Abstract
Spindle checkpoint proteins monitor the interaction of the spindle apparatus with the kinetochores, halting anaphase even if the microtubule attachment of only a single chromosome is altered. In this study, we show that Bub3p of Saccharomyces cerevisiae, an evolutionarily conserved spindle checkpoint protein, exhibits distinct interactions with an altered or defective kinetochore(s). We show for the first time that green fluorescent protein-tagged S. cerevisiae Bub3p (Bub3-GFP) exhibits not only a diffuse nuclear localization pattern but also forms distinct nuclear foci in unperturbed growing and G2/M-arrested cells. As Bub3-GFP foci overlap only a subset of kinetochores, we tested a model in which alterations or defects in kinetochore or spindle integrity lead to the distinct enrichment of Bub3p at these structures. In support of our model, kinetochore-associated Bub3-GFP is enriched upon activation of the spindle checkpoint due to nocodazole-induced spindle disassembly, overexpression of the checkpoint kinase Mps1p, or the presence of a defective centromere (CEN). Most importantly, using a novel approach with the chromatin immunoprecipitation (ChIP) technique and genetically engineered defective CEN [CF/CEN6(Δ31)], we determined that Bub3-GFP can associate with a single defective kinetochore. Our studies represent the first comprehensive molecular analysis of spindle checkpoint protein function in the context of a wild-type or defective kinetochore(s) by use of live-cell imaging and the ChIP technique in S. cerevisiae.
In Saccharomyces cerevisiae, chromosome segregation involves the controlled separation and movement of 32 chromosomes inside the confines of the nuclear envelope. The process requires spindle pole bodies (SPBs) that are connected to the kinetochore of each chromosome (a complex of centromere [CEN] DNA and kinetochore proteins) through spindle microtubules (MTs). The connection between MTs and chromosomes is maintained by large numbers of core and accessory kinetochore proteins (10). Complexes of kinetochore proteins are clustered throughout the cell cycle (22, 25, 41). Centromeres of individual chromosomes remain connected to MTs through most of the cell cycle (55), except when centromeres are replicated or kinetochore structure is compromised. In the latter case, spindle checkpoint proteins provide a fail-safe mechanism allowing extra time to retrieve misaligned chromosomes (for a review, see reference 39).
The interplay of at least six evolutionarily conserved spindle checkpoint proteins (Mad1p, Mad2p, Mad3p, Bub1p, Bub3p, and Mps1p) (26, 32, 33, 53) monitors the integrity of the kinetochore-MT complex and mediates a signal to halt anaphase even if the association of only a single chromosome with the MT is altered or compromised (for a review, see reference 39). A multiorganism approach to elucidating the functions of these proteins (38) revealed that individual checkpoint proteins functionally interact and form complexes containing various combinations of Mad1p, Mad2p, Bub1p, Bub3p, Mad3p (BubRI), and Cdc20p (4, 13, 19, 24, 43, 48). Complexes of Bub1p-Bub3p-Mad1p and Mad2p-Bub3p-Mad3p-Cdc20p may form after checkpoint activation in response to altered or compromised kinetochore-MT structures and are believed to constitute diffusible signals responsible for halting anaphase entry through inhibition of the anaphase-promoting complex (for a review, see reference 30).
Until recently, the majority of in vivo cell biological observations pertaining to checkpoint proteins were made with larger eukaryotic cells. These results suggest that checkpoint proteins are localized to one or several different cellular locations, including the cytosol (11), the nuclear pore complex (NPC) (8), and kinetochores (2, 8, 12, 34, 35, 50), and diffusely distributed within the nucleus (50). Localization of these proteins appears to be transient and to depend on the presence of other proteins as well as the status of chromosome segregation.
In contrast to the vast body of literature on the localization of checkpoint proteins in higher eukaryotes, there are very few reports pertaining to such studies in S. cerevisiae and Schizosaccharomyces pombe. These studies include immunofluorescence approaches for localizing epitope-tagged fusion constructs of Mps1p of S. cerevisiae (9), Mad3p of S. pombe (SpMad3p) (37), and SpBub3p (3) to kinetochores. Recent advances in cytological live-cell image analysis of ScMad2p, SpMad2p, and SpBub1p and biochemical approaches for ScBub1p provide powerful tools for correlating their kinetochore localizations with a wealth of information from previous genetic and biochemical approaches with these model organisms (28, 29, 31, 51).
This report describes the results of a comprehensive analysis of the in vivo localization and biochemical associations of S. cerevisiae Bub3p in the context of its functional role in the checkpoint pathway. We investigated the foci formed by green fluorescent protein (GFP)-tagged S. cerevisiae Bub3p (Bub3-GFP) during the unperturbed cell cycle, during G2/M arrest, and upon activation of the spindle checkpoint. We determined that Bub3-GFP foci colocalize predominantly with kinetochores in vivo and with CEN DNA in chromatin immunoprecipitation (ChIP) experiments. Bub3-GFP foci appear to associate with a subset of kinetochores, and this association is dependent on the presence of checkpoint protein Bub1p and functional kinetochore protein Ndc10p. Finally, in a novel approach with ChIP assays and genetically engineered defective CEN [CF/CEN6(Δ31)], we determined that Bub3-GFP associates in vivo with altered kinetochores. The latter technique should prove widely applicable to study of the interactions between checkpoint proteins and a single defective kinetochore in the context of normally segregating chromosomes. These studies are especially important because budding yeast Bub3p may provide important clues to the functional roles of these proteins in chromosome segregation in yeasts (52) and humans.
MATERIALS AND METHODS
Yeast media, strains, and plasmids.
