Short abstract
A comparative fluorescence in situ hybridization approach, using probes selected from a combination of physical mapping, genomic sequence, and segmental duplication analyses has been employed to narrow the breakpoint interval of a pericentric inversion in chimpanzee involving the orthologous human 15q11-q13 region.
Abstract
Background
Pericentric inversions are the most common euchromatic chromosomal differences among humans and the great apes. The human and chimpanzee karyotype differs by nine such events, in addition to several constitutive heterochromatic increases and one chromosomal fusion event. Reproductive isolation and subsequent speciation are thought to be the potential result of pericentric inversions, as reproductive boundaries form as a result of hybrid sterility.
Results
Here we employed a comparative fluorescence in situ hybridization approach, using probes selected from a combination of physical mapping, genomic sequence, and segmental duplication analyses to narrow the breakpoint interval of a pericentric inversion in chimpanzee involving the orthologous human 15q11-q13 region. We have refined the inversion breakpoint of this chimpanzee-specific rearrangement to a 600 kilobase (kb) interval of the human genome consisting of entirely duplicated material. Detailed analysis of the underlying sequence indicated that this region comprises multiple segmental duplications, including a previously characterized duplication of the alpha7 neuronal nicotinic acetylcholine receptor subunit gene (CHRNA7) in 15q13.3 and several Golgin-linked-to-PML, or LCR15, duplications.
Conclusions
We conclude that, on the basis of experimental data excluding the CHRNA7 duplicon as the site of inversion, and sequence analysis of regional duplications, the most likely rearrangement site is within a GLP/LCR15 duplicon. This study further exemplifies the genomic plasticity due to the presence of segmental duplications and highlights their importance for a complete understanding of genome evolution.
Background
The karyotypes of humans and the African and Asian great apes are remarkably well conserved, with relatively few large-scale chromosomal changes among these species despite the considerable phenotypic and biological differences between hominoids [1-3]. This conservation is particularly relevant in comparisons between the human (Homo sapiens; HSA) and common chimpanzee (Pan troglodytes; PTR) genomes, for in order to completely ascertain the evolution of our own lineage it is necessary to understand what differentiates us at the genomic level from our closest relatives. Furthermore, insight into the mechanism(s) underlying primate chromosomal evolution can be obtained from the molecular characterization of species-specific rearrangement breakpoints. Recent analyses of synteny disruptions between man and mouse, although valuable, do not provide the level of detail afforded by the comparison of closely related species such as chimpanzee and human [4,5].
Several mechanisms of genetic change are thought to lead to speciation, including gene evolution via coding sequence mutation, gene expression variation by regulatory element mutation and chromosomal rearrangement - which has the potential to create reproductive barriers and induce genetic isolation within an existing population [6]. Although a thorough determination of all genetic differences between humans and our closest non-human primate relatives will not be possible until the genomic sequences of great ape species have been determined, many of the chromosomal rearrangements between great apes have been previously characterized at the cytogenetic level.
Primarily examined through the use of G-banding cytogenetic techniques, the human and common chimpanzee karyotypes differ by only 10 euchromatic rearrangements: a telomere fusion between PTR chromosomes 12 and 13, resulting in HSA chromosome 2, and 9 pericentric inversions (HSA 1, 4, 5, 9, 12, 15, 16, 17, 18) [1,2]. The predominance of pericentric inversions between chimps and humans highlights their potential importance in the divergence of human from non-human primate species, and provides an opportunity to investigate the mechanism facilitating these rearrangements. Recent studies have characterized several evolutionary breakpoints in common chimpanzee and other great ape species including pericentric inversions and a chromosome translocation [7-9]. We present here a refinement of the breakpoint associated with a previously identified pericentric inversion of the human 15q11-q13 orthologous region in common chimpanzee (XVp) to a region containing multiple segmental duplications [1,2,10].
Results
A panel of bacterial artificial chromosome (BAC) and P1 artificial chromosome (PAC) clones spanning approximately 10 megabases (Mb) of human genome sequence was initially used to characterize the pericentric inversion of human 15q11-q13 in chimpanzee. Probes were selected that flank known sites of genomic rearrangement based upon previous yeast artificial chromosome (YAC) and BAC/PAC mapping studies, together with the human sequence map available from the University of California at Santa Cruz (UCSC) Genome Browser [11-15]. Probes flanking the Prader-Willi/Angelman syndrome breakpoints, in addition to a characterized inv dup(15) rearrangement were selected to test the possibility that the pericentric inversion evolutionary breakpoint corresponded to a site of known human genomic instability (Figure 1). Initially, the cytogenetic map position of a total of 18 BAC/PAC probes were comparatively mapped using fluorescence in situ hybridization (FISH) (data not shown).
