Abstract
Despite the potentially important roles of untranslated RNAs in cellular form or function, genes encoding such RNAs have until now received surprisingly little attention. One such gene encodes BC1 RNA, a small non-mRNA that is delivered to dendritic microdomains in neurons. We have now eliminated the BC1 RNA gene in mice. Three independent founder lines were established from separate embryonic stem cells. The mutant mice appeared to be healthy and showed no anatomical or neurological abnormalities. The gross brain morphology was unaltered in such mice, as were the subcellular distributions of two prototypical dendritic mRNAs (encoding MAP2 and CaMKIIα). Due to the relatively recent evolutionary origin of the gene, we expected molecular and behavioral consequences to be subtle. Behavioral analyses, to be reported separately, indicate that the lack of BC1 RNA appears to reduce exploratory activity.
Relatively few novel genes appear to have arisen in diverse mammalian orders after they diverged from a common ancestor about 80 to 100 million years ago (1). In fact, many differences between two genomes could just as easily be explained by gene loss in one lineage as opposed to acquisition in the other. Truly novel mammalian genes that are specifically expressed in neurons should, therefore, be of particular relevance in dissecting the molecular basis of higher brain functions. One such candidate is dendritic BC1 small non-mRNA (snmRNA). This RNA arose 60 to 110 million years ago in a common ancestor of all rodents by a process termed retroposition, which involves reverse transcription of cellular RNA (2) followed by random integration of the resulting DNA into chromosomes. Most of the time these events lead into evolutionary dead-ends. On rare occasions, however, the retronuon (a nuon is any discrete sequence of DNA or RNA) (4, 5) encounters regulatory elements at its locus of integration and is expressed. For BC1 RNA, a tRNAAla served as the template for reverse transcription. The retronuon acquired transcriptional regulatory elements as well as a unique 3′ region (3, 11). The ∼150-nucleotide (nt) RNA is transcribed from a single gene by RNA polymerase III (24), and it lacks an open reading frame and hence does not encode a protein or peptide. It is expressed in a unique pattern in the rodent nervous system (42). In association with specific proteins (28), BC1 RNA is transported into the cytoplasm and dendritic processes of neurons as a component of an 8.7S RNP particle (7, 10, 42). BC1 RNA has been suggested to operate in the regulation of dendritic protein synthesis or, alternatively, as a mediator of dendritic mRNA transport (18, 39, 40, 42, 43). Both hypotheses, in conjunction with the unique expression pattern of BC1 RNA in the rodent nervous system, would therefore imply the existence of corollaries in higher-brain functions and behavioral phenotypes.
MATERIALS AND METHODS
Isolation of isogenic DNA for embryonic stem (ES) cell-targeting experiment.
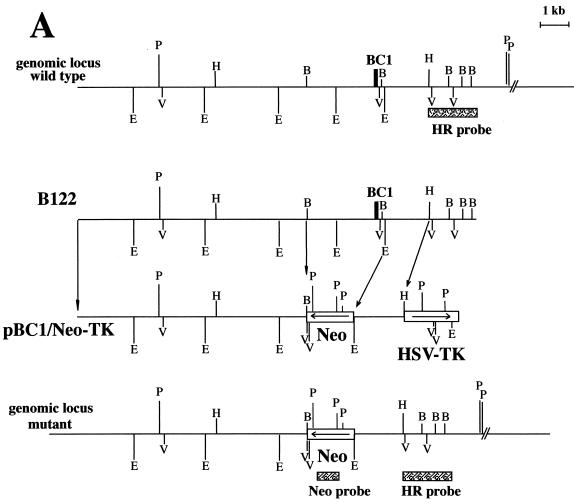
We isolated two clones from a genomic bacteriophage λ library, B122 and B142, that contained the mouse BC1 RNA gene locus. After detailed mapping and sequencing, clone B122 was used to design the gene replacement-targeting construct (Fig. 1A). The bacteriophage λ library was constructed with DNA from the 129SvJ mouse strain (Stratagene). As a probe to screen the genomic library, we used a 1.1-kb EcoRI-BglII fragment located immediately 5′ to the 1.7-kb EcoRI fragment harboring the BC1 RNA gene. The insert size of clone B122 was 14 kb, including 10.5 kb 5′ and 3.5 kb 3′ to the BC1 RNA gene.
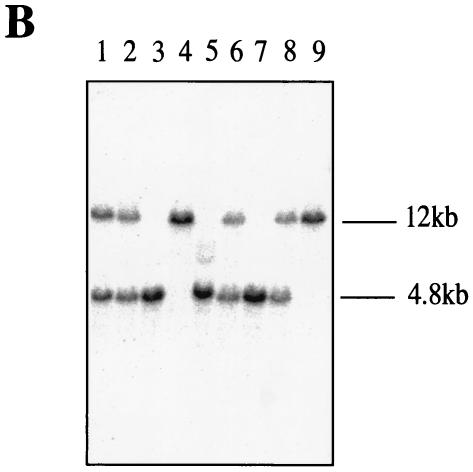
FIG. 1.