Media used for yeast growth, sporulation, and manipulation were described previously (1, 45), except where indicated. YEPD is yeast extract-peptone-dextrose. Benomyl-containing plates were made as described previously (27). All yeast strains are listed in Table 1. The GAL-MPS1-containing plasmid (pAFS120MPS1+) was a generous gift from Mark Winey (University of Colorado, Boulder). The bub1Δ::LEU2 strain with BUB1 in pRS316 (YKH481) was a generous gift from Kathy Hyland and Phil Hieter (University of British Columbia, Vancouver, British Columbia, Canada). The ndc10-1 strain JK421 was a generous gift from John Kilmartin (MRC-LMB, Cambridge, United Kingdom). The diploid strain YPH630, containing the CF/CEN6(Δ31) chromosome fragment (CF) CFIII(D8B.d.YPH429)URA3 SUP11, and the bub3Δ::LEU2 deletion strain (YFS1100) were generous gifts from Forrest Spencer (School of Medicine, The Johns Hopkins University, Baltimore, Md.). Plasmids containing fusions of MTW1 and the genes encoding the green, cyan, and yellow fluorescent proteins (GFP, CFP, and YFP, respectively) in vectors pRS315 and pRS316 were described previously (29).
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference, source, or derivation |
|---|---|---|
| YPH278 | MATα ura3-52 lys2-80l ade2-101 his3-200 leu2Δ1 CFIII (CEN3L.YPH278)URA3 SUP11 | 46 |
| YMB1293 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB2-GFP/HIS5 CFIII (CEN3L.YPH278)URA3 SUP11 | This study |
| YMB1296 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 MAD2-GFP/HIS5 CFIII (CEN3L.YPH278)URA3 SUP11 | 29 |
| YMB1302 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB3-GFP/HIS5 CFIII (CEN3L.YPH278)URA3 SUP11 | This study |
| YMB3098 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB3-CFP/HIS5/KAN CFIII (CEN3L.YPH278)URA3 SUP11 | This study |
| YMB4025 | MATaura3-52 lys2-801 ade2-101 trp1Δ1 his3Δ200 leu2Δ1 ΔCEN6::(CEN11/LEU2)/CEN6 CFIII(D8b.d.YPH429)URA3 SUP11 | This study |
| YMB4033 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB3-GFP/HIS5 | This study |
| YMB4105 | MATaura3-52 lys2-801 ade2-101 trp1Δ1 leu2Δ1 BUB3-GFP/HIS5/his3Δ200 CEN6 CFIII(D8b.d.YPH429)URA3 SUP11 | This study |
| YMB4115 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB3-GFP/HIS5(pAFS120MPS1+/LEU2) (pMTW1/CFP/URA3) | This study |
| YMB4116 | MATaura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB3-GFP/HIS5 | This study |
| YMB4119 | MATaura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB3-GFP/HIS5 (pAFS120MPS1+/LEU2) | This study |
| YMB4140 | MATα ura3-52 lys2-801 ade2-101 his3-200 leu2Δ1 BUB3-GFP/HIS5 bub1::LEU2 | Meiotic progeny of YKH481 × YMB4116 |
| YMB4147 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB2-CFP/HIS5/KAN CFIII (CEN3L YPH278)URA3 SUP11 | This study |
| YMB4155 | MATα ura3 leu2 his3 BUB3-GFP/HIS5 ndc10-1 | Meiotic progeny of YMB4033 × JK421 |
| YMB4192 | MATα ura3 leu2 his3 MAD2-GFP/HIS5 ndc10-1 | Meiotic progeny of YMB1296 × YKH481 |
| YMB4204 | MATaura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB3-GFP/HIS5 BUB2-CFP/HIS5/KAN | Meiotic progeny of YMB4116 × YMB4147 |
| YMB4225 | MATaura3-52 lys2-801 ade2-101 trp1Δ1 leu2Δ1 his3Δ200 CEN6 CFIII (D8b.d.YPH429)URA3 SUP11 | This study |
| YMB4245 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 BUB3-GFP/HIS5 SPC29-CFP/KAN CFIII (CEN3L YPH278)URA3 SUP11 | This study |
| JK421 | MATaade2-1 ura3-1 his3-11,1 trp1-1 leu2-3,112 can1-100 ndc10-1 | 21 |
| YKH481 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 bub1Δ::LEU2 BUB1/URA3 | Isogenic to YFP2 (27) |
| YPH630 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1Δ1/trp1Δ1 HIS3/his3Δ200 leu2Δ1/leu2Δ1 ΔCEN6::(CEN11/LEU2)/CEN6 CFIII(D8b.d.YPH429)URA3 SUP11 | 47 |
| YFS1100 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 bub3Δ::LEU2 CFIII (CEN3.L.YPH278)URA3 SUP11 | 52 |
Construction of tagged BUB3, BUB2, SPC29, and MAD2 genes.
BUB3, BUB2, and MAD2 were tagged with the GFP gene (GFP) following their last amino acid codon by use of an integrative PCR-based transformation procedure (54). Primers and the GFP-HIS5 template plasmid pGFP (36) were generously supplied by Dan Burke (University of Virginia, Charlottsville). Briefly, GFP and the HIS5 marker were PCR amplified from plasmid pGFP with a sense primer containing a region of BUB3, BUB2, or MAD2 just before the termination codon and an antisense primer containing a region of BUB3, BUB2, or MAD2 just past the termination codon. PCR products were transformed into a wild-type strain (YPH278). His+ transformants were screened for in-frame fusions of BUB3-GFP, BUB2-GFP, and MAD2-GFP by PCR and tested for functional complementation by assaying the tagged strain for growth in media containing sublethal doses of the MT-depolymerizing drug benomyl and the ability to maintain the CF. Strains YMB1302, YMB4033, and YMB4116 (BUB3-GFP/HIS5), YMB1293 (BUB2-GFP/HIS5), and YMB1296 (MAD2-GFP/HIS5) (29) were used for subsequent studies.
