Abstract
Histone acetylation, regulated by two antagonistic enzymes — histone acetyltransferases (HATs) and histone deacetylases (HDACs) — results in transcriptional changes and also plays a critical role in cardiac development and disease. A new study shows that overexpression of the atypical transcriptional corepressor homeodomain-only protein (Hop) causes cardiac hypertrophy via recruitment of a class I HDAC. In contrast to the body of work on transcriptional mechanisms that drive cardiac hypertrophy, including class II HDACs, this report elucidates a novel growth-suppressing transcriptional pathway in cardiac muscle that opposes hypertrophic growth.
Cardiac mechanical dysfunction poses a therapeutic challenge that is not effectively addressed by existing therapy. Heart failure represents the final common pathway for diverse disorders intrinsic to or impinging on the myocardium, including myocardial infarction and hypertension-induced cardiac hypertrophy. Pathological hypertrophy comprises defects in myocyte performance, fibrosis, and myocyte loss, compounded by insufficient replacement (1). Finding the signals and effectors that execute this phenotype has been a unifying theme for two decades of research into the molecular underpinnings of heart failure, encapsulated as the transcriptional activation of an ultimately adverse gene program, whose elements encompass contractile proteins, ion transport proteins, growth factors, cytokines, and regulators of survival (1). Among the best-proven nodal control points for gene regulation in hypertrophic myocardium is histone acetylation (2). Together with other histone modifications, the consequent change in chromatin structure is a prerequisite for access of transcription factors to their target DNA (3–5).
The status of histone acetylation, at a given promoter, is determined by the balanced action of histone acetyltransferases (HATs) and histone deacetylases (HDACs). The essential role of a HAT protein in cardiac muscle was first proven by deletion of the coactivator p300, which perturbed heart development and cell proliferation (6). But p300 is an especially complex platform for factor-factor interactions, and it remains ambiguous which of its effects are contingent on HAT activity, which on recruitment of p300/CBP-associated factor — also a HAT protein — and which on other properties (7).
Three classes of mammalian HDACs have been discovered. Class I HDACs are expressed ubiquitously and are composed largely of the catalytic domain. Class II HDACs are highly expressed in striated muscle and contain extensions at the N- or C-terminus (2). A third class comprises homologues of yeast Sir2 (8). Notably, class II HDACs can act as signal-responsive repressors of cardiac hypertrophy, inhibiting gene expression that is dependent on myocyte enhancer factor-2 (1, 2). Consistent with this repressive role, mutant mice lacking the class II HDAC9 are hypersensitive to hypertrophic signals (2).
The Hop-less heart
Homeodomain-only protein (Hop) is a new, unusually short homeodomain protein that was discovered by screening of expressed sequence tag databases for cardiac transcripts encoding novel homeobox genes (9, 10). Hop is specific to the heart in mid-gestation and, although it is a nuclear protein, does not bind DNA. The phenotype resulting from germline deletion of Hop is incompletely penetrant. As many as half of the null embryos have ventricular hypocellularity, thinning, and perforation, bloody pericardial effusions, and death before birth (9, 10). Blocking Hop expression in zebrafish had similar effects (9). In surviving mice, though, myocyte proliferation, myocyte number, and ventricular wall thickness were increased, a seeming paradox (10).
How can the null mutation of Hop cause antithetical phenotypes? The first hint was obtained from studies on the mode of action of Hop. Hop-dependent endogenous genes and promoters were found often to contain binding sites for serum response factor (SRF). Hop binds SRF, inhibits SRF-dependent gene expression, and interferes with the cooperation of SRF with its coactivator myocardin (9, 10). The dual role of Hop in cardiac growth control is reminiscent of the dichotomous roles of SRF in proliferation versus muscle differentiation and potentially contingent on differing signals and factors converging on SRF at different developmental stages or contexts (11–13). How, then, does Hop inhibit SRF activity? Hop was shown to associate with SRF directly and prevent DNA binding by SRF in vitro (9, 10). However, this may not be the sole or universal mechanism.
Hop recruits a class I HDAC
In this issue of the JCI, Kook and colleagues shed light on the potential functions of Hop in adult myocardium and provide penetrating insights into how this protein may work (14). Biological consequences of Hop were revealed via gain-of-function analysis, using forced expression of Hop via the heart-specific α-myosin heavy chain (α-MHC) promoter. Increased Hop expression caused concentric ventricular hypertrophy followed by later dilatation, myocardial fibrosis, and sudden death. The authors are alert to the possibility that expression at a supraphysiological level could have untoward consequences, and a strength of their design is the use of an inactivated Hop as a control, mutated in the second α-helix and incapable of binding SRF. It is not immediately possible, though, to know whether hypertrophy caused by overexpression of Hop is a physiological function of Hop at its native level. For instance, hypertrophy could result from the impairment in pump function and the fibrosis that are present, rather than inherent growth-promoting effects.
The conceptual breakthrough in the present report is the demonstration that Hop’s inhibit SRF-regulated transcription by recruiting an HDAC to SRF (14). Specifically, Hop associated with HDAC2, a class I HDAC. It should be noted that the involvement of other HDACs has not been excluded. Consistent with the biochemical function of HDAC2, Hop prevented the ability of myocardin to cause histone acetylation. Treatment of Hop transgenic mice with trichostatin A, a pharmacological HDAC inhibitor, prevented hypertrophy, with no effect in wild-type animals. In short, Hop recruits HDACs, including HDAC2, to SRF target promoters, and the deacetylase activity of HDAC proteins is likely necessary for Hop-induced hypertrophy (Figure 1).