Map of the replacement targeting vector and Southern blot of mouse tail DNA. (A) Wild-type genomic locus, genomic clone B122, targeting vector pBC1/Neo-TK, and modified genomic locus lacking the BC1 RNA gene. Neo, neomycin phosphotransferase gene; HSV-TK, TK gene under control of the simian virus 40 promoter. The bars indicate probes for Southern blot hybridization: HR, homologous recombination probe; Neo, probe specific for the neomycin phosphotransferase gene. B, BglII; E, EcoRI; V, EcoRV; H, HindIII; P, PstI. (B) Southern blot analysis of PstI-digested genomic DNAs from nine different pups after crossing of two heterozygous parents. Mice 4 and 9 possess both wild-type alleles (12 kb); mice 1, 2, 6, and 8 are heterozygous; and mice 3, 5, and 7 have both gene deletion alleles (4.8 kb).
In order to increase the odds for homologous recombination, we used approximately 8 kb of flanking region 5′ (to the left) of the desired deletion in the targeting construct pBC1/Neo-TK, which included the thymidine kinase (TK) gene for negative counterselection. The negative selection procedure decreased the number of negative colonies by at least fourfold. PCR screening of 288 colonies identified 24 positive colonies. Twenty colonies could be expanded and characterized.
Construction of the gene-targeting vectors.
The gene replacement vector pBC1/Neo was generated by exchanging the 2.8-kb BglII-EcoRI DNA fragment containing the BC1 RNA gene and promoter elements with the PgkNeo cassette (35). As a result, the left flanking region consisted of a NotI-BglII DNA fragment from the BC1 RNA locus and the right flanking region consisted of a 1.5-kb EcoRI-HindIII fragment. This construct was cloned between the NotI and HindIII sites of pBluescript KS(+) (Stratagene).
In order to select against insertions at nonhomologous loci, the TK gene was added by releasing the whole insert from pBC1/Neo as an 11.3-kb NotI-XhoI fragment. This was combined with a 1.9-kb XhoI-HindIII TK gene-containing cassette (35) between the NotI and HindIII sites of pZERO 2.1 (Invitrogen) to yield the targeting construct, pBC1/Neo-TK.
ES cell transfection and selection of targeted clones.
RI ES cells (passage 13 [129/Sv × 129/Sv-CP; both derived from noninbred strains]) (13, 30) were expanded in HEPES-buffered Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum (HyClone), nonessential amino acids, l-glutamine, β-mercaptoethanol, 1,000 U of recombinant leukemia inhibitory factor (Chemicon) per ml, and antibiotics (penicillin [100 U/ml] and streptomycin [100 μg/ml]). For electroporation, 2 × 107 cells were resuspended in 20 mM HEPES (pH 7.4)-173 mM NaCl-5 mM KCl-0.7 mM Na2HPO4-6 mM dextrose-0.1 mM β-mercaptoethanol (38). The replacement targeting vector pBC1/Neo-TK was linearized with NotI, and 55 μg of DNA was electroporated at 25 μF and 400V (Gene Pulser; Bio-Rad). After electroporation, cells were plated onto 100-mm-diameter culture dishes containing a gamma-irradiated monolayer of primary G418-resistant fibroblast feeder cells. Thirty-eight hours later, 350 μg of G418 (Invitrogen) per ml and 2 μM ganciclovir (Syntex Corp.) were added to the culture medium. The medium was replaced every day, and colonies were picked and analyzed 8 days after plating.
PCR analysis of ES cell clones.
Positively targeted ES cell clones were analyzed by using a nested PCR approach. Colonies were digested in 50 mM Tris HCl (pH 7.9)-0.05 mM Triton X-100-100 μg of proteinase K per ml. After heat inactivation, DNA released from the cells was subjected to two rounds of PCR analysis. For the first round, the primer pair BCB1 (5′-TGAGGCTTGCCTAGAGTTCC-3′)-NF3 (5′-CTTCAGTGACAACGTCGAGC-3′) was employed. The PCR was with 50 mM Tris HCl (pH 9.5), 20 mM (NH4)2SO4, 1 mM dithiothreitol, 1.5 mM MgCl2, 0.005% NP-40, 10% dimethyl sulfoxide, 200 μM deoxynucleoside triphosphates, and a 0.5 μM concentration of each primer; annealing was done at 93°C for 2 min, and 40 cycles of 93°C for 45 s, 55°C for 30 s, and 65°C for 4 min were then performed. The PCR product was subjected to a second round of 20 cycles as described above with internal PCR primers BCB2 (5′-GGATGACAGGTAAGGAGCACC-3′) and NF4 (5′-CATAGCCGAATAGCCTCTCC-3′). The resulting PCR products were analyzed by agarose gel electrophoresis.
DNA blot analysis.
