Abstract
The tumor suppressor p53 regulates diverse biological processes primarily via activation of downstream target genes. Even though many p53 target genes have been described, the precise mechanisms of p53 biological actions are uncertain. In previous work we identified by microarray analysis a candidate p53 target gene, FLJ11259/DRAM. In this report we have identified three uncharacterized human proteins with sequence homology to FLJ11259, suggesting that FLJ11259 is a member of a novel family of proteins with six transmembrane domains. Several lines of investigation confirm FLJ11259 is a direct p53 target gene. p53 siRNA prevented cisplatin-mediated up-regulation of FLJ11259 in NT2/D1 cells. Likewise in HCT116 p53+/+ cells and MCF10A cells, FLJ11259 is induced by cisplatin treatment but to a much lesser extent in isogenic p53-suppressed cells. A functional p53 response element was identified 22.3kb upstream of the first coding exon of FLJ11259 and is shown to be active in reporter assays. In addition, chromatin immunoprecipitation assays indicate that p53 binds directly to this element in vivo and that binding is enhanced following cisplatin treatment. Confocal microscopy showed that an FLJ-GFP fusion protein localizes mainly in a punctate pattern in the cytoplasm. Overexpression studies in Cos-7, Saos2, and NT2/D1 cells suggest that FLJ11259 is associated with increased clonal survival. In summary, we have identified FLJ11259/DRAM as a p53-inducible member of a novel family of transmembrane proteins. FLJ11259/DRAM may be an important modulator of p53 responses in diverse tumor types.
Keywords: FLJ11259, DRAM, p53, target gene
1. Introduction
An important area of investigation is to unravel the complex network of target gene regulation surrounding the tumor suppressor p53 [1,2]. p53 is involved in the regulation of cell cycle arrest and apoptosis following DNA damage or cellular stress. Other p53 regulated biological processes include maintenance of genomic stability, inhibition of angiogenesis, DNA repair and cytoskeletal function [3,4]. p53 regulation and subsequent downstream target gene activation may be cell type-specific and context-dependent adding to the difficulty in identifying p53 transcriptional targets. Computational methods predict that thousands of genes possess p53 DNA binding elements. However, functional p53 enhancer identification requires experimental validation [5].
Our prior work utilizing microarray analysis uncovered a p53-dominant transcriptional response to cisplatin in the human embryonal carcinoma derived cell line, NT2/D1 [6]. In addition to many well-known direct p53 target genes, a previously uncharacterized open reading frame designated FLJ11259 was uncovered as the fifth-highest gene induced with cisplatin [6]. Here we confirm that FLJ11259 is a direct p53 target gene. FLJ11259 is highly conserved throughout species and predicted to have six transmembrane domains. FLJ11259 is regulated by DNA damaging agents in various cell lines in a p53-dependent manner, and p53 directly binds to an upstream element in the FLJ11259 gene. FLJ11259 was observed to localize in a punctate pattern throughout the cytoplasm. Stable overexpression of FLJ11259 in Cos-7, Soas2, and NT2/D1 cells resulted in increased clonagenicity, demonstrating that FLJ11259 may play a role in cell survival under certain conditions.
2. Materials and methods
2.1 Cell Culture and Drug Treatments
NT2/D1, Cos7 and Saos2 cells (ATCC) were cultured in DMEM (Gibco) with 10% FBS supplemented with glutamine and antibiotics. HCT116 colon cancer cell lines, p53+/+ and p53−/− were a gift from Dr. B. Vogelstein (Johns Hopkins University). HCT116 cells were cultured in McCoys 5A media (Gibco) supplemented with 10% FBS, antibiotics and glutamine. MCF10A immortalized breast epithelium cells and the MCF10A/Δp53 cell line stably transfected with p53 shRNA were previously described and generously provided by Dr. A. Eastman (Dartmouth Medical School) [7]. MCF10A cells were cultured in DMEM/F12 supplemented with 10% FBS, 8 μg/ml insulin, 20 ng/ml epidermal growth factor and 500 ng/ml hydrocortisone. The MCF10A/Δp53 cell line was cultured in the same media in the presence of G418 which was removed prior to experimental conditions. Cisplatin (Bristol Laboratories) treatments were performed in the respective media at the concentrations and time points indicated.
2.2 siRNA
Two independent siRNAs designated p53 siRNA-1 and p53 siRNA-2 were designed to human p53, targeting sequences AAGACUCCAGUGGUAAUCUAC and CGGCAUGAACCGGAGGCCCAU, respectively. The siRNA control was the Scramble II sequence (Dharmacon). A final concentration of 75 nM siRNA was transfected using OligofectAMINE reagent (Invitrogen), as described previously [8].
2.3 Northern, RT-PCR and Western Analysis
Total RNA was harvested using TriReagent (Invitrogen). Northern hybridizations were performed with 5 μg RNA as previously described [9]. Expression levels of FLJ11259 were measured by semi quantitative RT-PCR. The cDNA was synthesized using Superscript II RT from 5 μg total RNA. PCR analysis was performed with Taq polymerase (Invitrogen) to varying cycle numbers within the linear range as previously described [9]. RT-PCR expression analysis was also performed on normal (Human II MTC panel, cat# 636743) and tumor (Human multiple tumor panel, cat# K1422-1) cDNA samples (BD Bioscience). Primer sequences are available on request. For Western analysis cells were lysed in RIPA buffer and protein concentrations determined by Bradford technique. Proteins were analyzed by SDS-PAGE as previously described [10]. A GFP antibody was employed (JL-8 from BD Biosciences).
2.4 Chromatin Immunoprecipitation Assays (ChIP)
Cells were plated at a density of 3 × 106 per 15 cm dish. Cisplatin treatment of NT2/D1 cells was 2 μM for 6 hours prior to harvesting 16 hours later. In HCT116 cells the duration of 24 hours at 20 μM cisplatin treatment was employed. Subsequent steps were in accordance with the Chromatin Immunoprecipitation kit (Upstate Biotech). Briefly, cells were fixed with 1% formaldehyde for 10 minutes at 37 °C. Following cell lysis in the presence of protease inhibitors, cells were sonicated for 3 × 10 second pulses at setting 3.5 on a Vibra Cell sonicator (Danbury, CT). Diluted samples were then pre-cleared with agarose beads. Lysates were incubated overnight with antibodies specific for p53 (DO-1, Santa Cruz and DO-7, Neomarkers) and a control antibody for the estrogen receptor (Ab-10, Neomarkers). Antibodies were used at a concentration of 2 μg/ml of lysate. Following incubation with agarose beads, and subsequent washing steps, the antibody complexes were eluted and incubated at 65°C for 4 hours to reverse cross-linkages. DNA was eluted with the Qiagen PCR purification kit (according to the manufacturers directions) prior to PCR amplification. PCR primers surrounding the p53 response element of FLJ11259 were GGGAGGTCTGCAC-TGTTGAATT (sense) and CCAGCTTGCTTTA-GGCAGACA (anti-sense). Primers surrounding the p53RE in p21 were CGAGGCAGGCCAAGGG (sense) and GCAGAGGATGGATTGTTCA (anti-sense).