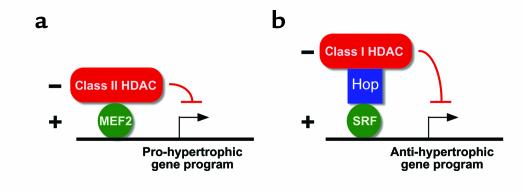
Figure 1.
Complementary functions of class I and class II HDACs in cardiac hypertrophy. (a) The myocyte enhancer factor-2–based transcriptional program that triggers cardiac hypertrophy is repressed by signal-responsive class II HDACs, which are exported from the nucleus as the result of hypertrophic cues (1, 2). (b) A class I HDAC (HDAC2) is recruited to SRF — via Hop as an intermediary — thereby blocking a transcriptional program that opposes hypertrophy (14). MEF2, myocyte enhancer factor-2.
HDAC inhibitors as potential therapy for cardiac hypertrophy
In certain respects, this work is consistent with an equally counterintuitive report that HDAC inhibitors can block hypertrophic growth and associated fetal gene induction in cultured cardiomyocytes (15), contrary to the expectation that HDACs suppress a pro-hypertrophic pathway (2). Trichostatin A inhibits both class I and class II HDACs, and growth suppression by HDAC inhibitors need not be specific to Hop and its partners. Nevertheless, the present study is highly noteworthy, from both a mechanistic and a translational point of view. HDAC inhibitors are well tolerated in humans, and clinical trials investigating their efficacy as anticancer agents are currently underway (16). If cardiac hypertrophy is construed to be an adverse adaptation, as supported by the preponderance of evidence (1), and if cardiac function can be preserved even in the face of stress when hypertrophy is blocked (17), the therapeutic merit of HDAC inhibitors in heart failure is potentially important.
The apparent conflict between results with HDAC inhibitors and the HDAC9-null mutation would be reconciled most easily if different HDACs have opposing effects on cardiac hypertrophy. If so, development of inhibitors specific to a particular HDAC isoform could be of particular benefit. Much also needs to be learned about the interaction of HDAC-sensitive pathways with other known mechanisms for hypertrophic growth, such as translational control (18) and transcription elongation (19).
At a more fundamental level, several questions also remain to be addressed to understand better the roles of Hop in cardiac growth and development. If Hop is recruited to SRF, what are the signals or circumstances that promote Hop’s recruitment? SRF is subject to both stimulatory (9, 10, 12, 13, 20) and inhibitory binding partners (9, 10, 21), thereby forming either transcriptionally active or inactive assemblies. Phosphorylation of SRF may also be germane. Events that control the conversion between these two presumptive forms of the SRF complex will need to be clarified in order to understand Hop-regulated cardiac growth. It is even more important to determine what SRF target genes are directly regulated by Hop. Given the opposing phenotypes in embryonic versus neonatal hearts of Hop-deficient mice, different target genes are presumed to be regulated by Hop, depending on developmental stage. It is, of course, possible that Hop regulates factors beyond SRF alone.
Footnotes
See the related article beginning on page 863.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: histone acetyltransferase (HAT); histone deacetylase (HDAC); homeodomain-only protein (Hop); serum response factor (SRF).
References
- 1.Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes Dev. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 2.Zhang CL, et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 5.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 6.Yao TP, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 7.Baluchamy S, Rajabi HN, Thimmapaya R, Navaraj A, Thimmapaya B. Repression of c-Myc and inhibition of G1 exit in cells conditionally overexpressing p300 that is not dependent on its histone acetyltransferase activity. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9524–9529. doi: 10.1073/pnas.1633700100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 9.Chen F, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 10.Shin CH, et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 11.Wang DZ, et al. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 13.Chang DF, et al. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell. 2003;4:107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 14.Kook H, et al. Cardiac hypertrophy and histone deacetylase–dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Invest. 2003;112:863–871. doi:10.1172/JCI200319137. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antos CL, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J. Biol. Chem. 2003;278:28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 16.Marks PA, Richon VM, Breslow R, Rifkind RA. Histone deacetylase inhibitors as new cancer drugs. Curr. Opin. Oncol. 2001;13:477–483. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 17.Sano M, Schneider MD. Still stressed out but doing fine: normalization of wall stress is superfluous to maintaining cardiac function in chronic pressure overload. Circulation. 2002;105:8–10. [PubMed] [Google Scholar]
- 18.Shioi T, et al. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation. 2003;107:1664–1670. doi: 10.1161/01.CIR.0000057979.36322.88. [DOI] [PubMed] [Google Scholar]
- 19.Sano M, et al. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 20.Sepulveda JL, Vlahopoulos S, Iyer D, Belaguli N, Schwartz RJ. Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J. Biol. Chem. 2002;277:25775–25782. doi: 10.1074/jbc.M203122200. [DOI] [PubMed] [Google Scholar]
- 21.Davis FJ, Gupta M, Camoretti-Mercado B, Schwartz RJ, Gupta MP. Calcium/calmodulin-dependent protein kinase activates serum response factor transcription activity by its dissociation from histone deacetylase, HDAC4. Implications in cardiac muscle gene regulation during hypertrophy. J. Biol. Chem. 2003;278:20047–20058. doi: 10.1074/jbc.M209998200. [DOI] [PubMed] [Google Scholar]