Figure 1.
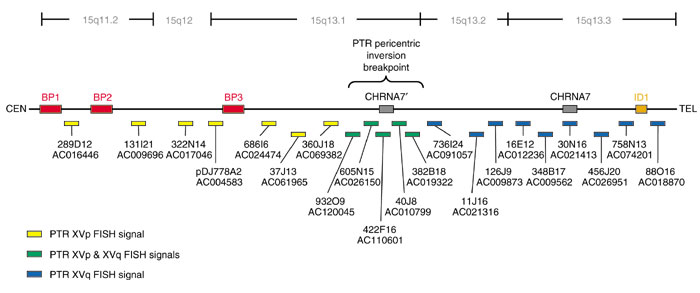
Map of Human 15q11-q13. Probes used to localize the chimpanzee (PTR) pericentric inversion are shown color-coded as to whether they hybridized to PTR XVp (yellow) or PTR XVq (blue). The breakpoint spanning clones are indicated in green. The three major PWS/AS common deletion breakpoints are indicated in red as BP1, BP2 and BP3. The previously characterized inv dup(15) breakpoint is indicated in orange as ID1. The duplicate CHRNA7 sequence is designated CHRNA7'. Accession numbers for all clones are indicated, all clones were obtained from the RPCI-11 BAC library. The map is not drawn to scale.
During this analysis, chimpanzee, pygmy chimpanzee, gorilla and orangutan chromosomes were assayed for the species-specificity of the pericentric inversion (Figure 2). Previous cytogenetic analyses of human, chimpanzee, gorilla and orangutan G-banded metaphase chromosomes demonstrated that the pericentric inversion was a chimpanzee-specific event, with humans maintaining the ancestral state of this region when compared to gorilla and orangutan [1,2]. Two-color FISH experiments utilizing distal probe RP11-88O16, and the PWS/AS common deletion breakpoint-marking probe pDJ-778A2 clearly show that the chimpanzee lineage is the sole great ape species to harbor this rearrangement (Figure 2).
Figure 2.
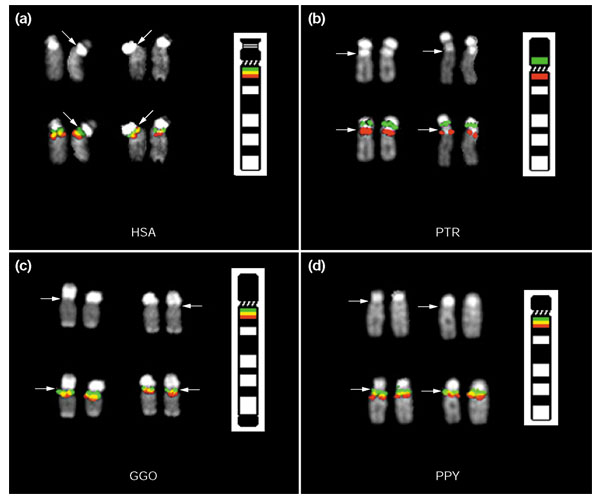
Pericentric inversion of HSA 15q11-q13 in chimpanzee two-color FISH. Probe RP11-88O16 (red), located in distal human 15q13.3, and probe pDJ-778A2 (green), which hybridizes to the HERC2-related PWS/AS breakpoint clusters in 15q11.2 and 15q13.1 clearly demonstrate the inversion of HSA 15q proximal material in chimpanzee (b). Humans (a) share the ancestral state with gorilla (c) and orangutan (d). The DAPI image for each chromosome is shown above the hybridization image and centromeres are indicated by the arrows.
On the basis of the appearance of dual FISH signals on PTR XVp and XVq, the general location of the pericentric inversion breakpoint was identified with probe RP11-40J8 (Figures 1,3). Using the most current assembly of the human genome available, build 31 - released November 2002 - probes adjacent to and overlapping RP11-40J8 were used in further FISH experiments to characterize and more precisely define the breakpoint interval (Figure 3). Clones selected for the array were represented as either completely finished or working draft sequence within the human genome assembly. These clones were chosen because of their presence in at least working-draft status in the human genome assembly (RP11-932O9, RP11-605N15, RP11-422F16, RP11-382B18, and RP11-736I24). Overall, near-complete sequence coverage in the form of working draft and finished BAC clones is available for this region of 15q13, although there are several discontiguous placements of working draft sequences, termed 'warping', within the assembly around RP11-40J8 and the overlapping clone RP11-382B18 (see for example, Figure 4, RP11-422F16 and RP11-605N15). We believe that this warping, or fragmented arrangement, of working draft sequence contigs that span a distance exceeding the total sequence length within an accession is likely to be due to the presence of segmental duplications, as discussed below. In this second round of FISH experiments, all but one of these clones, RP11-736I24, displayed the dual signal on PTR XVp and XVq (Figure 3).