For the examination of Bub3-GFP, Mad2-GFP, or Mtw1-GFP in wild-type or other strains, we mated the appropriate strains, followed by tetrad analysis and characterization of representative haploid meiotic progeny. For a bub1Δ strain, we mated a bub1Δ::LEU2 strain (YKH481) and a BUB3-GFP strain (YMB4116), resulting in the haploid bub1Δ::LEU2 BUB3-GFP strain (YMB4140). For the examination of Bub3-GFP or Mad2-GFP in the ndc10-1 strain, we mated the ndc10-1 strain (JK421) with either a BUB3-GFP strain (YMB1302), yielding the haploid ndc10-1 BUB3-GFP strain (YMB4155), or a MAD2-GFP strain (YMB1296), resulting in the ndc10-1 MAD2-GFP strain (YMB4192). The CF/CEN6(Δ31)-containing strain (YMB4025) was derived from the dissection of a diploid strain containing CF/CEN6(Δ31) (YPH630). For the examination of Bub3-GFP in a strain containing CF/CEN6(Δ31), we crossed a CF/CEN6(Δ31) strain (YMB4025) with a BUB3-GFP strain (YMB1302), resulting in the CF/CEN6(Δ31) BUB3-GFP strain (YMB4105).
We generated a BUB2-CFP strain (YMB4147) for the visualization of SPBs. This strain was equivalent to a BUB2-GFP/HIS5 strain (YMB1293), except that the GFP portion was replaced with CFP by use of the open reading frame carried on plasmid pDH3/KAN/CFP (The Yeast Resource Center, Seattle, Wash.). Similarly, GFP in a BUB3-GFP/HIS5 strain (YMB1302) was replaced with CFP to generate a BUB3-CFP-expressing strain (YMB3098). A BUB2-CFP strain (YMB4147) was mated with a BUB3-GFP strain (YMB4116), and the haploid BUB2-CFP BUB3-GFP strain (YMB4204) was used for further analysis. We generated an SPC29-CFP fusion in a BUB3-GFP strain (YMB1302) to visualize SPBs. Briefly, SPC29-CFP/KAN was PCR amplified from strain SLJ940 (a generous gift from Mark Winey) and transformed into YMB1302 to form the BUB3-GFP/HIS5 SPC29-CFP/KAN strain (YMB4245). All constructs were analyzed by PCR, followed by sequence analysis of the PCR products, functional complementation, and fluorescence microscopy to confirm expression.
Cell cycle arrest.
For cell cycle arrest, we used early-logarithmic-phase cultures grown at 30°C. For G1 arrest, cells were incubated with 1 μg of α-factor (T-6901; Sigma, St. Louis, Mo.)/ml for 90 min (5). For nocodazole treatment (G2/M arrest), cells arrested with α-factor were incubated with 15 μg of nocodazole (M-1404; Sigma)/ml for 90 min. Cell cycle arrest was monitored by examining cells with a microscope (nuclear and spindle morphology) and by flow cytometry analysis (29). G2/M arrest resulting from Mps1p overexpression from the GAL-MPS1-containing plasmid was induced by the addition of galactose (4%) to sucrose (2%)-grown cultures for 3 h at 30°C. DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI).
In vivo cross-linking and ChIP.
Yeast strains were grown in selective medium to an optical density at 600 nm of 1.2 to 1.6 and cross-linked with 1% paraformaldehyde for 30 min at room temperature. Fixed cells were induced to form spheroplasts with Zymolyase 20T and sonicated to fragment chromosomal DNA to an average size of 200 to 1,000 bp. Chromatin solutions were immunoprecipitated as described previously (38) with anti-GFP (1814460; Roche, Indianapolis, Ind.) or anti-hemagglutinin (HA) (mock) antibodies at a concentration of 4 μg/ml. Sonicated λDNA (4 μg) was added, and immune complexes were harvested by incubation with protein A-Sepharose CL-4B beads (Sigma) for 2 h at 25°C. Beads were washed, and immunoprecipitated material was eluted with an appropriate buffer containing 1% sodium dodecyl sulfate at 65°C for 5 h and precipitated with ethanol. The precipitates were treated with proteinase K (Roche), extracted with organic solvents, and ethanol precipitated.
Aliquots of total input DNA (3-μl chromatin solution) and immunoprecipitated DNA (30-μl chromatin solution) were analyzed by PCR with primers specific for CEN1, CEN3, CEN6, CEN16, and ACT1. Primer pairs used were as follows: for CEN1, PM57 (5′-CTCGATTTGCATAAGTGTGCC-3′) and PM58 (5′-GTGCTTAAGAGTTCTGTACCAC-3′); for CEN3, PM22 (5′-GATCAGCGCCAAACAATATGG-3′) and PM48 (5′-AACTTCCACCAGTAAACGTTTC-3′); for CEN6, OMB CEN6F (5′-GATGGCTCAAAACAAATTACC-3′) and OMB CEN6R (5′-GATCTCTAATTTATGGTTTGC-3′); for CEN16, PM55 (5′-GCAAAGGTTGAAGCCGTTATG-3′) and PM56 (5′-GCTTTGCCGATTTCGCTTTAG); and for ACT1, OMB444 (5′-ACAACGAATTGAGAGTTGCCCCAG) and OMB445 (5′-AATGGCGTGAGGTAGAGAGAAACC).
Amplification products of CEN6(Δ31) and CEN6 (wild type) were cloned into Topo TA2.1 vectors according to the manufacturer's instructions (Stratagene, La Jolla, Calif.). To quantitate CEN6 (wild-type) and CEN6(Δ31) amplification products, individual DNA fragments were separated on 5% polyacrylamide gels, stained with SYBR green, and analyzed with a Fuji phosphorimager. In order to better separate the CEN6 (wild-type) and CEN6(Δ31) species, one-half of the total amplification products was digested with XhoI prior to electrophoresis. Reported values and errors correspond to the results of at least two independent experiments.
Fluorescence microscopy.
Strains expressing GFP-, CFP-, and YFP-tagged genes were grown overnight in synthetic dextrose-containing medium supplemented with 0.16 mM adenine. Untreated and nocodazole-treated cells were examined for GFP-, CFP-, and YFP-tagged proteins with an Axioscope 2 microscope (Carl Zeiss Inc., Thornwood, N.Y.) fitted with a Sensicam (Cooke, Auburn, Mich.), a GFP filter set (CZ909; Chroma Technology Corp., Brattleboro, Vt.), a CFP filter set (XF114-2; Omega Optical Inc., Brattleboro, Vt.), and a Uniblitz (Rochester, N.Y.) shutter assembly.