2.5 Expression Constructs
A clone of FLJ11259 in pCMV-SPORT6 was purchased from Open Biosystems. Restriction digestion with EcoR1 and Sal1 removed the entire coding sequence which was inserted into pcDNA 3.1 Myc-His (−) (BD Bioscience) containing a G418 resistance cassette. To facilitate in-frame ligation into the pEGFP-N2 vector (BD Bioscience), Xho1 and Kpn1 sites were engineered at the 5' and 3' ends respectively of the FLJ11259 insert by PCR. The 3' primer was designed to mutate the stop codon of FLJ11259 to allow continuous translation between FLJ and GFP to create a fusion protein. Sequence analysis confirmed wild-type sequence and the correct codon usage of the FLJ-GFP fusion. A 600 bp fragment of the FLJ11259 gene containing the identified p53 response element was isolated from BAC clone RP11-9F22 by PCR and cloned into pCR 2.1-TOPO vector. Restriction digestion with Xho1 and Kpn1 facilitated insertion of this DNA fragment into the pGL3-promoter luciferase reporter vector (Promega). A version of FLJ-luc in which the p53 was mutated from AGGCATGTTTAGGCAAGCTC to AGGaATaTTTAGGaAAaCTC was constructed using QuikChange Site-Directed Mutagenesis (Stratagene), and was DNA sequence confirmed. The tk-luc vector was a kind gift from Dr. J. DiRenzo (Dartmouth Medical School). Oligonucleotides corresponding to the FLJ p53RE (sense – AGGCATGTTTAGGCAAGCTC) were annealed and ligated into the tk-luc vector to generate a 5x p53RE construct designated FLJp53RE-luc. The dominant-negative p53 (DNp53) construct has been previously described [10].
2.6 Confocal Microscopy
Cos-7 cells were plated on coverslips at a density of 0.125 × 106 cells per well of a 6 well dish. Transfection of 1 μg DNA per well was performed with Fugene according to the manufacturers directions. The presence of GFP was detected 24 hours later using fluorescent microscopy. For mitochondrial staining, Mitotracker Deep Red 633 (Molecular Probes) was prepared in DMEM at a working concentration of 500 nM and cells were incubated at 37°C for 30 minutes. Cells were washed with PBS, fixed with 3.7% formaldehyde at 37°C for 15 minutes prior to mounting with prolong containing DAPI (Molecular Probes). The plasma membrane was stained with 5 μg/mL Alexa Fluor 594 wheat germ agglutinin (Molecular Probes) in Hank's balanced salt solution (HBSS, Gibco) for 10 minutes at 37°C. Cells were washed with HBSS and fixed in 4% formaldehyde for 15 minutes at 37°C prior to mounting with prolong/DAPI. Endoplasmic reticulum (ER) was stained with ER-Tracker Red (glibenclamide BODIPY TR) at a final concentration of 1 μM in HBSS for 30 minutes at 37°C. Cells were then washed with PBS and fixed for two minutes at 37°C with 4% formaldehyde. Following washing cells were mounted with prolong/DAPI. Confocal microscopy was performed on the Zeiss-LSM-510 META point scanning confocal microscope. Cells were imaged using a 63x oil objective and a pinhole of 1 airy unit was the standard for image capture.
2.7 Clonal Growth Assay
Cos-7, Saos2 or NT2/D1 cells were plated at 0.175 × 106 cells per well in triplicate in 6 well dishes. Transfection with Fugene was performed according to the manufacturers directions with either empty pcDNA 3.1, or pcDNA 3.1 containing the FLJ11259 insert (FLJ-1 and FLJ-2). In the p53 null, Saos2 cell line, transfection of a p53 expression vector (pC53-SN3) and empty vector control was also performed. Following transfection, 2 × 105 cells were replated into T75 flasks and treated with media containing 500 μg/ml G418. No colonies formed on plates of non-transfected cells following G418 selection. Surviving transfected cells formed colonies following 7-14 days depending on the cell line. Cells were stained with Giemsa and colonies were counted manually.
2.8 Reporter Assays
Cells were plated at 0.175 × 106 cells per well of a 6 well dish. Fugene transfections were performed in triplicate for each condition with 0.8 μg reporter, 0.2 μg pRL-TK and 0.5 μg of either p53 expression plasmid, dominant-negative p53 (DNp53) or empty vector control. Cells were harvested 40 hours following transfection and Dual Luciferase Reporter analysis was performed (Promega). Luciferase activity was normalized against renilla activity and represented as the average of triplicate transfections.
3. Results
3.1 In Silico Analysis of FLJ11259 Uncovers a Predicted Transmembrane Protein