Figure 3.
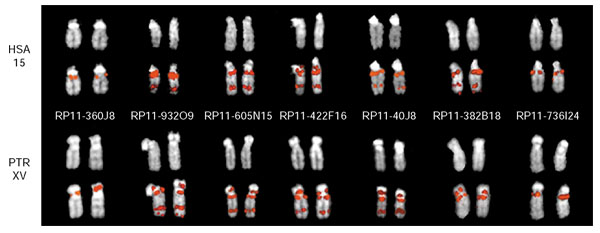
FISH analysis of breakpoint-spanning clones. FISH hybridization results for probes spanning the PTR inversion breakpoint interval on human chromosome 15 (HSA) and chimpanzee chromosome XV (PTR) are shown. The DAPI image for each chromosome is shown above the hybridization image. PTR probes RP11-360J18 and RP11-736I24 flank the breakpoint interval on the PTR XVp and XVq side, respectively, and map uniquely. All clones tested that map between RP11-360J18 and RP11-736I24 demonstrated dual FISH signals on PTR XVp and XVq. The clones are ordered from left to right according to their starting position within the November 2002 human genome assembly (see Figure 4).
Figure 4.
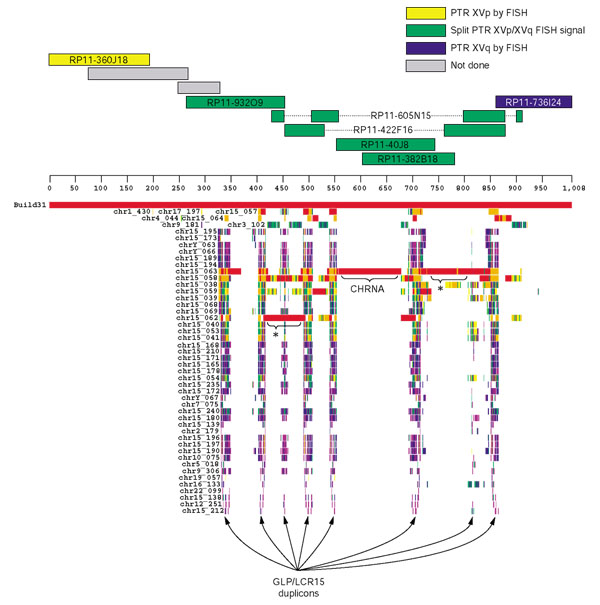
Segmental duplications in the inversion breakpoint interval. The human genomic sequence encompassing the most proximal PTR XVp probe (RP11-360J18) and most distal PTR XVq probe (RP11-736I24) was extracted from the genome assembly (November 2002) and sequence-similarity searches performed against the entire human genome (see Materials and methods). The output was displayed using the program Parasight (Jeff Bailey, unpublished work), which shows pairwise alignments as colored horizontal boxes below the heavy black line, which represents the 1 Mb interval. The color-coding of each horizontal box is a reflection of the sequence similarity of the alignment: 100-99% is indicated in red, 99-97% in orange, 97-95% in yellow, 95-93% in green, 93-91% in blue and 91-89% in purple. *Additional putative duplicons; these may, however, be artifacts of assembly and they require further verification.
We then defined the inversion breakpoint interval as the region between the most distal probe on HSA 15q to display a PTR XVp signal, RP11-360J18 and the most proximal probe on HSA 15q to display a PTR XVq signal, RP11-736I24 (Figure 3). This interval encompasses approximately 1 Mb of genomic sequence, including all clones which produced dual FISH signals in PTR hybridizations. Interestingly, all of the clones which produced the dual FISH signals in PTR were confined to a 600 kb segment of this interval. These results imply that either the pericentric inversion involved the duplication of a large chromosomal segment, or that the inversion involved a sequence common to all clones in the 600 kb interval.