DNA, either from the expanded individual PCR-positive ES cell colonies or from mouse tails, was extracted as described previously (20). Approximately 5 μg of genomic DNA was digested with PstI, fractionated on 0.8% agarose gels, and transferred to GeneScreen nylon membranes (NEN DuPont) by positive-pressure blotting. The membranes were hybridized with a 32P-labeled 1.6-kb probe that contained sequences 3′ to the targeted homology (HR probe [Fig. 1A]) and washed with (final concentrations) 0.5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1mM EDTA [pH 7.7])-0.5% sodium dodecyl sulfate at 65°C.
Blastocyst injection.
ES cells from three independent clones with appropriate (and identical) deletions (clones 24, 156, and 179) were injected into 3.5-day C57BL6/J blastocysts. Routinely, we injected 10 to 14 ES cells into one blastocoele. After injection, blastocysts were kept in M16 medium and subsequently transferred into the uteri of 2.5-day pseudopregnant CD-1 foster mice. The mice carried the pups to term. Chimeras were identified by their agouti coat color.
Animals.
The subjects were male and female BC1-deficient mice. The lines were established by breeding male chimeras 6, 13, and 15 from three independent mutant ES cell lines with female C57/Bl6J mice to produce heterozygous mice. Subsequently, heterozygous mice were interbred to BC1−/− homozygocity. In order to minimize genetic drift, for each line we bred at least 5 to 7 males and 10 to 21 females with different coat colors. We tried to avoid breeding littermates whenever possible. A control line was analogously established by breeding BC1+/+ homozygous siblings from all three chimeric lines. Establishment of three separate control lines would have exceeded the capacity of our animal facilities. Both the three established BC1-deficient lines and the single control line thus featured a heterozygous genetic background with different relative contributions of alleles from the strains 129Sv and C57BL/6, with an average 50% contribution of each. Mice were bred at the central animal facility of the University Clinics, Münster, Germany, in a temperature-controlled (21°C) room with a 12-h, 12-h light-dark cycle and housed under nonenriched, standard conditions.
Pups were weaned at 19 to 23 days after birth, and females were kept separately from males. The mice were housed in standard cages of 27 (length) by 21 (width) by 15 (height) cm or 42 by 27 by 15 cm for up to three or up to seven littermates, respectively. General health checks were performed for all lines of control and knockout mice to ensure that any findings were not the result of deteriorating physical conditions of the animals.
General health check.
Health and neurological statuses were assessed by using a 10-min protocol including tests as described in standard check lists, such as SHIRPA (33) and the Fox battery (14). Animals were inspected for physical appearance and underwent neurological testing, including acoustic startle, visual placing, grip strength, and reflex functions. Additionally, life spans and fertility of mice were recorded. Sperm counts for several individual males of all three BC1-deficient lines and control mice was carried out.
Histological analysis of brain and testes.
Coronal, sagittal, and horizontal sections of brains from BC1-deficient and control mice were stained with cresyl violet and compared at different magnifications (×5 to ×400; Nikon Eclipse E600), with emphasis on major brain structures. Likewise, testes from BC1-deficient and control mice were analyzed with respect to their size and microscopically after staining with hematoxylin-eosin and periodic acid-Schiff base (Zeiss Axioskop at object magnifications of ×10 to ×100).
RNA blot analysis.
Total RNA was extracted from different mouse tissues by using TRIzol reagent (Invitrogen). RNA was separated on 6% polyacrylamide-7 M urea gels and electrotransferred to positively charged nylon membranes (Qiagen). After UV-cross-linking, the membranes were hybridized with 32P-labeled oligonucleotides in 1 M sodium phosphate (pH 6.2)-7% sodium dodecyl sulfate. The blots were washed several times in 2× SSPE-0.05% sodium dodecyl sulfate at room temperature and exposed to X-ray film (Kodak BioMax MS-1).
Oligonucleotide SB358 was complementary to the 5′ portion of BC1 RNA(5′-atccGACCGAACCCAGGGCCTTGCGCTTGCTAGGCAAGCGCTCTACCACTGAGCTAAATCgg-3′); oligonucleotide 90 was complementary to the 3′ unique portion of BC1 RNA (5′-cAAAAGGTTGTGTGTGCCAGTTACCTTGTTTTTggtac-3′), and oligonucleotide 7SL36B (control) was complementary to SRP RNA (5′-cGAGGTCACCATATTGATGCCGAACTTAGTGggtac-3′). Nucleotides in lowercase are unrelated to BC1 or SRP snmRNA sequences.
In situ hybridization.