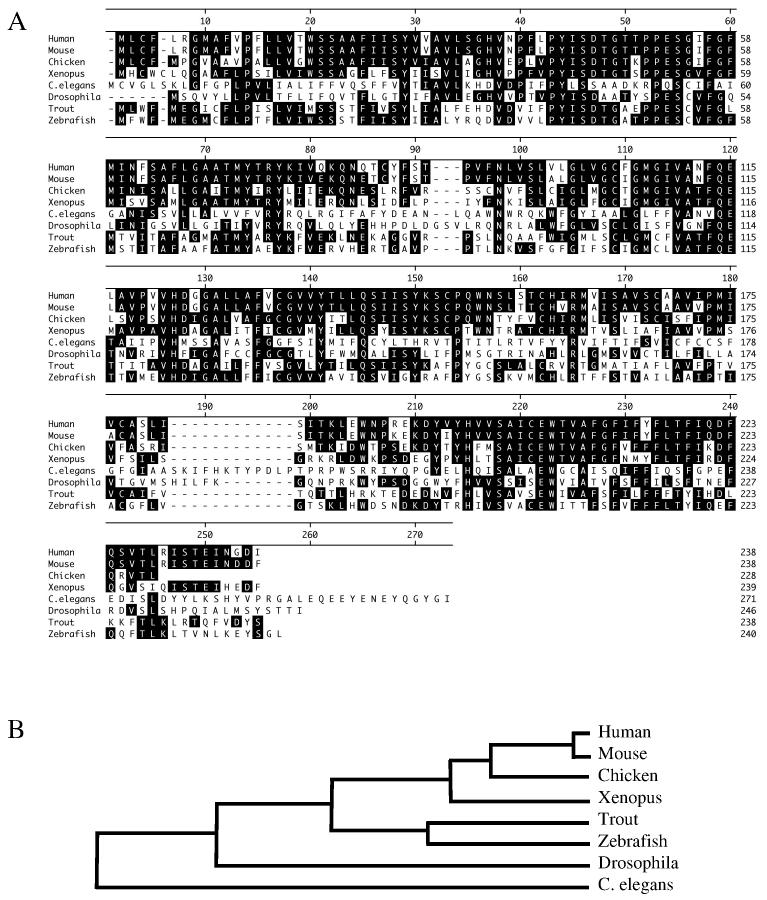
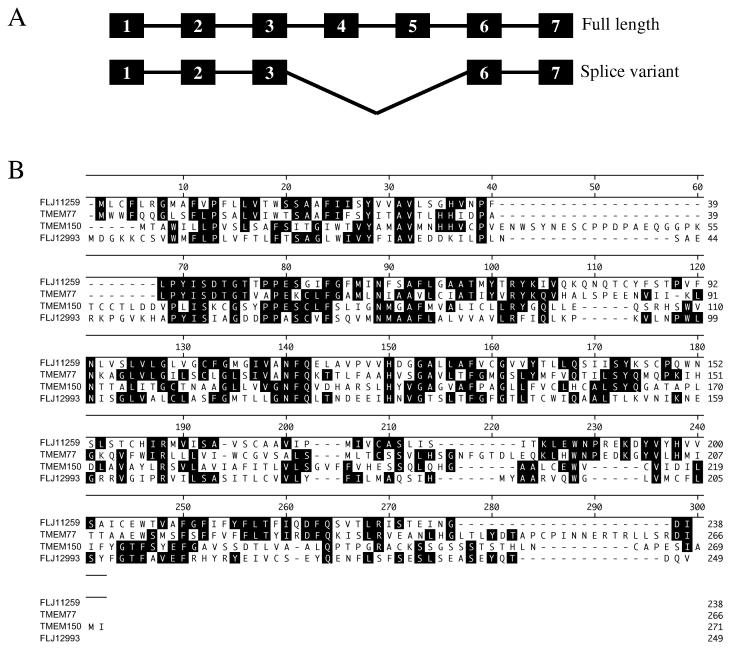
FLJ11259 was identified by microarray analysis of cisplatin treated NT2/D1 embryonal carcinoma cells [6]. The Affymetrix microarray probe-set (218627_at) for FLJ11259 is located within the 3' UTR of this gene. Searches of the NCBI website identified the complete sequence as represented by BC018435. This gene has 7 exons and is located on chromosome 12q23. The protein product is estimated to be 26 kD in size. Blast alignment of the protein sequence throughout species shows that FLJ11259 is a highly conserved protein in many vertebrates (Fig. 1A). There is a high degree of homology between human FLJ11259 and that of the mouse (97%) and rat (83%), as well as trout (63%) and other species. Phylogenetic analysis shows a high degree of conservation of FLJ1159 throughout evolution. Evolutionary similarity is highest between human and mouse sequences, with C. elegans being the most evolutionary diverse (Fig. 1B). Using the tools on the Expasy website (http://us.expasy.org/tools/), FLJ11259 is predicted to be a 6-transmembrane protein with both the N- and C-termini located in the intracellular compartment of the cell. FLJ11259 does not have significant homology to any other characterized proteins in the Genbank database, and does not contain any known conserved functional domains. RT-PCR analysis with primers designed to amplify the entire transcribed region of FLJ11259 identified a potential splice variant (Fig. 2A). Sequence analysis of the alternative PCR product demonstrated a seamless deletion of exon 4 and exon 5, resulting in conservation of the reading frame with a predicted internal deletion of the fourth and fifth predicted transmembrane domains of FLJ11259. Both of these FLJ11259 variants are equally inducible by cisplatin in NT2/D1 cells (data not shown). The functional significance of this alternative splicing has yet to be determined.
Fig. 1.
FLJ11259 is highly conserved throughout species. (A) Protein alignment of FLJ11259 showing homology between species, residues in the black boxes match the consensus. The sequence for chicken has an additional 13 amino acids upstream of that reported in this alignment, which was removed for clarity. (B) Phylogenetic tree representing conservation of FLJ11259 sequence throughout evolution. Both the protein alignment and phylogenetic analysis were performed with Lazergene software.
Fig. 2.
Potential FLJ11259 splice variants and family members. (A) Potential splice variants of FLJ11259, the longer version contains all 7 exons, whereas the shorter, truncated version does not contain exons 4 and 5. (B) Alignment of FLJ11259 and novel human family members. The four proteins contain both sequence and structural homology and are represented by FLJ11259 (Hs.525634), TMEM77 (Hs.485606), TMEM150 (Hs.591599) and FLJ12993 (Hs.507676).
3.2 Human FLJ11259 Family Member Expression in Normal and Tumor cDNA
In-silico analysis has identified three additional uncharacterized human proteins with sequence and structural homology to FLJ11259. Thus FLJ11259 appears to be one member of a novel family of proteins. There are multiple conserved amino acid residues between the four family members which are over 40% homologous with each other (Fig. 2B). Hydropathy analysis, using the dense alignment surface method (DAS) transmembrane prediction server from Stockholm University (http://www.sbc.su.se/~miklos/DAS/), shows that the four proteins each contains 6 hydrophobic alpha helices and the location of these predicted transmembrane regions is relatively well conserved (data not shown). The expression profile of this novel family of genes was investigated (Fig. 3). In general the four transcripts appear to be expressed in a high percentage of the tissue samples investigated. Normal tissue expression of FLJ11259 is highest in the ovary, with the lowest expression in colon and leukocytes (Fig. 3A). The tumor panel suggests that FLJ11259 is expressed in the majority of tumor tissues analyzed (Fig. 3B), however direct comparison of matched normal and tumor samples has not been performed. FLJ11259 is also expressed in clinical germ cell tumor samples to a similar extent as the basal level of FLJ11259 in cultured NT2/D1 cells (Fig. 3C).
Fig. 3.