We extracted the 1 Mb interval from the November 2002 human genome assembly and performed a sequence-similarity search against the entire human genome (build 31 (Figure 4) and the non-redundant nucleotide (NT) and high throughput genomic sequence (HTGS) databases (data not shown)) using MEGABLAST. The output was parsed and displayed in the graphical program Parasight (Jeff Bailey, unpublished work), which allows for the visual identification of duplicated sequences (Figure 4). Interestingly, the segmental duplications identified in this analysis were confined to the 600 kb covered by the clones which produced the dual FISH signals in PTR. We identified two known duplications from this analysis, which is in agreement with previous segmental duplication analyses available from the UCSC Genome Browser [14,16]. The first is derived from the a duplication of the CHRNA7 gene, characterized in detail by Riley et al. [17]. This sequence was duplicated from the intact CHRNA7 locus in 15q13.3 and is highly homologous (> 99% similar) to the ancestral locus. The CHRNA7 duplication involves at least 125 kb of material (indicated in Figure 4), however because of the fragmented nature of the assembly in this region at this time, that number could increase with future versions of the human genome assembly. The second duplicon recognized within the 600 kb sub-interval was originally described as the Golgin-Linked-to-PML (promyelocytic leukemia) duplicon, or GLP duplicon, by Gilles et al. [18] and subsequently described as LCR15 by Pujana et al. [19], Gratacos et al. [20] and Pujana et al. [21]. The roughly 30 kb GLP/LCR15 duplicon is present in approximately eight copies throughout the 600 kb duplication-rich sub-interval (Figure 4).
Although the CHRNA7 duplication had, on the basis of sequence identity, previously been postulated to be human-specific, the location of this duplicon within the 600 kb interval associated with the pericentric inversion in chimpanzee necessitated determination of the copy number of CHRNA7 within the chimpanzee genome [17]. First, chimpanzee BAC library hybridizations (8.7 times coverage from RPCI-43 and CHORI-251 segment 1 BAC library filters) were performed with a probe derived from CHRNA7-related sequence, yielding a total of four positives, which was consistent with a single-copy locus (data not shown). Second, comparative FISH analysis of RP11-30N16, a clone which contains a substantial portion of the ancestral 15q13.3 CHRNA7 locus, produced a single FISH signal on PTR XVq only (summarized in Figure 1). Finally, Southern blot analysis was performed using a probe designed to a PstI restriction fragment length polymorphism (RFLP) which distinguished the donor and duplicate copies of human CHRNA7 (Figure 5). Upon hybridization of this probe to a panel of human, pygmy chimpanzee, common chimpanzee and gorilla genomic DNAs, two bands were observed in the human sample and a single band was noted in all great ape species, providing further evidence that the CHRNA7 duplication was specific to humans and was not present at the time of the pericentric inversion event in the chimpanzee genome.
Figure 5.
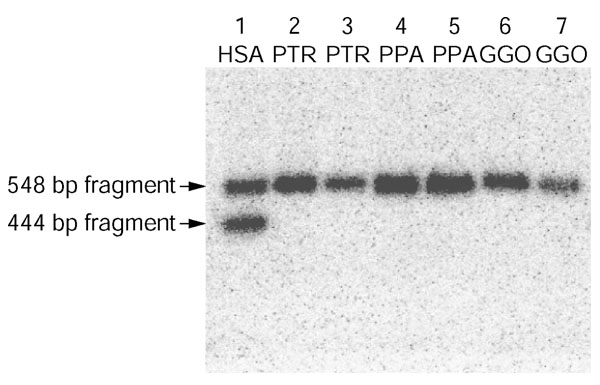
Southern analysis of the CHRNA7 duplication. Probe CHRNA7-PstI was used to demonstrate a 104 bp RFLP distinguishing the intact (548 bp band) and duplicated (444 bp band) CHRNA7 loci in the human genome (lane 1). A single band - corresponding to the intact CHRNA7 locus - is seen in PTR, PPA and GGO (lanes 2 and 3, 4 and 5, 6 and 7, respectively), indicating that the duplication of CHRNA7 was a recent human-specific event and that therefore the gene is single copy in the PTR genome.
Discussion
Using a panel of FISH probes spanning several known sites of rearrangement within 15q11-q13 we were able to narrow the region containing the PTR pericentric inversion breakpoint to a complex cluster of segmental duplications approximately 600 kb in length, according to the most recent assembly of the human genome. Comparative analysis using material from other great apes including gorilla and orangutan indicated the inversion event was restricted to the chimpanzee lineage. It should be noted that the inversion, although primarily characterized in the common chimpanzee (Pan troglodytes) was also observed in the pygmy chimpanzee (Pan paniscus) thus dating the inversion event more precisely to an interval of 2-5 million years ago (data not shown). Although human chromosome 15q11-q13 is noted for its exceptional instability, associated with human disease and chromosomal rearrangement, it is noteworthy that the orthologous site of rearrangement in the human genome did not correspond to one of the three major previously described common disease rearrangement breakpoints [11,12,21,22]. Similarly, probe RP11-88O16, distal of the previously characterized inv dup(15) breakpoint, and probes RP11-456J20 and RP11-758N13, which flank an additional inv dup(15) rearrangement breakpoint (S. Schwartz, unpublished data), also appeared to have no role in this evolutionary breakpoint between man and chimpanzee [23,24].