RESULTS
S. cerevisiae Bub3-GFP localizes to the nucleus and forms distinct nuclear foci.
To understand the molecular role of Bub3p, the chromosomal copy of the BUB3 open reading frame was tagged with either the gene encoding GFP or the gene encoding CFP. We determined that strains expressing Bub3-GFP or Bub3-CFP were checkpoint proficient and grew in a manner similar to that of a wild-type strain on medium containing benomyl, an MT-depolymerizing agent. Defects in checkpoint function cause inhibition of the growth of a bub3Δ strain on benomyl-containing medium (Fig. 1A). Also, chromosome loss assays failed to detect an increase in the loss of a nonessential CF in the BUB3-GFP-expressing strain compared to the wild-type strain (data not shown). This finding is in contrast to the increased loss of the CF in an isogenic bub3Δ strain (52).
FIG. 1.
Bub3-GFP localizes to the nucleus and forms distinct foci in S. cerevisiae. (A) Strains expressing Bub3-GFP or Bub3-CFP are not sensitive to the MT-destabilizing drug benomyl. Growth of the parental wild-type (WT) strain (YPH278) was compared to that of BUB3-GFP (YMB1302), BUB3-CFP (YMB3098), and bub3Δ (YFS1100) strains. All strains were grown to logarithmic phase in YEPD at 30°C, diluted, spotted in 10-fold increments on YEPD and YEPD containing benomyl (BEN, 15 μg/ml), and incubated for 4 days at 27°C. (B) Bub3-GFP exhibits diffuse nuclear staining and can localize to distinct nuclear foci. The strain expressing Bub3-GFP (YMB1302) was grown to logarithmic phase at 30°C and examined under a fluorescence microscope (Bub3-GFP). The same strain was also stained with DAPI to determine whether Bub3-GFP foci (green spots indicated by arrows) overlap nuclear DNA (red) (Bub3-GFP/DAPI). (C) In another experiment, a strain expressing Bub3-GFP (YMB4116) was synchronized in G1 by treatment with α-factor and released into fresh medium without α-factor. Samples were analyzed by fluorescence microscopy. Prevalent cell morphologies, the time period after release from arrest (minutes), and the occurrence of Bub3-GFP foci (percentages) are indicated in the lower panel; at least 100 cells were counted for each time point in two independent experiments. Scale bars, 5.0 μm.
We next analyzed the subcellular localization of Bub3-GFP by fluorescence microscopy of live cells. In logarithmically growing cells, Bub3-GFP could be visualized as a diffuse intranuclear GFP signal. We noted, however, that a small fraction (about 10%) of Bub3-GFP-expressing cells consistently showed one or two distinctly staining foci (Fig. 1B, upper left panel). Furthermore, Bub3-GFP foci overlapped nuclear DNA in DAPI-stained nuclei (Fig. 1B, upper right panel) and could be observed throughout the cell cycle of logarithmically growing cells, with the exception of cells in late mitosis (Fig. 1B, lower panel). In order to test whether the occurrence of the Bub3-GFP foci overlapped a defined phase of the cell cycle, we examined cells after α-factor arrest (G1) followed by synchronous release into the cell cycle. From this arrest-release experiment, we determined that nuclear Bub3-GFP foci were almost absent in cells in G1 phase, were increased in small budded cells (7 to 18%), and were increased no further in G2/M phase (9%). Again, Bub3-GFP foci were rarely, if ever, observed in cells in late mitosis (Fig. 1C).
Activation of the spindle checkpoint by nocodazole treatment leads to the enrichment of Bub3-GFP foci.
Our data suggest that a subpopulation of cells contain nuclear Bub3-GFP foci. Consistent with a role in the spindle assembly checkpoint, we reasoned that altered kinetochore-spindle integrity might lead to an enrichment of Bub3p foci at such sites in these cells. Hence, we examined the localization and frequency of Bub3-GFP after α-factor arrest (G1) and at various times after release into nocodazole-containing medium. Nocodazole is an MT-depolymerizing agent that causes the activation of the spindle assembly checkpoint and G2/M arrest. Our results showed that after the release of G1-synchronized cells into nocodazole-containing medium, very few cells exhibited Bub3-GFP foci at early times (15 to 30 min) but increased to ∼60% after 90 min (Fig. 2A and B). The cell cycle progression and efficacy of nocodazole arrest were monitored by flow cytometry analysis of the samples at each time point (Fig. 2C). These results suggest that Bub3-GFP foci are enriched in response to activation of the spindle checkpoint.
FIG. 2.
Activation of the spindle checkpoint by nocodazole leads to enrichment of Bub3-GFP foci. (A) A strain expressing Bub3-GFP (YMB4116) was grown to logarithmic phase in YEPD at 30°C, synchronized in G1 by treatment with α-factor, and then released into medium containing nocodazole (15 μg/ml). Images of cells were recorded after release from α-factor arrest (G1) into nocodazole at various times from zero minutes (0), at 15-min intervals, for 1.5 h (90 min). Scale bar, 5.0 μm. (B) Graphic representation of the percentage of cells showing Bub3-GFP foci, corresponding to the image data shown in panel A, at various times. (C) Layered fluorescence-activated cell sorting images corresponding to samples collected at the times indicated in panel A. Cell cycle arrest with a prominent G2 peak at 90 min is indicated. Depolymerization of the mitotic spindle was confirmed by immunofluorescence analysis of fixed cells (data not shown).
Bub3-GFP foci localize to a subset of kinetochores.
