Abstract
Insulin-dependent diabetes mellitus is usually caused by the autoimmune destruction of pancreatic β cells by T cells. Methodologies to track the development, migration, and functional activation of one class of such T cells (CD4 T cells) have been limited. However, it now appears that this limitation has been overcome .
Type 1 diabetes, or insulin-dependent diabetes mellitus (IDDM), in humans and rodent models results from the T cell–mediated autoimmune destruction of pancreatic islet β cells. Both MHC class I–restricted CD8 T cells and MHC class II–restricted CD4 T cells contribute to this pathogenesis. Methodologies to identify, quantify, and track such autoreactive T cells have been lacking, especially for the CD4 subset. This is an important issue for studies designed to monitor the efficacy of potential intervention protocols. Over the past several years, tetramer reagents have been introduced and utilized for the quantification and tracking of CD8 T cells. These tetramers are synthesized as fluorochrome-labeled complexes of MHC class I molecules to which the appropriate peptide antigen has been bound. However, it has been difficult to fold complexes of MHC class II molecules for similar identification of antigen-specific CD4 T cells.
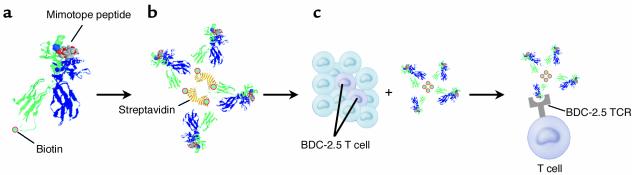
In this issue of the JCI, an article by Stratmann and colleagues (1) introduces an exciting new tetramer technology that allows a particular set of diabetogenic CD4 T cells to be quantified and tracked. Specifically, these authors utilized the nonobese diabetic (NOD) mouse model of IDDM wherein the unusual murine MHC class II Ag7 molecule is a known mediator of β cell–autoreactive CD4 T cell responses. One important β cell autoantigen targeted by CD4 T cells in NOD mice is that recognized by the previously identified BDC-2.5 clone. The BDC-2.5 clone recognizes a peptide derived from an unknown β cell granule protein. The authors used the BDC-2.5 clone to screen a recombinatorial peptide library in order to identify a ligand that mimics the natural antigen (designated as a “mimotope”). A strong agonist peptide was selected solely on the basis of its biologic activity without regard for the affinity of the peptide/Ag7 complex for the BDC-2.5 T cell receptor (TCR). The availability of soluble Ag7 molecules then allowed the authors to construct a fluorochrome-labeled tetramer complex containing a selected agonist peptide (Figure 1). As described earlier, the staining of naive CD4 T cells with MHC multimers has been highly problematic. Hence, it was significant that the tetramer technology employed by Stratmann et al. overcame these problems, facilitating an accurate, quantitative, and qualitative characterization of how the BDC-2.5 clonotype contributes to IDDM.
Figure 1.
Tetramer technology used to enumerate a diabetogenic CD4+ T cell clonotype (designated BDC2.5) in autoimmune diabetes–prone NOD mice. A mimotope peptide structurally similar to the native peptide antigen processed from a pancreatic β cell granule protein is presented by the NOD H2-Ag7 MHC class II molecule (a). The MHC class II α chain is depicted in green, the β chain in blue, and the mimotope peptide in grey/red space fill representation. The α chain of the peptide-associated H2-Ag7 complexes are biotinylated, allowing tetramer formation by binding four such molecules to streptavidin (b). Such a reagent allows detection of T cells with the BDC2.5 specificity in the anatomical site of choice (c). Tetramer crystal structure courtesy of Luc Teyton (The Scripps Research Institute, La Jolla, California, USA).