Our FISH results using probe RP11-40J8, which demonstrated dual signals on PTR XVp and XVq, indicated we had reached the site of the pericentric inversion, or alternatively a sequence within RP11-40J8 had been duplicated to both arms of PTR XV. Consequently, we performed comparative FISH in human and chimpanzee using multiple BAC clones that flanked RP11-40J8 in the November 2002 build of the human genome (Figures 3,4). From on its position in the human assembly, clone RP11-360J18 was the most distal to show a FISH signal on a single PTR chromosome arm (XVp). Conversely, probe RP11-736I24 hybridized to HSA and PTR XVq, indicating the pericentric inversion breakpoint likely lies proximal of this position in the human assembly. All probes tested between RP11-360J18 and RP11-736I24 produced signals on PTR XVp and PTR XVq near the centromere. In addition, several probes that yielded dual signals flanking the PTR centromere also produced a signal in the region orthologous to HSA 15q22-24 (Figure 3).
We next investigated the nature of the sequence underlying the pericentric inversion breakpoint by extracting the 1 Mb interval encompassing the sequence between the most distal PTR XVp probe, RP11-360J18, and the most proximal PTR XVq probe, RP11-736I24, from the November 2002 human genome assembly. This interval includes approximately 600 kb of sequence spanning FISH probes RP11-932O9, RP11-605N15, RP11-422F16, RP11-40J8 and RP11-382B18, all of which produced dual FISH signals flanking the centromere in PTR XVq and XVp (Figures 3,4). Sequence similarity searches of the 1 Mb interval against both the November 2002 assembly and the NT and HTGS databases were in agreement with previous global analyses of the human genome, which indicated multiple segmental duplications in the interval (Figure 4) [16,25].
We were able to discern two major duplicons from this analysis: one segment derived from the CHRNA7 locus in 15q13.3 previously described by Riley et al. [17], and several copies of a duplicon described as Golgin-linked-to-PML (GLP) [18] and as LCR15 [19-21]. A third, previously undetected, potential segmental duplication was identified in the proximal third of the 600 kb region (Figure 4, indicated by the asterisk). However, considering the assembly of the region, which involves significant warping or artifactual fragmentation of accessions AC110601 (RP11-422F16) and AC026150 (RP11-605N156), and the high sequence identity of the duplications, further validation is warranted.
In Riley et al. [17], the authors theorize that the duplication of the CHRNA7 locus is a human-specific event because of the high sequence homology of the duplicated segment to the donor segment in 15q13.3 and the fact the CHRNA7 duplication appeared polymorphic in the human population [17]. This hypothesis deserved consideration in this study, as the site of duplication of the CHRNA7 locus in the human genome may correspond to the pericentric inversion breakpoint, and the high sequence homology noted between the donor and duplicated CHRNA7 loci does not establish the human-specificity of the CHRNA7 duplication, as gene conversion may also maintain high sequence homology of duplicated segments [26,27]. Thus, we tested the chimpanzee genome by Southern genomic analysis for the presence of the CHRNA7 duplication which revealed this duplication is most likely human-specific and therefore did not contribute to the pericentric inversion in chimpanzee (Figure 5). Our BAC library hybridization results also support this assertion. In addition, FISH results with probe RP11-30N16, which contains the intact donor CHRNA7 locus, did not produce dual FISH signals on PTR XVp and XVq, further demonstrating this sequence is not involved in the evolutionary rearrangement. Finally, the association of the dual FISH signal with clones that did not contain the CHRNA7 duplication (RP11-422F16) indicates the split signal is caused by a duplicated sequence within the 600 kb interval other than the CHRNA7-related sequence. Barring other duplication events that may have been lost through subsequent deletion, we estimate that the breakpoint region in the ancestral genome was significantly smaller (around 400 kb or potentially smaller).
Interestingly, the extent of the segmental duplications in the 1 Mb region examined was restricted to the clones that showed the dual FISH signal in PTR XVp and XVq and each clone that produced dual FISH signals contained a GLP/LCR15 duplicon by sequence analysis (Figure 4). With our results indicating the CHRNA7 sequence is single copy in the chimpanzee genome and thus unlikely to be involved in the inversion, the GLP/LCR15 duplicon is therefore the best candidate sequence for being present at the site of the pericentric inversion. Unfortunately, the highly duplicated nature of the region prevents precise narrowing of the breakpoint to a specific sequence; however, we believe large-scale sequence analysis of this region between human and chimpanzee will be able to reveal the precise nature of the breakpoint sequence.