Studies with metazoans (2, 35) have localized Bub3p to kinetochores of individual chromosomes. Our results showing the enrichment of Bub3-GFP foci upon spindle checkpoint activation prompted us to examine whether these foci localized to S. cerevisiae kinetochores by using CFP- or YFP-tagged kinetochore marker protein Mtw1p (22). In the first experiment, using logarithmically growing cells, we analyzed a strain coexpressing Bub3-CFP and Mtw1-YFP. Overall, Bub3-CFP foci were observed in about 10% of the Mtw1-YFP-stained cells. Both proteins colocalized with a single kinetochore cluster (Fig. 3A, upper panels). As the cells progressed through the cell cycle, the kinetochore clusters of sister chromatids were separated from each other, as evidenced by two Mtw1-YFP foci (Fig. 3A, lower panels). Our data for two Mtw1-YFP foci are consistent with previous observations of “double dots” of kinetochore clusters visualized with the CEN15 (1.*)-GFP and Mtw1-GFP markers (22). We observed that 5 to 10% of large budded logarithmically growing cells with double dots of kinetochore clusters also contained Bub3-GFP. In greater than 95% of these cells, Bub3-GFP foci coincided with only one of the two observed kinetochore clusters.
FIG. 3.
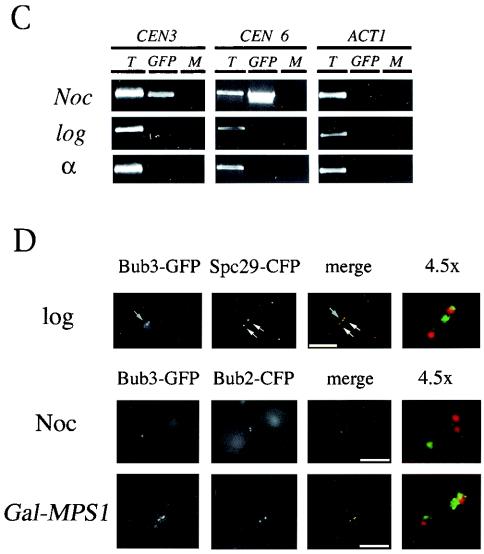
Bub3-GFP foci colocalize predominantly with kinetochores and associate with CEN DNA. (A) Localization of Bub3-CFP and Mtw1-YFP in unperturbed cells. A strain coexpressing Bub3-CFP and the kinetochore marker Mtw1-YFP (YMB3098 containing pOKMTW1-YFP) was grown to logarithmic phase (log) in YEPD at 30°C and examined by fluorescence microscopy. Representative images of small budded cells (upper panels) and large budded cells (lower panels) stained for both Bub3-CFP (green) and Mtw1-YFP (red) foci are shown. Diagrams of yeast cells indicate the nuclear positions of clustered kinetochores overlapping Bub3-CFP (○) or without Bub3-CFP foci (•). Note that Bub3-CFP foci always overlap Mtw1-YFP foci. Furthermore, Bub3-CFP foci are rarely, if ever, observed in G1 and late mitotic cells. (B) Localization of Bub3-GFP and Mtw1-CFP foci in nocodazole-arrested cells. A strain coexpressing Bub3-GFP and the kinetochore marker Mtw1p-CFP (YMB4116 containing pOKMTW1-CFP) was grown to logarithmic phase in YEPD at 30°C, arrested in G1 with α-factor, and then treated with nocodazole (Noc). Bub3-GFP (green)- and Mtw1-CFP (red)-stained kinetochores are shown (arrows). Depolymerization of the mitotic spindle was confirmed by immunofluorescence analysis of fixed cells (data not shown). (C) ChIP assay with CEN-specific primers and chromatin derived from a nocodazole-arrested strain expressing Bub3-GFP (YMB4116). The ChIP assay was performed with chromatin prepared from untreated cells (log), cells synchronized with α-factor (α), or cells arrested with nocodazole (Noc). Total chromatin (T), chromatin precipitated with anti-GFP antibodies (GFP), or mock-precipitated chromatin (M) was used in PCR amplifications with centromere-specific primer pairs for CEN3 and CEN6. Actin primers (ACT1) not related to CEN sequences were used as a negative control. Control experiments involving immunoprecipitation of these samples with anti-HA antibody did not show PCR amplification products (data not shown). (D) Localization of Bub3-GFP foci in comparison to the SPB markers Spc29-CFP and Bub2-CFP. A strain coexpressing Bub3-GFP (green) and Spc29-CFP (red) (YMB4245) was grown to logarithmic phase at 30°C (log; upper panel) and observed under a fluorescence microscope. Indicated are Spc29-CFP-labeled SPBs (double white arrows) and prominent Bub3-GFP foci (single gray arrow). In a second experiment, a strain coexpressing Bub3-GFP (green) and Bub2-CFP (red) (YMB4204) was used. This strain was either grown to logarithmic phase in YEPD at 30°C and treated with nocodazole (Noc) (middle panel) or transformed with pAFS120MPS1+ (GAL-MPS1) to induce the overexpression of MPS1 with galactose (lower panel). Localization of SPBs and Bub3-GFP foci after nocodazole treatment or Mps1p overexpression is also shown in overlays (merge) and magnified images (4.5×). Note that Bub3-GFP foci may exist as separate entities away from or in close proximity to SPBs. The images shown were chosen to display well-separated SPBs and Bub3-GFP foci. All strains were examined by fluorescence microscopy. Scale bars, 5.0 μm.
In the second experiment, we examined the localization of Bub3-GFP and Mtw1-CFP in cells coexpressing both proteins after α-factor arrest (G1) and release into nocodazole-containing medium for 90 min. Consistent with our previous observations (Fig. 2B), up to ∼60% of the cells contained Bub3-GFP foci (Fig. 3B). Surprisingly, we found that simple nocodazole treatment of unsynchronized, logarithmically growing cells caused G2/M arrest with single Mtw1-CFP-stained kinetochore clusters (80%), but these cells rarely, if ever, showed visible Bub3-GFP foci (data not shown) (2). We noticed, however, that both single and double dots of Mtw1-CFP-stained kinetochore clusters were present when cells were first synchronized with α-factor and subsequently treated with nocodazole. Under the latter experimental conditions, we observed that a subset of Mtw1-CFP foci also stained for Bub3-GFP (compare arrows marking four Bub3-GFP foci and eight Mtw1-CFP foci in the upper panels of Fig. 3B; see Discussion). These results suggest that Bub3-GFP associates with a subset of kinetochores. However, we cannot exclude the possibility that low levels of Bub3-GFP associate with all kinetochores and are below the detection limit.