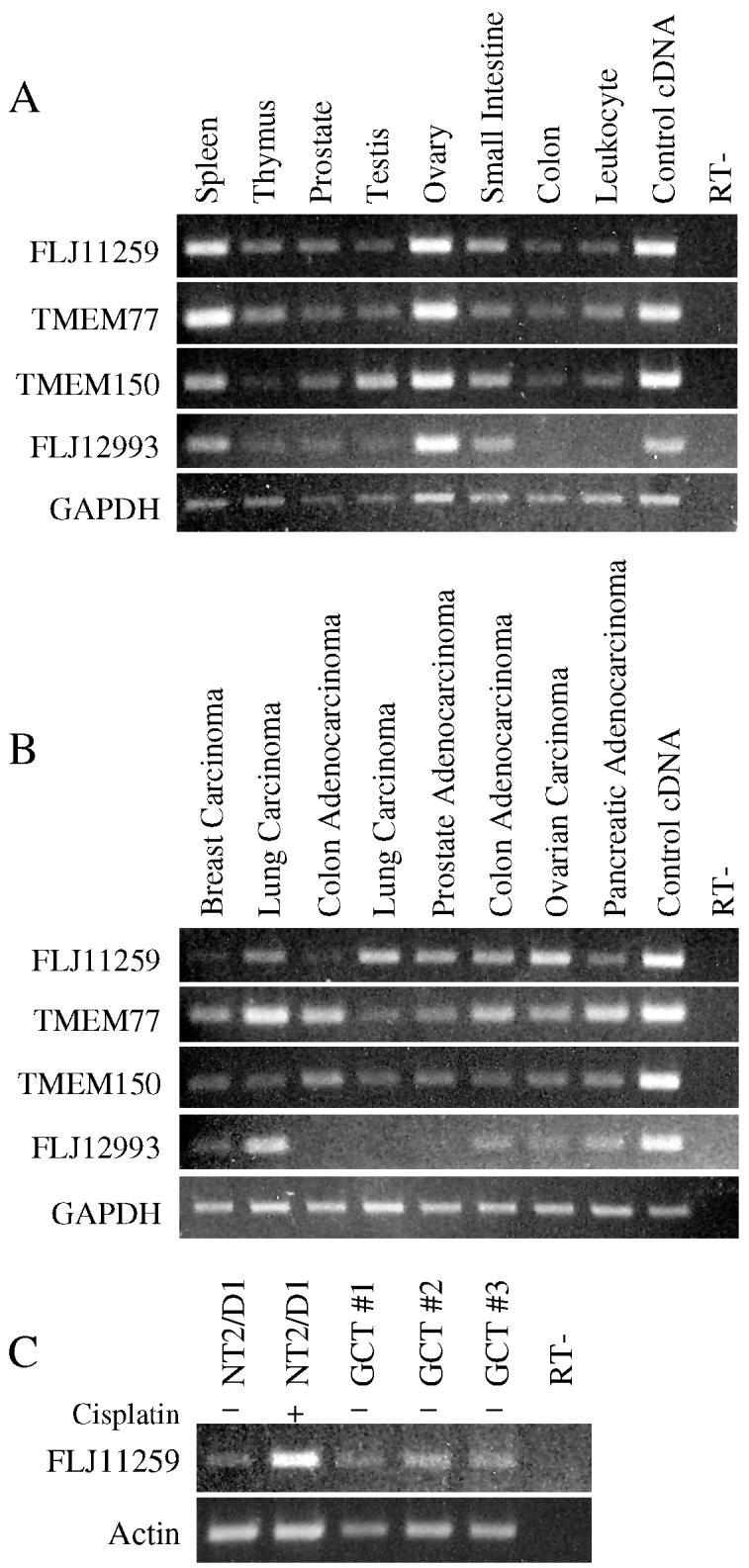
FLJ11259 family expression profiles in normal and tumor samples. (A) RT-PCR expression analysis of family members in normal cDNA samples from the Human II multiple tissue cDNA (MTC) panel (BD Bioscience). (B) Expression analysis of family members in tumor cDNA samples from the Human tumor MTC panel (BD Bioscience). (C) FLJ11259 is expressed in clinical germ cell tumor samples (GCT#1, 2 and 3). For comparison untreated and cisplatin treated (2 μM for 6 hours with a 24 hour recovery) NT2/D1 cells are shown. FLJ11259 primers are located in a region that does not distinguish between potential splice variants. GAPDH and actin primers were used for normalization. Data is representative of two independent experiments.
3.3 FLJ11259 Induction by DNA Damaging Agents Occurs in a p53-Dependent Manner
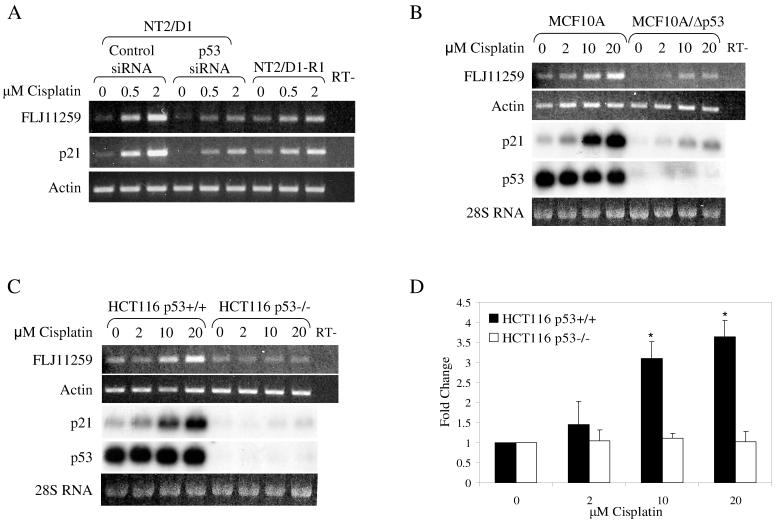
Preliminary evidence utilizing p53 siRNA indicated that cisplatin regulation of FLJ11259 in NT2/D1 cells was partially dependent on p53 [6]. Thus we sought to further characterize the importance of p53 in FLJ11259 induction by utilizing additional cell culture models (Fig. 4). FLJ11259 induction by cisplatin in NT2/D1 cells occurs in a dose-dependent manner similar to that of p21. As previously shown, NT2/D1 cells treated with p53 siRNA had diminished induction of FLJ11259 [6]. Further, a cisplatin resistant cell line, NT2/D1-R1, which was previously shown to have a defect in p53 signaling [10] also shows diminished FLJ11259 induction (Fig. 4A). Dose-response studies in HCT116 and MCF10A cell lines indicated a decreased sensitivity to cisplatin compared with NT2/D1 cells, hence a 10-fold higher dose range of cisplatin was chosen. FLJ11259 was induced following 24 hours of cisplatin treatment at concentrations of 2 μM to 20 μM in the colon cancer cell line HCT116 p53+/+, however induction of FLJ11259 by cisplatin in the p53 deleted cell line (HCT116 p53−/−) did not occur (Fig. 4C and D). Similar results suggesting FLJ11259 induction is p53-dependent were obtained in MCF10A cells, either expressing wild-type p53 or stably expressing shRNA to p53 (MCF10A/Δp53) (Fig. 4B). Northern analysis confirmed the absence of p53 expression in the HCT116 p53−/− and MCF10A/Δp53 cell lines and confirmed that p21 induction by cisplatin was also p53-dependent (Fig. 4B and C). Taken together these data suggest that FLJ11259 is induced by cisplatin, largely in a p53-dependent manner. Since FLJ11259 was the only gene in this family that was regulated by cisplatin according to our microarray analysis, it was of interest to determine if the newly identified family members were also cisplatin regulated. A cisplatin dose-response analysis determined that only FLJ11259 was cisplatin-inducible in NT2/D1 cells, and in contrast to FLJ11259, the expression of the other family members was not altered by p53 siRNA (data not shown).
Fig. 4.
FLJ11259 is induced by cisplatin in a p53-dependent manner. (A) RT-PCR analysis of FLJ11259 in cisplatin sensitive NT2/D1 cells transfected with either control or p53 siRNA, compared with resistant NT2/D1-R1 embryonal carcinoma cells. RNA was harvested 24 hours following a 6-hour cisplatin treatment. Induction of FLJ11259 occurs in a similar manner to p21. Prominent induction is seen in cisplatin treated NT2/D1 cells transfected with control siRNA. This induction is reduced in the p53 siRNA treated NT2/D1 cells and the cisplatin resistant NT2/D1-R1 cells. (B) FLJ11259 is up-regulated following 24 hours of cisplatin treatment in MCF10A cells, but this is suppressed in MCF10A/Δp53 cells stably expressing shRNA to p53. (C) FLJ11259 is up-regulated following 24 hours of cisplatin treatment in wild-type HCT116 cells (HCT116 p53+/+), but not in the isogenic p53 deleted line (HCT116 p53−/−). Northern analysis (B and C) confirms that p21 and FLJ11259 regulation by cisplatin is p53-dependent. (D) Densitometry of the RT-PCR analysis in part C was performed using Scion image. Data is represented as fold-change compared to the untreated condition in each cell line. Error bars are standard deviation. A 1-tailed, 2 sample student t-test was performed comparing FLJ11259 expression between HCT116 p53+/+ and HCT116 p53−/− at each concentration of cisplatin used. * p<0.001. Data is representative of at least two independent experiments.