Pericentric inversions are the most common rearrangement differentiating humans and the great ape species at the karyotypic level [1,2]. In addition, it has been proposed that multiple inversions within a single chromosome could potentially lead to speciation [28]. The mechanism underlying their origin, however, remains poorly understood. It has been shown in multiple studies that regions of segmental duplication can be associated with evolutionary rearrangements [8,29]. The pericentric inversion of chimpanzee XVp, having been localized to a site of multiple segmental duplications in the human genome, indicates that the evolutionary rearrangement is likely to involve such sequences. On the basis of the findings of Pujana et al., the GLP/LCR15 duplicon is present at multiple regions of human genomic instability [21]. In addition, the GLP/LCR15 duplicon was shown by Gilles et al. to be polymorphic in the human population - showing that these sequences have considerable intraspecific plasticity [18]. The GLP/LCR15 duplication family has expanded over the last 20 million years of primate evolution, before the divergence of the great ape lineages, resulting in a multitude of copies (estimated at 27) present at multiple sites along chromosome 15, providing the conditions required for rearrangements driven by nonallelic homologous recombination (NAHR) [18,19,21,27]. We have begun an investigation of the organization of this region within the chimpanzee lineage. Both genomic library characterization and partial sequence characterization (AC119799) indicate the presence of both LCR15 and HERC2 duplicons in this region. No evidence of CHRNA7 was found either by sequence-similarity searches or through sequence-tagged site (STS)-content hybridization mapping (data not shown). Large-scale sequence analysis of this region between human and chimpanzee will be necessary to refine the exact position of the pericentric inversion. For NAHR to have had a role, however, one must presume the presence of a GLP/LCR15-related sequence on the ancestral 15p. Alternatively, this site, which contains a recent human-specific segmental duplication of the CHRNA7 locus, may be a preferential site of breakage, which allowed the pericentric inversion to occur. The association of two independent chromosomal rearrangement events at one site: a pericentric inversion in the chimpanzee genome, and the insertion of a novel segmental duplication within the human lineage, may be a consequence of the overall genomic instability of this region.
Materials and methods
FISH probe selection
We selected probes to refine the chimpanzee XVq11-q13 pericentric inversion breakpoint on the basis of YAC maps of human 15q11-q13, in addition to the genome assemblies produced by the Human Genome Project [30,31]. We chose BACs targeted for complete sequencing (Figure 1) so that the underlying sequence of each probe would be available for further analysis pending comparative FISH results. Particular attention was paid to the position of each probe with respect to the three large low-copy repeat clusters found within 15q11-q13; as these clusters contain both inter- and intrachromosomal segmental duplications, and correspond to the common deletion breakpoints of Prader-Willi and Angelman syndrome deletions [12,13,21,32]. The duplication track of the UCSC Genome Browser, August 2001 assembly, displays information obtained from a global analysis of segmental duplications in the human genome, and allows for the selection of informative unique clones [14,16]. In this manner, we selected probes flanking the human 15q11-q13 rearrangement breakpoints associated with common PWS/AS deletions. To mark the regions of low-copy repeats in human 15q11-q13, we utilized a probe derived from the donor locus of a major component of the PWS/AS breakpoint duplication clusters, PAC clone pDJ-778A2 which contains a significant portion of the HERC2 gene in 15q13 [33]. In addition, we selected probes corresponding to sites distal to the PWS/AS domain flanking a large inverted-duplicated 15 (inv dup(15)) supernumerary marker chromosome [23]. Specifically, probe RPCI-11-88O16 (abbreviated RP11-88O16) was selected for FISH analysis because it lies distal to the inversion-duplication event breakpoint and is linked to STS marker D15S1010 [23,24]. Upon reaching the PTR inversion breakpoint interval with probe RP11-40J8, flanking BACs were chosen from the November 2002 human genome assembly to establish the extent of the interval and define the boundaries of the interval. These additional hybridizations were performed on human and chimpanzee material only.
Comparative FISH
Previous reports indicated that the inversion of human 15q11-q13 in the chimpanzee genome occurred solely in the chimpanzee lineage, whereas humans share the ancestral state with the gorilla (Gorilla gorilla; GGO) and orangutan (Pongo pygmaeus; PPY). To confirm these observations, metaphase chromosome preparations were generated from lymphoblast-derived cell lines for human, common chimpanzee, pygmy chimpanzee (Pan paniscus; PPA), gorilla and orangutan. BAC and PAC DNAs, isolated from bacterial cultures (Nucleobond, Clontech, Franklin Lakes, NJ), were labeled by nick-translation with biotin and digoxigenin. FISH experiments were performed under standard conditions [34]. For each FISH experiment at least 20 independent metaphase chromosome preparations were examined.
