossypii ORF translations were searched against a locally built yeast inter-ORF sequence database using BLAST 2.0. A cut-off threshold E-value of 1e-2 was used to filter the results. RNA genes were automatically filtered out and the remaining hits were manually checked for synteny. Regions of discrepancy were carefully checked in A. gossypii and in all cases matched good-quality consensus sequence. The current S. cerevisiae genome annotation release was re-annotated with proposed changes or novel sequence using the Artemis annotation tool [44] and the Sequin submission tool [32,33].
Experimentally verified sequence corrections are available in GenBank under the accession numbers: AY260888, AY260892, AY260889, AY260891, AY260879, AY260880, AY227894, AY260898, AY227895, AY260881, AY260893, AY260894, AY260895, AY260896, AY260882, AY260897, AY260883, AY260884, AY260885, AY22789, AY260886, AY260887, AY245791, AY245792 and AY245793.
Additional data files
The following files are available: a list of the novel S. cerevisiae ORFs proposed by Blandin et al. and Wood et al. [17,18] for which a syntenic homolog was found in the A. gossypii genome (Additional data file 1); a list of all S. cerevisiae hypothetical ORFs for which a homolog was found in the A. gossypii genome (Additional data file 2); a list of all S. cerevisiae hypothetical ORFs for which no homolog was found in the A. gossypii genome (Additional data file 3). A list of all class C overlaps together with gene sizes, gene functions and overlap directions can be found in Additional data file 4; these genes are suggested to be spurious based on our criteria. A list of class D overlaps: none of the two overlapping genes in S. cerevisiae has a homolog in A. gossypii can be found in Additional data file 5; the size and function classifications were used to predict spurious genes in some of these cases. A graphical display of the proposed S. cerevisiae annotation changes based on comparison with A. gossypii can be found in Additional data file 6. GenBank files for each of the S. cerevisiae chromosomes that take account of the proposed modifications (prior to confirmation of the sequence) can be found in Additional data file 7. GenBank files for each A. gossypii genomic locus used to infer annotation corrections in S. cerevisiae can be found in Additional data file 8.
Supplementary Material
A list of the novel S. cerevisiae ORFs for which a syntenic homolog was found in the A. gossypii genome
A list of all S. cerevisiae hypothetical ORFs for which a homolog was found in the A. gossypii genome
A list of all S. cerevisiae hypothetical ORFs for which no homolog was found in the A. gossypii genome
A list of all class C overlaps together with gene sizes, gene functions and overlap directions
A list of class D overlaps: none of the two overlapping genes in S. cerevisiae has a homolog in A. gossypii
A graphical display of the proposed S. cerevisiae annotation changes based on comparison with A. gossypii
GenBank files for each of the S. cerevisiae chromosomes that take account of the proposed modifications (prior to confirmation of the sequence)
GenBank files for each A. gossypii genomic locus used to infer annotation corrections in S. cerevisiae
Acknowledgments
Acknowledgements
We thank Philippe Luedi and Amy Gladfelter for supporting discussions, and Arndt Brachat and Mike Primig for critical reading of the manuscript. This work was supported by grants from the University of Basel and Duke University.
References
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Mewes HW, Albermann K, Bahr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maierl A, Oliver SG, et al. Overview of the yeast genome. Nature. 1997;387:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- Sequence updates at SGD http://genome-www.stanford.edu/Saccharomyces/sequenceupdates.shtml
- Chromosome III resequencing information at MIPS http://mips.gsf.de/cgi-bin/proj/yeast/THREE
- Philippsen P, Kleine K, Pohlmann R, Dusterhoft A, Hamberg K, Hegemann JH, Obermaier B, Urrestarazu LA, Aert R, Albermann K, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications. Nature. 1997;387:93–98. [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Dietrich FS. Verification of a new gene on Saccharomyces cerevisiae Chromosome III. Yeast. 2003;20:731–738. doi: 10.1002/yea.996. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Miura F, Yada T, Nakai K, Sakaki Y, Ito T. Differential display analysis of mutants for the transcription factor Pdr1p regulating multidrug resistance in the budding yeast. FEBS Lett. 2001;505:103–108. doi: 10.1016/s0014-5793(01)02792-2. [DOI] [PubMed] [Google Scholar]
- Blandin G, Durrens P, Tekaia F, Aigle M, Bolotin-Fukuhara M, Bon E, Casaregola S, de Montigny J, Gaillardin C, Lepingle A, et al. Genomic exploration of the hemiascomycetous yeasts: 4. The genome of Saccharomyces cerevisiae revisited. FEBS Lett. 2000;487:31–36. doi: 10.1016/s0014-5793(00)02275-4. [DOI] [PubMed] [Google Scholar]