In situ hybridization on brain sections of BC1 RNA-negative mice and control animals was performed as described previously (42). Probes specific for dendritic MAP2 mRNA and CaMKIIα mRNA were in vitro transcribed and 35S labeled, using clones pMAP2-19a (16) and pCaMKIIα (8), respectively. Following hybridization, dried sections were dipped in 1:1 diluted NTB2 nuclear track emulsion (Eastman Kodak) and counterstained with cresyl violet. Slides were examined under dark-field and bright-field optics (Nikon Microphot-FXA) at magnifications of ×20, ×100, and ×200. Images were digitally recorded by using dark-field optics (Nikon Eclipse E600) with a magnification of ×20. The signal was quantified with the aid of the Openlab version 3 program (Improvision Ltd.). Signal intensities were quantified in the stratum pyramidale, stratum radiatum, and lacunosum moleculare layers of the hippocampal CA3 area. For background assessment we measured the values of adjacent brain areas devoid of signal.
RESULTS AND DISCUSSION
Health, anatomy, and morphology: the general health of BC1-deficient animals appeared to be normal.
The assessment of the general appearance, health state, gross sensory functions, reflexes, and motor abilities did not reveal any significant differences between BC1-deficient and control mice. In one of the three BC1-deficient lines (line 13), we observed cataracts in about 5% of the animals. These mice were excluded from further analyses. In general, all three lines of BC1-deficient animals appeared to be healthy. The absence of neurological abnormalities (normal acoustic-startle response, eyelid reflex, and grip reflex) was a further indication of an overall healthy phenotype. Like control mice, BC1-deficient mice lived, if permitted to, for more than 1 year.
In addition to brain, BC1 RNA is expressed in testes, predominantly in spermatogonia (29). This organ system was therefore a focus of attention in regard to potential organic changes and/or dysfunctions. Size, weight, and microscopic features were not significantly different between BC1-deficient and control mice. No aspermia or oligospermia was observed in BC1-deficient animals. The number or mobility of spermatozoa did not differ between BC1-deficient and control animals. Males and females from all lines could be successfully bred for at least 1 year. Breeding data over 2 years revealed no differences in fertility and viability between BC1-deficient and control animals. The average numbers of pups and standard deviations per breeding pair were 6.6 ± 3.9, 7.4 ± 4.6, and 8.5 ± 4.3 for the BC1-deficient mouse lines 6, 13, and 15, respectively, and 10.0 ± 4.5 for control animals. Statistical analysis showed no significant difference between BC1-deficient and control animals. To further assess the fertility of BC1-deficient animals, we have outbred males and females to CD-1 and B2D2F1 mice. There was no deviation in sex ratio among offspring with any of the BC1-deficient mouse lines.
Neuroanatomical features of BC1-deficient animals: gross neuroanatomical features in BC1-deficient and control animals are indistinguishable.
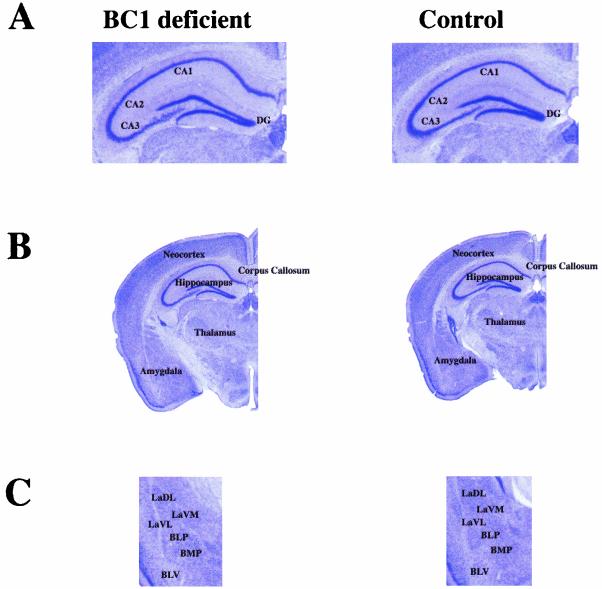
For all experiments we used three independent mouse lines that were derived from three separate ES cells. Some 129 substrains are known to exhibit brain abnormalities such as lack of a corpus callosum (22). However, C57BL/6 × 129SV hybrids do not show callosal dysgenesis (23). Coronal, sagittal, and horizontal sections of brains from both BC1-deficient and control mice were analyzed, and the integrity, shape, and size of major brain regions, with emphasis on those with high levels of BC1 RNA expression (e.g., the CA1 to CA3 pyramidal cells of the hippocampus and their corresponding dendritic fields, the amygdala, and discrete regions of the mesencephalon and diencephalon) (21, 42) were compared. These histological analyses revealed that the BC1-deficient mice had normal brain morphology and lacked the Probst bundles (23) present in acallosal mice (Fig. 2). At this point we cannot exclude subtle morphological differences at the ultrastructural level.
FIG. 2.
Coronal sections of brains from BC1-deficient and normal mice show no apparent morphological differences. (A) Hippocampal CA1 to -3 fields and dentate gyrus (DG) are indicated. (B) Major brain structures are identified. (C) Amygdala. LaDL, lateral amygdaloid nucleus, dorsolateral; LaVM, lateral amygdaloid nucleus, ventromedial; LaVL, lateral amygdaloid nucleus, ventrolateral; BLP, basolateral amygdaloid nucleus, posterior; BMP, basomedial amygdaloid nucleus, posterior; BLV, basolateral amygdaloid nucleus, ventral. All abbreviations are according to Franklin and Paxinos (15).