Stratmann et al. (1) demonstrated that, not surprisingly, the BDC-2.5 clonotype was positively selected in the thymus of NOD mice and expanded to higher levels in the periphery. However, the highest numbers of this clonotype were observed in the pancreatic lymph nodes. The proportion of CD4 T cells in peripheral lymph nodes with the BDC-2.5 antigenic specificity (0.25%) was several logs higher than the usual frequency of T cells with a given antigen specificity. Importantly, a small but definable population of BDC-2.5 cells was detected in the pancreatic islets. This new generation tetramer reagent also allowed the authors to isolate a series of T cells with BDC-2.5 antigen specificity. It also revealed that these cells are diverse in terms of their TCR repertoire and vary in their strength of antigen recognition.
The result that was most surprising to these commentators was that the tetramer revealed the presence of BDC-2.5 T cells in the B6.H2g7 and nonobese resistant (NOR) mouse strains, which are both IDDM resistant, but which express the diabetogenic NOD H2g7 haplotype (1). Although the H2g7 haplotype is the strongest contributor to IDDM development in NOD mice, it has been well established that multiple other non-MHC genes (described by Stratmann et al. as “background genes”) are also essential in the development of this disease. Based upon their findings, the authors concluded that the H2g7 haplotype is the only genetic element required for selection and emergence of autoreactive T cells without requiring other diabetogenic loci from the NOD genome. Because the B6.H2g7 and NOR mice remain IDDM-free, the authors concluded that the role of non-MHC genes is to peripherally regulate the activation of pathogenic T cells following their emergence. However, we suggest that caution should be employed in extrapolating results based on a single CD4 clonotype to the overall issues of how MHC and non-MHC genes interactively contribute to IDDM susceptibility or resistance.
Many other CD4 T cells that respond to antigens differing from those recognized by the BDC-2.5 clone also contribute to IDDM in NOD mice. While the study by Stratmann et al. (1) provides insight into how MHC and non-MHC genes regulate the development of T cells with BDC-2.5–like reactivity, several issues must be taken into account when considering the global applicability of their findings. Indeed, another recent study found that non-MHC genes in NOD mice contributed to defects that allowed CD4 T cells that recognize another pancreatic β cell protein to escape negative selection (2). Another question to consider is whether the native antigen recognized by T cells exhibiting a BDC-2.5 reactivity pattern is expressed intrathymically in the same way as some other diabetogenic antigens, such as insulin. Furthermore, the authors’ data indicate that while BDC-2.5 T cells are present in IDDM-resistant NOR mice, their levels are lower than in the closely related NOD strain. This suggests that some non-MHC genes present in NOR mice may contribute to a more limited selection of such effectors. As noted by Stratmann et al., autoreactive CD8 T cells are also essential contributors to the spontaneous development of IDDM in NOD mice, and the NOR strain exerts a non-MHC gene function that limits the development of at least one of these pathogenic CD8 T cell clonotypes (3).
While we are still at an early stage in understanding the complexities of interactions between MHC and non-MHC genes that underlie IDDM development in both humans and rodent models, the authors have identified a powerful new means to monitor and characterize CD4 T cell populations contributing to this disease. Such reagents may ultimately prove to be of use in monitoring the efficacy of intervention protocols that may inhibit progression to overt IDDM in humans at risk for this debilitating disease.
Footnotes
See the related article beginning on page 902.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Insulin-dependent diabetes mellitus (IDDM); nonobese diabetic (NOD); T cell receptor (TCR); nonobese resistant (NOR).
References
- 1.Stratmann T, et al. Susceptible MHC alleles, not background genes, select an autoimmune T cell reactivity. J. Clin. Invest. 2003;112:902–914. doi:10.1172/JCI200318337. doi: 10.1172/JCI18337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lesage S, et al. Failure to censor forbidden clones of CD4 T cells in autoimmune diabetes. J. Exp. Med. 2002;196:1175–1188. doi: 10.1084/jem.20020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdaguer J, Amrani A, Anderson B, Schmidt D, Santamaria P. Two mechanisms for the non-MHC-linked resistance to spontaneous autoimmunity. J. Immunol. 1999;162:4614–4626. [PubMed] [Google Scholar]