- Wood V, Rutherford KM, Ivens A, Rajandream MA, Barrell B. A reannotation of the Saccharomyces cerevisiae genome. Comp Funct Genomics. 2001;2:143–154. doi: 10.1002/cfg.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Kessler MM, Zeng Q, Hogan S, Cook R, Morales AJ, Cottarel G. Systematic discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res. 2003;13:264–271. doi: 10.1101/gr.232903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmstad RH, Bell RM. The sn-1,2-diacylglycerol cholinephosphotransferase of Saccharomyces cerevisiae. Nucleotide sequence, transcriptional mapping, and gene product analysis of the CPT1 gene. J Biol Chem. 1990;265:1755–1764. [PubMed] [Google Scholar]
- Harrison P, Kumar A, Lan N, Echols N, Snyder M, Gerstein M. A small reservoir of disabled ORFs in the yeast genome and its implications for the dynamics of proteome evolution. J Mol Biol. 2002;316:409–419. doi: 10.1006/jmbi.2001.5343. [DOI] [PubMed] [Google Scholar]
- Morris DK, Lundblad V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- Asakura T, Sasaki T, Nagano F, Satoh A, Obaishi H, Nishioka H, Imamura H, Hotta K, Tanaka K, Nakanishi H, et al. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield T, Judson RS, Knight JR, Lockshon D, Narayan VA, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Gerads M, Ernst JF. Overlapping coding regions and transcriptional units of two essential chromosomal genes (CCT8, TRP1) in the fungal pathogen Candida albicans. Nucleic Acids Res. 1998;26:5061–5066. doi: 10.1093/nar/26.22.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccharomyces Genome Sequencing http://genome.wustl.edu/projects/yeast/
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2003;31:23–27. doi: 10.1093/nar/gkg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccharomyces Genome Database http://www.yeastgenome.org
- Stanford Genome Technology Center, Candida albicans sequence http://www-sequence.stanford.edu/group/candida/download.html
- Whitehead Institute/MIT Center for Genome Research http://www-genome.wi.mit.edu
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR. Using the FASTA program to search protein and DNA sequence databases. Methods Mol Biol. 1994;25:365–389. doi: 10.1385/0-89603-276-0:365. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Mewes HW, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenstern B, Munsterkotter M, Rudd S, Weil B. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Li Y, Kane T, Tipper C, Spatrick P, Jenness DD. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol Cell Biol. 1999;19:3588–3599. doi: 10.1128/mcb.19.5.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robben J, Hertveldt K, Volckaert G. Revisiting the yeast chromosome VI DNA sequence reveals a correction merging YFL007w and YFL006w to a single ORF. Yeast. 2002;19:699–702. doi: 10.1002/yea.870. [DOI] [PubMed] [Google Scholar]
- Belenkiy R, Haefele A, Eisen MB, Wohlrab H. The yeast mitochondrial transport proteins: new sequences and consensus residues, lack of direct relation between consensus residues and transmembrane helices, expression patterns of the transport protein genes, and protein-protein interactions with other proteins. Biochim Biophys Acta. 2000;1467:207–218. doi: 10.1016/s0005-2736(00)00222-4. [DOI] [PubMed] [Google Scholar]
- Horowitz DS, Abelson J. A U5 small nuclear ribonucleoprotein particle protein involved only in the second step of pre-mRNA splicing in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2959–2970. doi: 10.1128/mcb.13.5.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Egeland DB, Jarnik M, Strich R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci USA. 2000;97:14548–14553. doi: 10.1073/pnas.250351297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Mu J, Farkas I, Huang D, Goebl MG, Roach PJ. Requirement of the self-glucosylating initiator proteins Glg1p and Glg2p for glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6632–6640. doi: 10.1128/mcb.15.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A list of the novel S. cerevisiae ORFs for which a syntenic homolog was found in the A. gossypii genome
A list of all S. cerevisiae hypothetical ORFs for which a homolog was found in the A. gossypii genome
A list of all S. cerevisiae hypothetical ORFs for which no homolog was found in the A. gossypii genome
A list of all class C overlaps together with gene sizes, gene functions and overlap directions
A list of class D overlaps: none of the two overlapping genes in S. cerevisiae has a homolog in A. gossypii
A graphical display of the proposed S. cerevisiae annotation changes based on comparison with A. gossypii
GenBank files for each of the S. cerevisiae chromosomes that take account of the proposed modifications (prior to confirmation of the sequence)
GenBank files for each A. gossypii genomic locus used to infer annotation corrections in S. cerevisiae