Molecular features of BC1-deficient animals. (i) BC1-deficient animals appear to have a normal molecular phenotype except that BC1 RNA is not expressed.
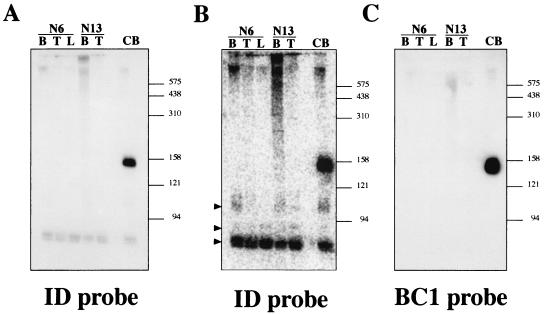
Northern blot analysis with a probe complementary to the unique 3′ domain confirmed that both BC1 RNA and other small RNAs were undetectable in brain tissue from all three homozygous BC1-deficient lines (Fig. 3A). With a second probe that was complementary to the 5′ domain (ID domain), we detected BC1 RNA in RNA from control brain but not in RNA isolated from brain and other tissues of the BC1-deficient animals (Fig. 3A and C). However, with the ID probe we observed weaker hybridization signals corresponding to smaller RNAs of ∼110, ∼90, and ∼75 nt (Fig. 3A and B). Their occurrence in RNA isolated from BC1-deficient mice rules out the possibility that they are BC1 RNA degradation products, as has been proposed for the smallest (occasionally termed T3) of the additional RNAs (26). This RNA is ubiquitously expressed, and reduction of the signal during increased washing stringency indicates that the band is likely to be caused by cross-hybridization with tRNAAla, progenitor of BC1 RNA (11). The weak signal at ∼90 nt may correspond to a tRNA precursor or a transcribed ID element. The additional signal of ∼110 nt in both control and BC1-deficient animals is, like BC1 RNA, prevalently expressed in brain and to a lesser degree in testes (Fig. 3B) and may correspond to the previously reported BC2 RNA (11, 12, 17, 26). However, compared to BC1 RNA, the signal is lower by at least an order of magnitude.
FIG. 3.
Northern blot analysis of RNAs extracted from different tissues of BC1-deficient mice. (A) Hybridization with a probe complementary to the 5′ ID domain of BC1 RNA (SB358). Lanes B, T, and L, RNA samples extracted from brain, testes, and liver, respectively, of homozygous BC1-deficient mouse lines 6 and 13. Lane CB, RNA extracted from brains of control mice which exhibits BC1 RNA of ∼150 nt. The size marker positions are shown in nucleotides on the right. (B) After exposure on a phosphorimager, additional hybridization signals corresponding to RNAs migrating faster than BC1 RNA are observed. Arrowheads identify signals at ∼110, ∼90, and ∼75 nt. (C) Hybridization with a probe complementary to the 3′ unique domain of BC1 RNA (probe 90).
We conclude that deletion of a single locus completely abolishes expression of ∼150-nt-long BC1 RNA. At the same time, small amounts of other snmRNAs with sequence similarity to ID retronuons were detectable. These results finally put to rest the initial hypothesis (27, 36, 37) that BC1 RNA is a heterogeneous transcript from many ID repetitive elements in rodent genomes. Such beliefs resurface from time to time to this day (9), although they have been thoroughly refuted in the past (11, 31, 34). Of course, we expect levels of RNA polymerase II transcripts (hnRNAs and mRNAs) that contain ID elements in untranslated regions to be unaltered. Such transcripts, however, are not resolved on polyacrylamide gels, blots of which are shown in Fig. 3.
(ii) Dendritic localization of other RNAs in neuronal dendrites is not affected by elimination of BC1 RNA.
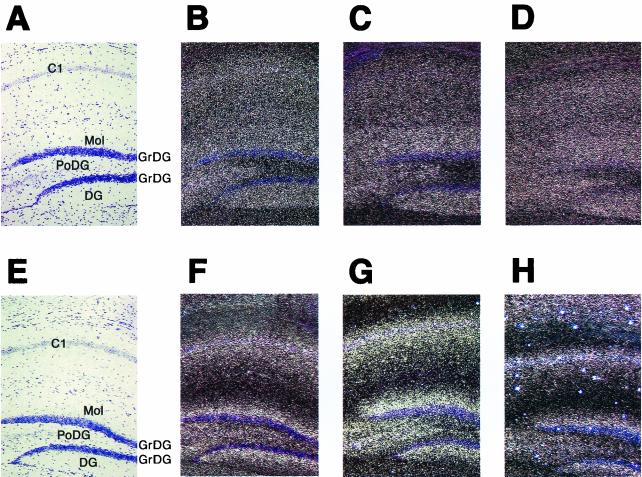
As representatives of dendritic mRNAs, CaMKIIα and MAP2 mRNAs were examined by in situ hybridization for potential changes in expression levels and subcellular distribution. Expression levels of neither mRNA were significantly altered compared with control animals (Fig. 4). CaMKIIα mRNA has been reported to extend to the full dendritic length in hippocampal neurons (32). Such a CaMKIIα mRNA expression pattern was observed in both BC1-deficient and control animals (Fig. 4A to D). Quantification of the signal intensity for CaMKIIα mRNA in the stratum pyramidale, stratum radiatum and lacunosum moleculare layers of the hippocampal CA3 area revealed no significant differences between BC1-deficient and control animals.