Duplication analysis
The approximately 1 Mb (1,008,705 base-pairs (bp)) interval encompassing the entire pericentric inversion breakpoint interval and the adjacent unique (that is, present on only one chromosome arm in PTR) FISH probes was extracted from the November 2002 human genome assembly and compared to the entire genome using MEGABLAST. Prior to sequence similarity searches, however, the 1 Mb of sequence was masked using RepeatMasker [35], and all identified high-copy repeats were filtered as lower-case letters. For efficient MEGABLAST analysis, the human genome sequence is fragmented into 400 kb pieces, each chromosome is fractioned separately and each fragment labeled consecutively from p-telomere to q-telomere. Thus, the most distal 400 kb segment of human chromosome 15 is labeled chr15_001, and so on. MEGABLAST parameters were set to take advantage of the lower-case masking, which allows for alignment extension (but not alignment seeding) through lower-case masked sequences. The MEGABLAST output was parsed into tab-delimited text and displayed using the graphical alignment display program Parasight (Jeff Bailey, unpublished work). The results were then compared to previous duplication analyses available through the UCSC Genome Browser [14-16].
Southern analysis
To determine the evolutionary age of the CHRNA7 duplication with respect to the human/chimpanzee divergence, we performed a genomic Southern blot using DNA extracted with the PureGene DNA isolation kit (Gentra Systems, Minneapolis, MN). DNA was obtained from cell lines and/or blood from a human (sample ID 02-0056), two unrelated common chimpanzees (sample IDs: NA03450B, NA03448A), two unrelated pygmy chimpanzees (sample IDs: BB501, LB502), and two unrelated gorillas (sample IDs: 9521, 9247). Probe CHRNA7-PstI was designed by identifying an approximately 100 bp RFLP between the CHRNA7-related sequence within accessions AC010799 (RP11-40J8) and AC111169 (RP11-20D7), using the web-based restriction mapping program Webcutter 2.0 [36]. Once a polymorphism distinguishing the two copies of CHRNA7 in Homo sapiens had been identified, probe CHRNA7-PstI was PCR amplified from BAC RP11-40J8 DNA using primers CHRNA7.probe2.1 (forward, 5'-TGAAACCTTGGGTGAGTTGG-3') and CHRNA7.probe2.2 (reverse, 5'-CAGATGAGACTGGGAAAGGC-3') at an annealing temperature of 55°C for 35 cycles. For Southern analysis, all DNAs were digested with PstI, blotted, and the resulting membranes were hybridized and washed according to standard protocols.
Filter hybridizations of the RPCI-43 and CHORI-251 Segment 1 libraries were performed according to previously established methods [37,38]. The PCR-amplified probe 40J8.Site2, amplified using primers 40J8.Site2.1 (forward, 5'-GTACTTTACAAGCAGGCGGC-3') and 40J8.Site2.2 (reverse 5'-CAGATGAGACTGGGAAAGGC-3') at an annealing temperature of 55 degrees Celsius for 35 cycles, was designed to the CHRNA7-related portion of BAC RP11-40J8 (accession AC010799).
Acknowledgments
Acknowledgements
We thank Juliann E. Horvath for critical reading of the manuscript and Sean D. McGrath for technical assistance. This work was supported by grants NIH GM58815, NIH HG002385 and DOE ER62862 to E.E.E., the financial support of Telethon, CEGBA (Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare), and MIUR (Ministero Italiano della Istruzione e della Ricerca) to N.A.
References
- Yunis JJ, Sawyer JR, Dunham K. The striking resemblance of high-resolution G-banded chromosomes of man and chimpanzee. Science. 1980;208:1145–1148. doi: 10.1126/science.7375922. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Dehal P, Predki P, Olsen AS, Kobayashi A, Folta P, Lucas S, Land M, Terry A, Ecale Zhou CL, Rash S, et al. Human chromosome 19 and related regions in mouse: conservative and lineage-specific evolution. Science. 2001;293:104–111. doi: 10.1126/science.1060310. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- White MJ. Models of speciation. New concepts suggest that the classical sympatric and allopatric models are not the only alternatives. Science. 1968;159:1065–1070. doi: 10.1126/science.159.3819.1065. [DOI] [PubMed] [Google Scholar]
- Nickerson E, Nelson DL. Molecular definition of pericentric inversion breakpoints occurring during the evolution of humans and chimpanzees. Genomics. 1998;50:368–372. doi: 10.1006/geno.1998.5332. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Park SS, Inoue K, Lupski JR. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 2001;11:1205–1210. doi: 10.1101/gr.181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Schreiner B, Tanzer S, Platzer M, Muller S, Hameister H. Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am J Hum Genet. 2002;71:375–388. doi: 10.1086/341963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke S, Verma RS. The genomic sequence for Prader-Willi/Angelman syndromes' loci of human is apparently conserved in the great apes. J Mol Evol. 1995;41:250–252. doi: 10.1007/BF00170680. [DOI] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- Ji Y, Rebert NA, Joslin JM, Higgins MJ, Schultz RA, Nicholls RD. Structure of the highly conserved HERC2 gene and of multiple partially duplicated paralogs in human. Genome Res. 2000;10:319–329. doi: 10.1101/gr.10.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- University of California Santa Cruz Genome Bioinformatics http://www.genome.ucsc.edu