3.4 FLJ11259 is a Direct p53 Target Gene
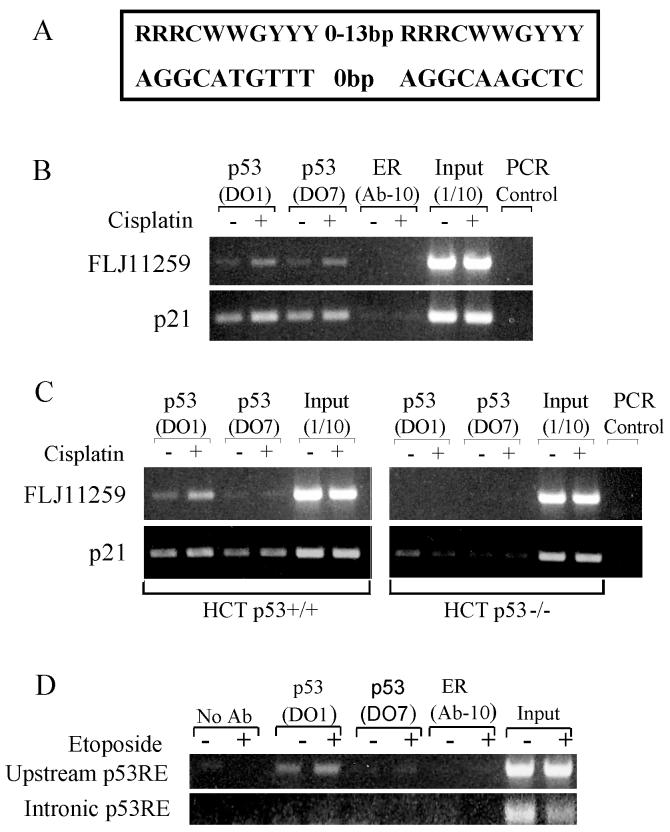
The majority of p53 target genes to date contain one or more p53 response elements (p53RE). A recent paper by Wei et al used a variation of the chromatin immunoprecipitation (ChIP)-chip technique to identify in a de novo manner genome-wide p53 binding sites [11]. Using HCT116 cells treated with 5-flurouracil they discovered p53 binding sites in 98 previously unidentified p53 target genes, including FLJ11259 [11]. The FLJ11259 site is a 100% consensus, bipartite p53 response element with 0-spacing between half-sites 22.3 kb upstream of the predicted transcriptional start site of FLJ11259 [11] (Fig. 5A). We performed ChIP assays in both NT2/D1 and HCT116 cells around this site. Immunoprecipitation was performed with two independent p53 antibodies and a control antibody to the estrogen receptor. The results indicate that in the absence of cisplatin stimulation there is binding of p53 to this p53RE at the endogenous FLJ11259 locus in NT2/D1 cells (Fig. 5B). Specific binding also occurs in HCT116 p53+/+ cells but not in HCT116 p53−/− cells (Fig. 5C). The addition of cisplatin further enhances basal binding approximately 2-fold in a p53-dependent manner, as compared to unstimulated cells (Fig. 5B and C). The p21 promoter contains a functional p53 response element and this site was used as a positive control demonstrating p53 specific binding and a further increase in p53 binding with cisplatin stimulation (Fig. 5B and C). During the preparation of the manuscript another report characterized an intronic p53RE between the first two coding exons of FLJ11259 [12]. We compared the upstream p53RE identified here with the intronic binding site by ChIP assay on the same samples using the primers and PCR conditions reported [12]. However, we were able to produce robust signals only in the upstream region using etoposide treated NT2/D1 cells and traditional PCR (Fig. 5D). It should be noted that Crighton et al. used different cells, drug treatments and the more sensitive real-time PCR method [12]. Further the input signal for the intronic p53RE is less than for the upstream p53RE suggesting less efficient amplification of this DNA segment even in the control.
Fig. 5.
p53 binds in vivo to an endogenous p53RE in FLJ11259. (A) Schematic showing the sequence of the p53 response element located upstream of the predicted transcriptional start site of FLJ11259. The p53RE has 100% homology with the consensus p53 element and contains no spacing between the 2 half sites. (B) Chromatin immunoprecipitation of NT2/D1 cells treated with vehicle or 2 μM cisplatin for 6 hours and harvested 16 hours later. Immunoprecipitation was performed with 2 independent p53 antibodies (DO-1 and DO-7) and a control antibody for the estrogen receptor (ER). PCR amplifications of FLJ11259 were performed around the p53RE and amplification of the p21 enhancer was performed surrounding its well-characterized p53 response element. (C) Chromatin immunoprecipitation of HCT116 p53+/+ and HCT116 p53−/− cells following 20 μM cisplatin treatment for 24 hours. Immunoprecipitation of p53 bound FLJ11259 is specific to p53 expressing cells. Data is representative of at least two independent experiments. (D) Chromatin immunoprecipitation of NT2/D1 cells treated with 1 μM etoposide for 6 hours and harvested 24 hours later. Immunoprecipitation was performed with antibodies described in Fig 5B. Upstream p53RE represents PCR amplification with primers used in Fig 5B and Fig 5C. Intronic p53RE represents PCR amplification primers previously reported [12].
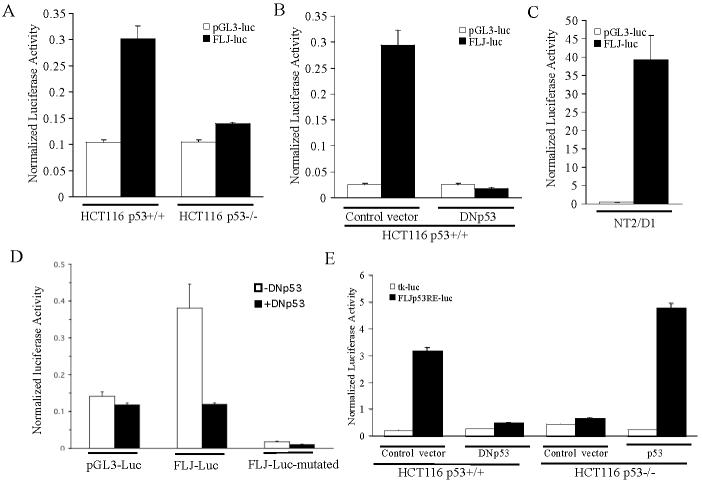
We cloned a 600-bp region of FLJ11259 surrounding the p53RE into the pGL3-promoter luciferase vector for reporter analysis. HCT116 p53+/+ and HCT116 p53−/− cells were transfected with either control reporter (pGL3-luc) or the FLJ11259 reporter construct (FLJ-luc). Only in HCT116 p53+/+ cells did FLJ-luc demonstrate activity above that of the control reporter (Fig. 6A). Co-transfection of dominant-negative p53 into HCT116 p53+/+ cells caused a dramatic decrease in FLJ-luc activity as compared to vector control (Fig. 6B). The same FLJ-luc construct transfected into NT2/D1 cells also had a high level of activity suggesting that this region of FLJ11259 contains a functional p53RE (Fig. 6C). Further, mutation of the p53RE within FLJ-luc decreased luciferase activity below the levels of the control vector and was not responsive to dominant-negative p53 (Fig 6D). We then generated an FLJp53RE-luc construct containing 5 copies of the p53RE sequence in front of the basal thymidine kinase (tk) promoter. HCT116 cells were transfected with FLJp53RE-luc or tk-luc control (Fig. 6E). FLJp53RE-luc activity occurs in a strictly p53-dependent manner since in HCT116 p53+/+ cells FLJp53RE-luc activity was abolished in the presence of DNp53. Likewise in HCT116 p53−/− cells activity was only observed following co-transfection of a p53 expression construct. The reporter data, together with the above ChIP results, establishes that there is a functional p53RE in the FLJ11259 gene.