FIG. 4.
In situ hybridization of frontal brain sections from BC1-deficient and normal mice with CaMKIIα and MAP2 mRNA probes. CaMKIIα and MAP2 mRNAs show a normal dendritic distribution in BC1-deficient mice. (A to D) Hybridization with a CaMKIIα antisense probe; (E to H) hybridization with a MAP2 antisense probe. All sections are counterstained with cresyl violet. (B and F) Sections from BC1-deficient mice (line 15); (C and G) sections from line 6. (A, E, D, and H) Sections from control mice. Dark-field (B, C, D, F, G, and H) and bright-field (A and E) micrographs are shown. C1, field CA1 of hippocampus; Mol, molecular layer dentate gyrus; PoDG, polymorph layer dentate gyrus; DG, dentate gyrus; GrDG, granular layer dentate gyrus. All abbreviations are according to Franklin and Paxinos (15).
In the hippocampi of BC1-deficient animals, MAP2 mRNA was observed extending into the stratum radiatum of Ammons horn (apical dendrites of areas CA1 to -3). The extent of labeling in this layer was indistinguishable from that in controls. In both experimental and control animals the MAP2 mRNA signal extended from the stratum pyramidale to the middle of the stratum radiatum (Fig. 4E to H). This reflects the typical subcellular distribution of MAP2 mRNA in the hippocampus, as previously reported (16, 32). Quantification of the signal intensity for the MAP2 mRNA probe in the stratum pyramidale, stratum radiatum and lacunosum moleculare layers of the hippocampal CA3 area revealed no significant differences between BC1-deficient and control animals. In summary, no clear differences between BC1-deficient and control animals were detectable in our analysis of the expression and subcellular distribution of two bona fide dendritic mRNAs.
Conclusions.
We have previously hypothesized that BC1 RNA and BC200 RNA operate as modulators of dendritic RNA functions (7, 40, 41, 42). Thus, BC1 RNA has been suggested to play a role in the transport of other dendritic components, (e.g., selected mRNAs). Alternatively, we speculated (6, 7, 42) that BC1 and BC200 RNAs function as modulators of translation in dendrites. We now find that the somatodendritic expression patterns of endogenous MAP2 mRNA and CaMKIIα mRNA were not significantly distinguishable from those in control animals. This result makes a requisite role of BC1 RNA in dendritic mRNA transport appear to be unlikely.
In contrast, the hypothesis of a functional role in translational regulation has been strengthened by recent experimental evidence (18, 43). Such a function may be expected to find phenotypic expression in animal behavior. We have now initiated behavioral analysis of BC1-deficient mice. Such analysis was conducted in parallel in three independent laboratories, including a naturalistic outdoor setting. BC1-deficient animals exhibit behavioral changes that are best interpreted in terms of reduced exploration and increased anxiety (unpublished data). In contrast, spatial memory appeared to be spared. The combined data indicate that neuron-specific BC1 non-mRNA may contribute to the modulation of animal behavior. These findings on the behavioral phenotype of BC1-deficient animals will be reported separately.
We submit that BC1 and similar small, untranslated synaptic RNAs are of relatively recent evolutionary origin (60 to 110 million years ago for BC1 RNA [25]), as they are present and conserved in narrow phylogenetic ranges, such as a mammalian order (e.g., rodents for BC1 RNA) or suborder (e.g., anthropoid primates for BC200 RNA [19]). As a consequence, such RNAs are not expected to perform obligatory roles in mammalian brain function but rather are expected to modulate or enhance neuronal form and function. They thereby confer a selective advantage during evolution of a mammalian order even though the observed changes in phenotype following targeted gene depletion are in fact subtle.
Acknowledgments
We thank Heidi Stuhlmann (Mount Sinai School of Medicine, New York, N.Y.) for R1 ES cells, Les Pennelly (Roche Pharmaceuticals) for ganciclovir, Alexander Kondrachov and Vladimir Kuryshev for help with blastocyst injections and DNA blot analysis, G. F. Weinbauer for morphological examination of testes, and Marsha Bundman and Ellen Hsu for critical reading and discussion of the manuscript. We thank Gideon D. F. Brosius for pointing out that some of the mice had cataracts.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Br 754/2 and Kr1624/3-1) and the Volkswagen Stiftung (I/74 040) to J.B., from the Deutsche Forschungsgemeinschaft (Sa 389/5) to N.S, and from the NIH (NS13458) to H.T.