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B, Williamson M, Collier D, Wilkie H, Makoff A. A 3-Mb map of a large segmental duplication overlapping the alpha7-nicotinic acetylcholine receptor gene (CHRNA7) at human 15q13-q14. Genomics. 2002;79:197–209. doi: 10.1006/geno.2002.6694. [DOI] [PubMed] [Google Scholar]
- Gilles F, Goy A, Remache Y, Manova K, Zelenetz AD. Cloning and characterization of a Golgin-related gene from the large-scale polymorphism linked to the PML gene. Genomics. 2000;70:364–374. doi: 10.1006/geno.2000.6388. [DOI] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Gratacos M, Peral B, Csiszar K, Gonzalez-Sarmiento R, Sumoy L, Estivill X. Additional complexity on human chromosome 15q: identification of a set of newly recognized duplicons (LCR15) on 15q11-q13, 15q24, and 15q26. Genome Res. 2001;11:98–111. doi: 10.1101/gr.155601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacos M, Nadal M, Martin-Santos R, Pujana MA, Gago J, Peral B, Armengol L, Ponsa I, Miro R, Bulbena A, et al. A polymorphic genomic duplication on human chromosome 15 is a susceptibility factor for panic and phobic disorders. Cell. 2001;106:367–379. doi: 10.1016/s0092-8674(01)00447-0. [DOI] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Guitart M, Armengol L, Gratacos M, Estivill X. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur J Hum Genet. 2002;10:26–35. doi: 10.1038/sj.ejhg.5200760. [DOI] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- Wandstrat A, Lena CJ, Jenkins L, Schwartz S. Molecular cytogenetic evidence for a common breakpoint in large (class III) inverted duplications of chromosome 15. Am J Hum Genet. 1998;62:925–936. doi: 10.1086/301777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandstrat AE, Schwartz S. Isolation and molecular analysis of inv dup(15) and construction of a physical map of a common breakpoint in order to elucidate their mechanism of formation. Chromosoma. 2000;109:498–505. doi: 10.1007/s004120000103. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Johnson ME, Alkan C, Tuzun E, Sahinalp C, Misceo D, Archidiacono N, Rocchi M. Divergent origins and concerted expansion of two segmental duplications on chromosome 16. J Hered. 2001;92:462–468. doi: 10.1093/jhered/92.6.462. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev. 2002;12:312–319. doi: 10.1016/s0959-437x(02)00304-0. [DOI] [PubMed] [Google Scholar]
- King M. Species Evolution: The Role of Chromosome Change. New York: Cambridge University Press; 1993. [Google Scholar]
- Nickerson E, Gibbs RA, Nelson DL. Sequence analysis of the breakpoints of a pericentric inversion distinguishing the human and chimpanzee chromosomes 12. Am J Hum Genet. 1999;65 (Supplement):A56. [Google Scholar]
- Christian SL, Bhatt NK, Martin SA, Sutcliffe JS, Kubota T, Huang B, Mutirangura A, Chinault AC, Beaudet AL, Ledbetter DH. Integrated YAC contig map of the Prader-Willi/Angelman region on chromosome 15q11-q13 with average STS spacing of 35 kb. Genome Res. 1998;8:146–157. doi: 10.1101/gr.8.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Amos-Landgraf J, Ji Y, Wandstrat A, Driscoll D, Schartz S, Nicholls R. Recombination between large, transcriptionally active repeated elements at the proximal and distal breakpoints in Prader-Willi and Angelman syndromes. Am J Hum Genet. 1997;61 (Supplement):A3. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Walkowicz MJ, Buiting K, Johnson DK, Tarvin RE, Rinchik EM, Horsthemke B, Stubbs L, Nicholls RD. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet. 1999;8:533–542. doi: 10.1093/hmg/8.3.533. [DOI] [PubMed] [Google Scholar]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- RepeatMasker http://ftp.genome.washington.edu/RM/RepeatMasker.html
- Webcutter http://www.firstmarket.com/cutter/cut2.html
- Horvath J, Schwartz S, Eichler E. The mosaic structure of a human pericentromeric segment: a strategy for characterizing complex regions of the human genome. Genome Res. 2000;10:839–852. doi: 10.1101/gr.10.6.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Children's Hospital Oakland Research Institute: BAC and PAC resources http://www.chori.org/bacpac