The apparent kinetochore localization of Bub3-GFP prompted us to examine whether this spindle checkpoint protein specifically associated with CEN DNA by using the ChIP technique (36). In our experiments, Bub3-GFP-expressing cells were (i) synchronized with α-factor and then released into nocodazole-containing medium, (ii) grown logarithmically, or (iii) synchronized with α-factor. Subsequently, chromatin was cross-linked, and Bub3-GFP- DNA complexes were immunoprecipitated. Precipitated DNA fragments were PCR amplified with CEN-specific and non-CEN-specific primers. Our results showed that Bub3-GFP from cells arrested with nocodazole associated specifically with CEN DNA (CEN3 and CEN6) and not with other loci, such as ACT1 (Fig. 3C). The association was specific to nocodazole-induced arrest, as control samples from cultures grown logarithmically or synchronized with α-factor failed to show enrichment of Bub3-GFP in CEN or non-CEN loci. These results support our localization data showing the enrichment of kinetochore-localized Bub3-GFP foci in spindle checkpoint-activated cells.
We next examined whether Bub3-GFP foci also colocalized with SPBs. These experiments were done with strains coexpressing Bub3-GFP and either Spc29-CFP or Bub2-CFP. Spc29p is an integral component of the SPB, while Bub2p is associated with the cytosolic phase of the SPB (15, 16, 18, 42). We determined that in logarithmically growing cultures, Bub3-GFP foci can be in close proximity to SPBs marked by Spc29-CFP or can exist as separate entities distinct from SPBs (Fig. 3D, upper panels). Next, we examined the localization of Bub3-GFP in a strain coexpressing Bub2-CFP. Our results indicated that some nocodazole-treated cells showed clearly separated Bub3-GFP and Bub2-CFP foci (Fig. 3D, middle panels). To rule out any nonspecific effects due to nocodazole treatment, we used the overexpression of MPS1 to activate the spindle checkpoint and examined the localization of Bub3-GFP and Bub2-CFP in the presence of intact MTs. The overexpression of MPS1 (GAL-MPS1) in a wild-type strain causes cells to arrest with a G2/M content of DNA (23). Strains coexpressing Bub3-GFP and Bub2-CFP with GAL-MPS1 when grown in galactose-containing medium showed an increase in the levels of double-dot or bipartite Bub3-GFP foci (∼85% of all arrested cells), with little or no overlap with Bub2-CFP-stained SPBs (Fig. 3D, lower panels). These results suggest that Bub3-GFP foci may exist as separate entities both away from SPBs and in close proximity to SPBs. These data support the kinetochore association of Bub3-GFP with CEN DNA in ChIP experiments (Fig. 3C).
Mps1p overexpression leads to the ubiquitous kinetochore association of Bub3-GFP.
The enrichment of Bub3-GFP foci in nocodazole-treated cells led us to examine whether activation of the spindle checkpoint in the presence of an intact spindle by the overexpression of MPS1 from a strong inducible GAL1 promoter would yield similar results (23). We determined that, in galactose-grown cultures, the overexpression of Mps1p led to evenly staining, bipartite Bub3-GFP foci in about 85% of G2/M-arrested cells; in comparison, only ∼10% staining foci were seen in sucrose-grown (control) cultures. Consistent with the kinetochore localization of Bub3-GFP, the majority of bipartite Bub3-GFP foci colocalized with Mtw1-CFP-stained kinetochore clusters (Fig. 4A). Most importantly, we observed that almost all of the Mtw1-CFP-stained clusters also stained for Bub3-GFP. Nearly identical results were obtained upon treatment of GAL-MPS1-arrested cells with nocodazole.
FIG. 4.
Activation of the spindle checkpoint by GAL-MPS1 leads to the association of Bub3-GFP with kinetochores and CEN DNA. (A) A strain coexpressing Bub3-GFP and Mtw1-CFP and containing a plasmid expressing GAL-MPS1 (YMB4115) was grown to logarithmic phase in sucrose-containing medium (GAL-MPS1 not induced), followed by the addition of galactose (GAL-MPS1 induced), and incubated for an additional 3 h. Induction of GAL-MPS1 led to activation of the checkpoint and G2/M arrest in ∼85% of the cells examined by fluorescence microscopy. All cells exhibiting Mtw1-CFP (red)-marked kinetochores also were stained for Bub3-GFP (green). The presence of the mitotic spindle was confirmed by immunofluorescence analysis of fixed cells (data not shown). Insets show complete overlap of kinetochore and Bub3-GFP staining. Scale bar, 5.0 μm. (B) Bub3-GFP associates with CEN DNA in a ChIP assay. The ChIP assay was performed with chromatin prepared from a strain (YMB4119) expressing Bub3-GFP and with GAL-MPS1 induced (Gal) or not induced (Suc) as described for panel A. Total chromatin (T), chromatin precipitated with anti-GFP antibodies (GFP), or mock-precipitated chromatin (M) was used in PCR amplifications with centromere-specific primer pairs for CEN3 and CEN16 as well as non-CEN-related control primers for actin (ACT1).