Fig. 6.
FLJ11259 contains a functional p53 responsive element. (A) Reporter assay of HCT116 p53+/+ and HCT116 p53−/− cells transfected with either control pGL3-promoter reporter (pGL3-luc) or FLJ11259 reporter (FLJ-luc: pGL3-promoter containing a 600 bp region of FLJ11259 surrounding the p53 response element). (B) Reporter activity of HCT116 p53+/+ cells transfected with the pGL3-luc or FLJ-luc reporter and either a control vector or dominant negative p53 (DNp53). FLJ-luc activity is diminished in the presence of DNp53. (C) FLJ-luc reporter activity in NT2/D1 cells. (D) Loss of FLJ-Luc reporter activity in HCT116 p53+/+ cells when the p53 RE is mutated. The core C and G in each half site of the p53RE was mutated in the context of the 600-bp upstream element as described in Methods. (E) HCT116 p53+/+ and HCT116 p53−/− cells were transfected with FLJp53RE or the corresponding empty tk-luc vector and DNp53 or p53 expression constructs. Activity of the FLJp53RE reporter occurs in a p53-dependent manner in both cell systems. Data is representative of three independent experiments each performed in triplicate, error bars are standard deviation.
3.5 Overexpression of FLJ11259 Results in Increased Clonagenicity
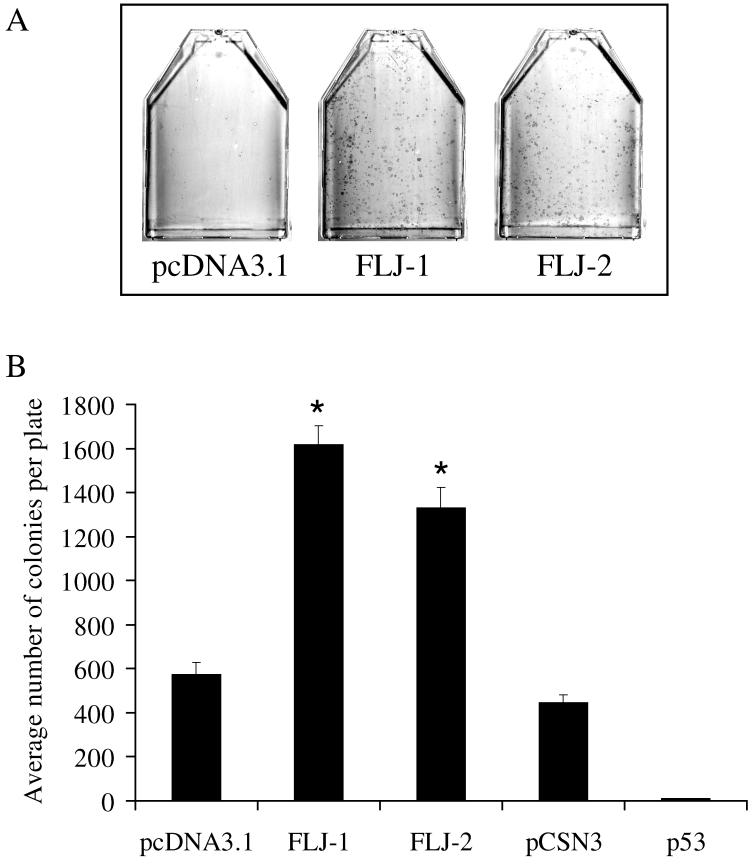
In order to gain insight into possible functions of FLJ11259, overexpression studies were performed. Cos-7, Saos2 and NT2/D1 cells were transfected with either insertless pcDNA3.1 or a pcDNA3.1-FLJ11259 expression vector each containing a G418 resistance cassette. In the case of Saos2 cells, a p53 expression plasmid (pC53-SN3) was also utilized. Following transfection, cells were cultured in 500 μg/ml G418 to select for stable integration. Table 1 summarizes the results of experiments in these cell lines. FLJ11259 transfection resulted in a substantial increase in colony formation when compared to controls (Table 1 and Fig. 7). As expected and shown by numerous groups, Saos2 cells transfected with a p53 expression plasmid resulted in substantially fewer colonies as compared to controls (Table 1 and Fig. 7B). These results suggest that FLJ11259 may have a positive effect on clonal survival under certain conditions.
Table 1.
Colony formation in FLJ11259 over-expressing cells*.
| Number of Colonies per T75 flask | ||||||
|---|---|---|---|---|---|---|
| Cell Line | Tx # | pcDNA3.1 | FLJ-1 | FLJ-2 | pCSN3 | p53 |
| Cos-7 | 1 | 7 | 297 | 256 | ND | ND |
| 2 | 34 | 463 | 565 | ND | ND | |
| NT2/D1 | 1 | 19 | 74 | 138 | ND | ND |
| 2 | 31 | 78 | 127 | ND | ND | |
| 3 | 26 | 77 | 102 | ND | ND | |
| Saos2 | 1 | 649 | 1645 | 1434 | 374 | 11 |
| 2 | 602 | 1461 | 1415 | 496 | 8 | |
| 3 | 467 | 1750 | 1145 | 466 | 11 | |
Cell lines were transfected with FLJ or p53 expression plasmids or corresponding empty vectors (pcDNA3.1 and pCSN3). Identical cell numbers were plated into G418 media. Colonies were counted approximately 2 weeks later. ND – Not determined. FLJ-1 and FLJ-2 are two independent plasmid preparations of pcDNA3.1-FLJ11259. Tx is transfection number.
Fig. 7.
Overexpression of FLJ11259 is associated with increased colony formation. (A) FLJ11259 overexpression increases colony formation after G418 selection of Saos2 cells transfected with FLJ expression vectors (FLJ-1 and FLJ-2 are two independent plasmid preparations of pcDNA3.1-FLJ11259) compared to empty vector (pcDNA3.1). Cells were stained with Giemsa to visualize colonies. (B) Number of colonies formed in Saos2 cells transfected with empty vectors (pcDNA3.1 or pCSN3) or FLJ expression vectors (FLJ-1 and FLJ-2) or p53. Triplicate plates were manually counted for each data point. Error bars are standard error of the mean. Statistical analysis performed with Student-Newman-Keuls multiple comparisons test, p<0.001 compared to pcDNA3.1 vector control.