REFERENCES
- 1.Brosius, J. 2003. The contribution of RNAs and retroposition to evolutionary novelties. Genetica, 118:99-116. [PubMed]
- 2.Brosius, J. 1999. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238:115-134. [DOI] [PubMed] [Google Scholar]
- 3.Brosius, J. 1999. Transmutation of tRNA over time. Nat Genet. 22:8-9. [DOI] [PubMed] [Google Scholar]
- 4.Brosius, J., and S. J. Gould. 1993. Molecular constructivity. Nature 365:102. [DOI] [PubMed] [Google Scholar]
- 5.Brosius, J., and S. J. Gould. 1992. On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA.” Proc. Natl. Acad. Sci. USA 89:10706-10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius, J., and H. Tiedge. 2001. Dendritic BC1 RNA: intracellular transport and activity-dependent modulation, p. 129-138. In D. Richter (ed.), Cell polarity and subcellular RNA localization. Springer Verlag, Berlin, Germany [DOI] [PubMed]
- 7.Brosius, J., and H. Tiedge. 1995. Neural BC1 RNA: dendritic localization and transport, p. 289-330. In H. D. Lipshitz (ed.), Localized RNAs. R. G. Landes, Austin, Tex.
- 8.Bulleit, R. F., M. K. Bennett, S. S. Molloy, J. B. Hurley, and M. B. Kennedy. 1988. Conserved and variable regions in the subunits of brain type II Ca2+/calmodulin-dependent protein kinase. Neuron 1:63-72. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D., K. Jin, K. Kawaguchi, M. Nakayama, X. Zhou, Z. Xiong, A. Zhou, X. O. Mao, D. A. Greenberg, S. H. Graham, and R. P. Simon. 2003. Ero1-L, an ischemia-inducible gene from rat brain with homology to global ischemia-induced gene 11 (Giig11), is localized to neuronal dendrites by a dispersed identifier (ID) element-dependent mechanism. J. Neurochem. 85:670-679. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, J. G., H. Tiedge, and J. Brosius. 1996. Identification and characterization of BC1 RNP particles. DNA Cell Biol. 15:549-559. [DOI] [PubMed] [Google Scholar]
- 11.DeChiara, T. M., and J. Brosius. 1987. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc. Natl. Acad. Sci. USA 84:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deininger, P. L., H. Tiedge, J. Kim, and J. Brosius. 1996. Evolution, expression, and possible function of a master gene for amplification of an interspersed repeated DNA family in rodents. Prog. Nucleic Acid Res. Mol. Biol. 52:67-88. [DOI] [PubMed] [Google Scholar]
- 13.Festing, M. F., E. M. Simpson, M. T. Davisson, and L. E. Mobraaten. 1999. Revised nomenclature for strain 129 mice. Mamm. Genome 10:836. [DOI] [PubMed] [Google Scholar]
- 14.Fox, W. M. 1965. Reflex-ontogeny and behavioural development of the mouse. Anim. Behav. 13:234-241. [DOI] [PubMed] [Google Scholar]
- 15.Franklin, K. B. J., and G. Paxinos. 1996. The mouse brain in stereotaxic coordinates. Academic Press, San Diego, Calif.
- 16.Garner, C. C., and A. Matus. 1988. Different forms of microtubule-associated protein 2 are encoded by separate mRNA transcripts. J. Cell Biol. 106:779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J., D. H. Kass, and P. L. Deininger. 1995. Transcription and processing of the rodent ID repeat family in germline and somatic cells. Nucleic Acids Res. 23:2245-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondrashov, A. V. 2001. Common principles of the regulatory systems of transcription and translation as revealed by module structure of preinitiation TATA-complex and BC1 RNA. Ph.D. dissertation. Russian Acadaemy of Sciences, St. Petersburg, Russia.
- 19.Kuryshev, V. Y., B. V. Skryabin, J. Kremerskothen, J. Jurka, and J. Brosius. 2001. Birth of a gene: locus of neuronal BC200 snmRNA in three prosimians and human BC200 pseudogenes as archives of change in the Anthropoidea lineage. J. Mol. Biol. 309:1049-1066. [DOI] [PubMed] [Google Scholar]
- 20.Laird, P. W., A. Zijderveld, K. Linders, M. A. Rudnicki, R. Jaenisch, and A. Berns. 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19:4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, Y., J. Brosius, and H. Tiedge. 2001. Neuronal BC1 RNA: co-expression with growth-associated protein-43 messenger RNA. Neuroscience 103:465-479. [DOI] [PubMed] [Google Scholar]
- 22.Lipp, H.-P., and D. Wahlsten. 1992. Absence of the corpus callosum, p. 217-252. In P. Driscoll (ed.), Genetically-defined animal models of neuro-behavioral dysfunction. Birkhaeuser, Boston, Mass.