The apparent kinetochore localization of Bub3-GFP after GAL-MPS1 arrest prompted us to examine whether Bub3-GFP specifically associated with CEN DNA in these cells by using the ChIP technique. In this experiment, Bub3-GFP-expressing cells transformed with a GAL-MPS1-expressing plasmid were either arrested by incubation in galactose-containing medium (Fig. 4B, upper panels) or allowed to grow logarithmically in sucrose-containing (control) medium (Fig. 4B, lower panels). Subsequently, chromatin was cross-linked, and Bub3-GFP- DNA complexes were immunoprecipitated as described in the legend to Fig. 3C. Our results showed that Bub3-GFP from cells arrested with GAL-MPS1 (Gal) associates specifically with CEN DNA (CEN3 and CEN16) but not with non-CEN loci, such as ACT1. The association is specific to GAL-MPS1-induced arrest, as control samples from cultures not induced with galactose (Suc) failed to show an association of Bub3-GFP with CEN or non-CEN loci. Again, these data support our localization (Fig. 3A and B) and ChIP (Fig. 3C) data regarding the association of Bub3-GFP with kinetochores in spindle checkpoint-activated cells.
Spindle checkpoint protein Bub1p and kinetochore protein Ndc10p are required for the kinetochore association of Bub3-GFP.
Bub3p has been shown to functionally interact in a complex with another checkpoint protein, Bub1p, a protein kinase that may phosphorylate Bub3p and other proteins required for spindle checkpoint function (17). Hence, we examined whether Bub1p is required for Bub3-GFP focus formation. First, we assessed nuclear Bub3-GFP foci in a bub1Δ strain with (pBUB1) or without (vector) plasmid-borne BUB1. Logarithmically growing Bub3-GFP-expressing cells containing pBUB1 showed the expected frequency of nuclear foci (∼10%). However, in Bub3-GFP-expressing cells without a functional copy of BUB1, Bub3-GFP foci were not visible or were absent (Fig. 5A, upper panels). Subsequently, we assessed the expression of Bub3-GFP foci in cells with or without BUB1 after activation of the spindle checkpoint by treatment with nocodazole. Again, the absence of BUB1 resulted in the absence of Bub3-GFP foci (Fig. 5A, lower panels). Our results are consistent with a similar requirement of Bub1p for the kinetochore association of Bub3-GFP in Drosophila melanogaster and Xenopus laevis (2, 44). Thus, we conclude that Bub3-GFP foci may represent the BUB1-dependent association of Bub3-GFP with one or several kinetochores and that the functional interaction between Bub3p and Bub1p may be conserved from yeasts to higher eukaryotes.
FIG. 5.
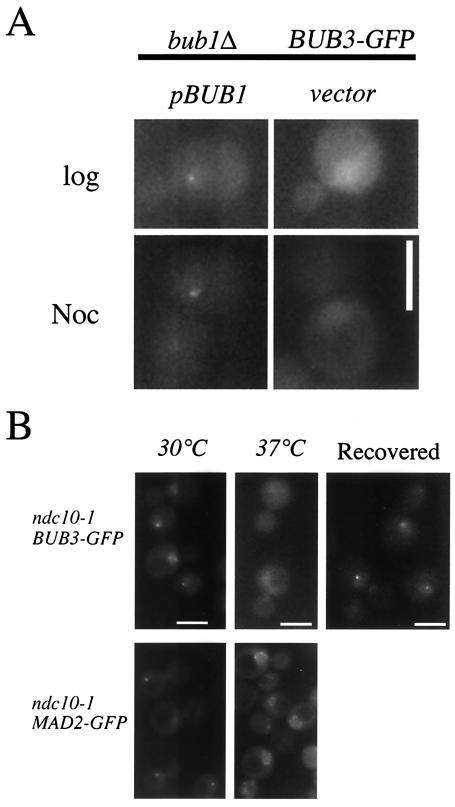
Checkpoint protein Bub1p and kinetochore protein Ndc10p are required for expression of Bub3-GFP foci. (A) A bub1Δ strain (YMB4140) expressing Bub3-GFP and containing a BUB1-expressing plasmid (pBUB1) or a control plasmid (vector) was grown to logarithmic phase in synthetic minimal medium lacking uracil in the absence (log) or presence (Noc) of nocodazole. Representative images of cells in the absence of BUB1 or in the presence of BUB1 showed that BUB1 is required for the expression of Bub3-GFP foci. (B) A temperature-sensitive ndc10-1 strain expressing Bub3-GFP (YMB4155) (upper panels) was grown to logarithmic phase at 30°C (left panel), shifted to 37°C for 3 h (middle panel), and then allowed to recover at 30°C for an additional 90 min (right panel). We observed bright Bub3-GFP foci in cells grown at 30°C and only faint diffuse nuclear staining at 37°C. The latter cells formed Bub3-GFP foci after they were shifted back to 30°C (recovered). As a control, we examined the kinetochore localization of checkpoint protein Mad2-GFP (29) in an ndc10-1 strain (YMB4192) (lower panels) grown to logarithmic phase, arrested with nocodazole for 90 min at 30°C (to elicit the kinetochore localization of this NPC-associated protein) (left panel), and then shifted to 37°C for an additional 3 h (right panel). After the temperature shift, Mad2-GFP was observed at the nuclear periphery in a pattern reminiscent of that of NPC staining. Bub3-GFP foci were observed in a wild-type strain shifted to 37°C for 3 h (data not shown). Scale bars, 5.0 μm.
A temperature-sensitive ndc10-1 strain containing a mutation in the essential kinetochore gene NDC10 disrupts both kinetochore structure and the ability to activate a spindle checkpoint response (14, 20, 21, 49). Hence, we examined the localization of Bub3-GFP in an ndc10-1 strain after incubation at 30°C (Fig. 5B, left column), after a shift to the nonpermissive temperature of 37°C for 3 h (second column), and after a shift back to 30°C for an additional 90 min (third column [Recovered]). We observed bright Bub3-GFP foci in cells grown at 30°C (∼30%) but only faint diffuse nuclear staining after a shift to 37°C. Bub3-GFP foci reappeared after cells were shifted from 37 to 30°C. These data suggest that in the absence of a functional kinetochore, Bub3-GFP fails to associate with or assemble on the CEN DNA-protein complex. The absence of a GFP signal in ndc10-1 cells at 37°C is not due to a thermolabile fusion protein, since Bub3-GFP foci can be observed in wild-type cells under the same experimental conditions (data not shown). As an additional control, we examined the kinetochore localization of the checkpoint protein Mad2-GFP (29) (Fig. 5B, lower panels) in an ndc10-1 strain after treatment with nocodazole (to elicit kinetochore localization of this NPC-associated protein). Shifting of the cells to 37°C results in a failure to recruit Mad2-GFP to the kinetochore and causes the localization of this checkpoint protein at the nuclear periphery in a pattern reminiscent of NPC staining. These data suggest that Ndc10p is required for the association of Bub3p and Mad2p with kinetochores.