3.6 FLJ11259 Localization
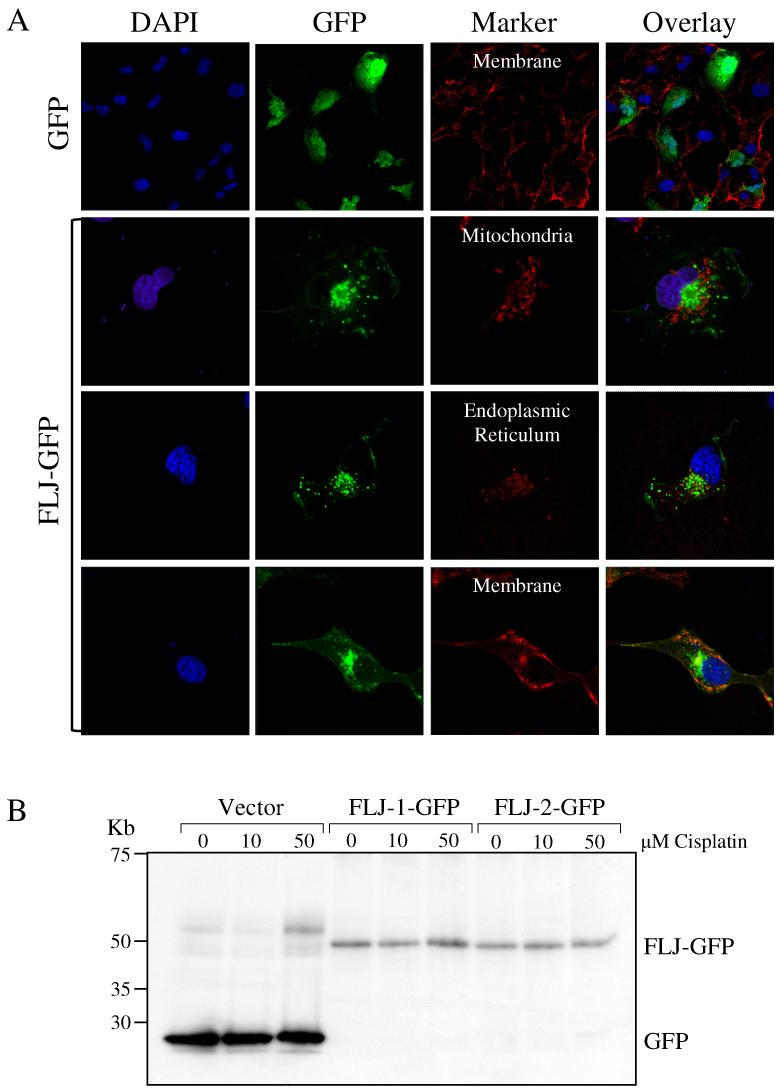
FLJ11259 is predicted to be a six-transmembrane protein and the TargetP prediction program (http://www.cbs.dtu.dk/services/TargetP/) suggests that FLJ11259 may contain a signal sequence and be involved in secretory pathways. The PSORTII program (http://www.psort.org/) predicted that the location of FLJ11259 with the highest probability was the endoplasmic reticulum (ER) (43.5%) followed by the plasma membrane (26.1%) and mitochondria (8.7%). We investigated the localization of an FLJ-GFP fusion protein to these three cellular compartments using confocal microscopy. Cos-7 cells were transfected with GFP alone or FLJ-GFP and then stained with either plasma membrane (WGA-Alexa 594), endoplasmic reticulum (ER tracker Red) or mitochondrial (Mitotracker red) markers. A series of images were taken along the z-axis through a single cell at various intervals, and through multiple cells, representative sections are shown. Control GFP expression demonstrated diffuse nuclear staining as indicated by co-localization of GFP fluorescence with DAPI stained nuclei (Fig. 8). In contrast, FLJ-GFP was largely dispersed in a punctate pattern throughout the cytoplasm with some plasma membrane localization. Localization of FLJ-GFP with the plasma membrane marker occurred in distinct cells at distinct regions as shown by the presence of yellow in the overlay images (Fig. 8). Differential interference contrast (DIC) images of cell morphology confirmed that the membrane marker was specific for the plasma membrane (data not shown). Co-localization of FLJ-GFP did not occur with the ER marker or the mitochondrial marker. These results suggest that FLJ11259-GFP is mainly localized to punctate cytoplasmic regions. There is currently no available antibody to FLJ11259, hence we were not able to ascertain the localization of endogenous FLJ11259.
Fig. 8.
FLJ-GFP localizes to punctate cytoplasmic areas with additional localization at the plasma membrane. (A) Confocal microscopy was performed in Cos-7 cells, transiently transfected with GFP or FLJ-GFP expression plasmids. GFP positive cells were analyzed for co-localization of GFP florescence with plasma membrane, endoplasmic reticulum and mitochondria stains. Nuclei are stained with DAPI. (B) In separate experiments, western analysis was performed using an anti-GFP antibody on Cos-7 stably transfected with GFP alone (vector) or FLJ-GFP. Western analysis confirms the expression of FLJ-GFP at the predicted molecular weight. Note: FLJ-GFP is not induced by cisplatin since expression is under the control of the constitutive CMV-promoter-enhancer.
4. Discussion
Testicular germ cell tumors (TGCTs) are one of the few solid tumors cured with conventional chemotherapy in advanced stages. The underling mechanism for this curability is unclear, although one proposed mechanism is linked to high levels of wild-type p53, characteristic of germ cell tumors (reviewed in [13]). Through gene expression profiling, we have previously identified a p53-dominant response to cisplatin and etoposide in the TGCT cell line, NT2/D1 [6]. In the current work a cisplatin inducible, p53 target gene has been characterized. FLJ11259 induction by cisplatin is highly dependent on p53 and occurs in a number of different cell systems (Fig. 4). Many genes are known to be regulated in a cell-specific and insult-specific manner [14,15], however the data here suggests that FLJ11259 is regulated in a p53-dependent manner in a variety of cell types and by at least two distinct DNA damaging agents, cisplatin and as previously shown, etoposide [6]. This supports a potential role for FLJ11259 in the biological response of tumors to chemotherapeutic agents.
p53 regulates gene transcription by binding to p53 response elements in target genes. We have validated an exact consensus p53RE upstream of the predicted transcriptional start site of FLJ11259. This site is bound basally by endogenous p53 in vivo, and p53 binding is increased with cisplatin treatment. Importantly, this site is functional and p53-responsive. This data supports FLJ11259 as a direct transcriptional target of p53. We have shown FLJ11259 mRNA is expressed in many tissues, however it will be important to determine whether FLJ11259 is induced at the protein level by cisplatin and other DNA damaging agents. In addition tissue specific post-translational modifications of FLJ11259 may be important for modulating its function following drug treatment. Using the ELM prediction program (http://elm.eu.org/), FLJ11259 is predicted to have 6 transmembrane regions, an intracellular N and C termini, and multiple sites of protein modification and protein-protein interactions. However, FLJ11259 does not contain any known protein motifs or possess a high degree of homology with any well-characterized proteins.