- 23.Magara, F., U. Muller, Z. W. Li, H. P. Lipp, C. Weissmann, M. Stagljar, and D. P. Wolfer. 1999. Genetic background changes the pattern of forebrain commissure defects in transgenic mice underexpressing the beta-amyloid-precursor protein. Proc. Natl. Acad. Sci. USA 96:4656-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martignetti, J. A., and J. Brosius. 1995. BC1 RNA: transcriptional analysis of a neural cell-specific RNA polymerase III transcript. Mol. Cell. Biol. 15:1642-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martignetti, J. A., and J. Brosius. 1993. Neural BC1 RNA as an evolutionary marker: guinea pig remains a rodent. Proc. Natl. Acad. Sci. USA 90:9698-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinnon, R. D., P. Danielson, M. A. Brow, F. E. Bloom, and J. G. Sutcliffe. 1987. Expression of small cytoplasmic transcripts of the rat identifier element in vivo and in cultured cells. Mol. Cell. Biol. 7:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milner, R. J., F. E. Bloom, C. Lai, R. A. Lerner, and J. G. Sutcliffe. 1984. Brain-specific genes have identifier sequences in their introns. Proc. Natl. Acad. Sci. USA 81:713-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muddashetty, R., T. Khanam, A. Kondrashov, M. Bundman, A. Iacoangeli, J. Kremerskothen, K. Duning, A. Barnekow, A. Hüttenhofer, H. Tiedge, and J. Brosius. 2002. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 321:433-445. [DOI] [PubMed] [Google Scholar]
- 29.Muslimov, I. A., Y. Lin, M. Heller, J. Brosius, Z. Zakeri, and H. Tiedge. 2002. A small RNA in testis and brain: implications for male germ cell. J. Cell Sci. 115:1243-1250. [DOI] [PubMed] [Google Scholar]
- 30.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens, G. P., N. Chaudhari, and W. E. Hahn. 1985. Brain “identifier sequence” is not restricted to brain: similar abundance in nuclear RNA of other organs. Science 229:1263-1265. [DOI] [PubMed] [Google Scholar]
- 32.Paradies, M. A., and O. Steward. 1997. Multiple subcellular mRNA distribution patterns in neurons: a nonisotopic in situ hybridization analysis. J. Neurobiol. 33:473-493. [DOI] [PubMed] [Google Scholar]
- 33.Rogers, D. C., J. Peters, J. E. Martin, S. Ball, S. J. Nicholson, A. S. Witherden, M. Hafezparast, J. Latcham, T. L. Robinson, C. A. Quilter, and E. M. Fisher. 2001. SHIRPA, a protocol for behavioral assessment: validation for longitudinal study of neurological dysfunction in mice. Neurosci. Lett. 306:89-92. [DOI] [PubMed] [Google Scholar]
- 34.Sapienza, C., and B. St. Jacques. 1986. ‘Brain-specific’ transcription and evolution of the identifier sequence. Nature 319:418-420. [DOI] [PubMed] [Google Scholar]
- 35.Skryabin, B. V., and C. Schmauss. 1997. Enhanced selection for homologous-recombinant embryonic stem cell clones with a neomycin phosphotransferase gene in antisense orientation. Transgenic Res. 6:27-35. [DOI] [PubMed] [Google Scholar]
- 36.Sutcliffe, J. G., R. J. Milner, J. M. Gottesfeld, and R. A. Lerner. 1984. Identifier sequences are transcribed specifically in brain. Nature 308:237-241. [DOI] [PubMed] [Google Scholar]
- 37.Sutcliffe, J. G., R. J. Milner, J. M. Gottesfeld, and W. Reynolds. 1984. Control of neuronal gene expression. Science 225:1308-1315. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, K. R., and M. R. Capecchi. 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51:503-512. [DOI] [PubMed] [Google Scholar]
- 39.Tiedge, H., F. E. Bloom, and D. Richter. 1999. RNA, whither goest thou? Science 283:186-187. [DOI] [PubMed] [Google Scholar]
- 40.Tiedge, H., and J. Brosius. 1996. Translational machinery in dendrites of hippocampal neurons in culture. J. Neurosci. 16:7171-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiedge, H., W. Chen, and J. Brosius. 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 13:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiedge, H., R. T. Fremeau, Jr., P. H. Weinstock, O. Arancio, and J. Brosius. 1991. Dendritic location of neural BC1 RNA. Proc. Natl. Acad. Sci. USA 88:2093-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, H., A. Iacoangeli, S. Popp, I. A. Muslimov, H. Imataka, N. Sonenberg, I. B. Lomakin, and H. Tiedge. 2002. Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 22:10232-10241. [DOI] [PMC free article] [PubMed] [Google Scholar]