During the characterization of FLJ11259 three homologous and evolutionarily conserved 6 transmembrane proteins were identified. These four gene products thus represent a new and novel family of proteins. The rat homolog of TMMEM150 is TM6P1, encoding a protein with a predicted mass of 29.5 kD [16]. TM6P1 has been reported to localize to the plasma membrane. TM6P1 was identified as being induced by fasting in rat liver cells, however its function is currently unknown [16]. Confirmation of the membrane topology of FLJ11259 and the related family of proteins will be necessary in order to fully understand their function. These additional FLJ11259 family members are non-responsive to cisplatin and were not expressed in a p53-dependent manner. Furthermore, we have identified a potential splice variant of FLJ11259 that lacks transmembrane spanning regions 4 and 5 but is also inducible with cisplatin. It would be interesting to study whether the two splice variants have distinct functions and localizations. Thus it appears that p53-regulates a single member of a novel four-member family of transmembrane proteins.
During the final preparation of this manuscript, identification of FLJ11259 as damage-regulated autophagy modulator (DRAM), a regulator of autophagy and p53-mediated death was reported by Crighton et al [12]. There are a number of important distinctions. We have identified a strong and distinct p53RE upstream of FLJ11259, in comparison to the intronic p53-binding site identified by Crighton et al. [12]. The p53RE reported here is an exact consensus with 0 base pairs separating the two p53 half-sites. Regulation of FLJ11259 is likely complex and characterization of the FLJ11259 promoter and transcriptional start site will be important for a complete description of FLJ11259 regulation.
We have not directly addressed whether FLJ11259 is involved in autophagy. However, the punctate pattern of expression is entirely consistent with the autosomal/lysosomal localization reported by Crighton et al [12]. Further studies will be required to address the precise localization of the endogenous FLJ11259 protein and whether a portion of endogenous FLJ11259 localizes to the plasma membrane, as reported for TM6P1 (rat homolog of TMEM77) [16]. We have shown that overexpression of FLJ11259 results in increased clonogenic survival. This could be consistent with a role in autophagy, as this process has been suggested to protect cells from stress under certain conditions [17]. Interestingly, TM6P1 was shown to be induced in fasting liver, consistent with the known induction of autophagy under nutrient starved conditions [16]. The clonagenic assays of Crighton et al, saw no difference in clonagenicity upon overexpression of FLJ11259/DRAM. This difference may be due to the differential stress of the transfection reagents employed or that the plating density differed. We have noted that heavy plating of cells in the assay diminishes the survival effect of overexpressed FLJ11259 (data not shown). In addition, it would be of importance to determine whether the other FLJ11259 family members described here, as well as the alternative spliced form of FLJ11259, have a likewise role in autophagy.
In summary we have further characterized FLJ11259 as a p53-inducible member of a novel class of 6-transmembrane proteins which may be important in the cellular response of tumor cells to stress. TGCTs express high levels of wild-type p53 protein, and as a result may have distinct or amplified patterns of p53-dependent gene regulation in response to chemotherapeutic agents. Hence TGCT models may be an important tool for identifying additional p53 transcriptional targets to enhance our understanding of the complex network of gene regulation controlled by p53.
Acknowledgements
We thank Dr. S. Hamilton (GlycoFi Inc.) for assistance with the in silico analysis and for insightful discussions. We thank Drs. A. Eastman (Dartmouth Medical School) and B. Vogelstein (Johns Hopkins University) for the MCF10A and HCT116 cell lines respectively, Dr. E. Dmitrovsky (Dartmouth Medical School) for the germ cell tumor samples, and Dr. J. DiRenzo (Dartmouth Medical School) for providing the tk-luc vector and technical guidance. This work was supported by the National Institute of Health Grant R01-CA104312 and a grant from the Lance Armstrong Foundation (MJS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.el-Deiry WS. Regulation of p53 downstream genes. Semin. Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 2.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 3.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura Y. Isolation of p53-target genes and their functional analysis. Cancer Sci. 2004;95:7–11. doi: 10.1111/j.1349-7006.2004.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spurgers KB, Coombes KR, Meyn RE, Gold DL, Logothetis CJ, Johnson TJ, McDonnell TJ. A comprehensive assessment of p53-responsive genes following adenoviral-p53 gene transfer in Bcl-2-expressing prostate cancer cells. Oncogene. 2004;23:712–1723. doi: 10.1038/sj.onc.1207293. [DOI] [PubMed] [Google Scholar]
- 6.Kerley-Hamilton JS, Pike AM, Li N, Direnzo J, Spinella MJ. A p53-dominant transcriptional response to cisplatin in testicular germ cell tumor-derived human embryonal carcinoma. Oncogene. 2005;24:6090–6100. doi: 10.1038/sj.onc.1208755. [DOI] [PubMed] [Google Scholar]
- 7.Levesque AA, Kohn EA, Bresnick E, Eastman A. Distinct roles for p53 transactivation and repression in preventing UCN-01-mediated abrogation of DNA damage-induced arrest at S and G2 cell cycle checkpoints. Oncogene. 2005;24:3786–3796. doi: 10.1038/sj.onc.1208451. [DOI] [PubMed] [Google Scholar]
- 8.White KA, Yore MM, Warburton SL, Vaseva AV, Rieder E, Freemantle SJ, Spinella MJ. Negative feedback at the level of nuclear receptor coregulation: self-limitation of retinoid signaling by RIP140. J. Biol. Chem. 2003;278:3889–43892. doi: 10.1074/jbc.C300374200. [DOI] [PubMed] [Google Scholar]
- 9.Freemantle SJ, Kerley JS, Olsen SL, Gross RH, Spinella MJ. Developmentally-related candidate retinoic acid target genes regulated early during neuronal differentiation of human embryonal carcinoma. Oncogene. 2002;21:2880–2889. doi: 10.1038/sj.onc.1205408. [DOI] [PubMed] [Google Scholar]
- 10.Curtin JC, Dragnev KH, Sekula D, Christie AJ, Dmitrovsky E, Spinella MJ. Retinoic acid activates p53 in human embryonal carcinoma through retinoid receptor-dependent stimulation of p53 transactivation function. Oncogene. 2001;20:2559–2569. doi: 10.1038/sj.onc.1204370. [DOI] [PubMed] [Google Scholar]
- 11.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 12.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 13.Giuliano CJ, Freemantle SJ, Spinella MJ. Testicular germ cell tumors: A paradigm for the successful treatment of solid tumor stem cells. Curr. Cancer Ther. Rev. 2006;2:255–270. doi: 10.2174/157339406777934681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kho PS, Wang Z, Zhuang L, Li Y, Chew JL, Ng HH, Liu ET, Yu Q. p53-regulated transcriptional program associated with genotoxic stress-induced apoptosis. J. Biol. Chem. 2004;279:21183–21192. doi: 10.1074/jbc.M311912200. [DOI] [PubMed] [Google Scholar]
- 15.Sax JK, El-Deiry WS. p53 downstream targets and chemotherapy. Cell Death Differ. 2003;10:413–417. doi: 10.1038/sj.cdd.4401227. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, D'Ercole AJ, Underwood LE. Identification of a new gene (rat TM6P1) encoding a fasting-inducible, integral membrane protein with six transmembrane domains. Biochim. Biophys. Acta. 2000;1492:280–284. doi: 10.1016/s0167-4781(00)00104-4. [DOI] [PubMed] [Google Scholar]
- 17.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]