Bub3-GFP associates with defective centromeres.
Consistent with a role in spindle checkpoint function, enrichment of Bub3-GFP at sites with defective kinetochore or spindle assembly or attachment may be expected. Therefore, we reasoned that mutations in a centromere DNA sequence that lead to altered kinetochore formation and spindle association may result in an enrichment of Bub3-GFP foci at these sites. Hence, we examined the localization of Bub3-GFP in a reporter strain that contains a nonessential CF with a 31-bp deletion in the central CEN6 element, CDEII [CF/CEN6(Δ31)] (40). The CEN6(Δ31) mutation leads to a high rate of loss of this CF and a delay in G2/M of the cell cycle compared to what is seen for strains containing a CF with wild-type CEN6 (47). When Bub3-GFP- and Mtw1-CFP-marked kinetochores were examined in the reporter strain, we observed an increased number of smaller, sharply delineated Bub3-GFP foci (∼40%) that consistently showed little or no overlap with the main Mtw1-CFP cluster (Fig. 6A). Therefore, it is possible that unattached CF/CEN6(Δ31) is recognized by Bub3-GFP and that the resulting foci “mark” the defective CF for possible retrieval prior to anaphase. Our observations are consistent with the results of studies with mammalian cells showing an enrichment of Bub3p on lagging chromosomes and on those without bound MTs (35).
FIG.6.
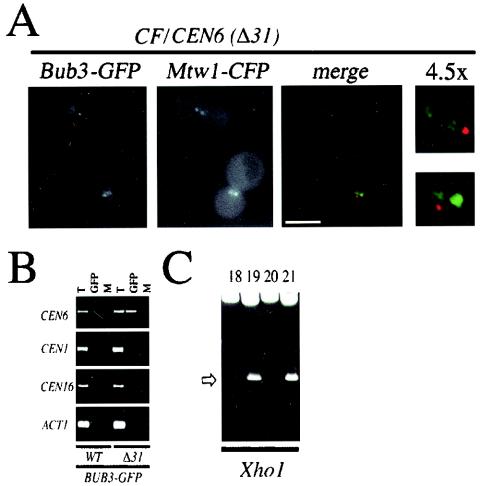
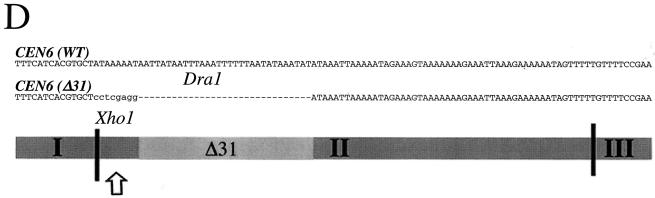
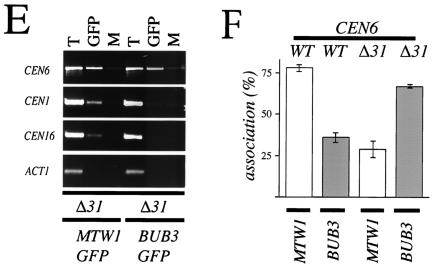
Bub3-GFP associates with a defective centromere. (A) A strain coexpressing Bub3-GFP and the kinetochore marker Mtw1-CFP harboring a CF with a defective centromere [CF/CEN6(Δ31)] (YMB4105 containing pOKMTW1-CFP) was grown to logarithmic phase in synthetic minimal medium lacking uracil at 30°C and examined under a fluorescence microscope. Bub3-GFP (red) and kinetochores stained with Mtw1-CFP (green) are shown. Localization of kinetochores (Mtw1-CFP) and Bub3-GFP foci is also shown as overlays (merge) and magnified images (4.5×). Note that Bub3-GFP foci can exist as separate entities away from the main kinetochore clusters. The signal intensities in the merged panel were adjusted to show the positions of the foci. The presence of the mitotic spindle was confirmed by immunofluorescence analysis of fixed cells (data not shown). Scale bar, 5.0 μm. (B) A strain expressing Bub3-GFP with mutant CF/CEN6(Δ31) (YMB4105) or the wild-type CF (WT) (YMB1302) was grown to logarithmic phase in synthetic minimal medium lacking uracil at 30°C, and chromatin was prepared. Total chromatin (T), chromatin precipitated with anti-GFP antibodies (GFP), or mock-precipitated chromatin (M) was used in PCR amplifications. Shown are PCR amplifications with centromere-specific primer pairs for CEN6, CEN1, and CEN16 and the non-CEN-related primer pair for actin (ACT1). (C) Pooled PCR amplification products from BUB3-GFP/CEN6(Δ31) chromatin anti-GFP ChIP assays (B, CEN6 primers, GFP lanes) were cloned into a Topo TA2.1 vector, and the resulting plasmids were digested with XhoI (shown) and DraI (not shown) restriction enzymes. Digests of representative clones 18, 19, 20, and 21 (of 33 independently analyzed clones) with restriction patterns indicative of CEN6(Δ31) (clones 19 and 21, arrow) and CEN6 (wild type) (clones 18 and 20) are shown. The identity of these clones was also confirmed by sequencing (see panel D). (D). Individual clones were sequenced. Shown is a sequence from CEN6 (wild type [WT]) (clone 19 or 21) or CEN6(Δ31) (clone 18 or 20) encompassing CDEI, CDEII, and CDEIII. The deleted Δ31 region in CDEII of CEN6(Δ31) is indicated by 31 dashes representing the deleted nucleotides. Also indicated are XhoI (arrow; lowercase nucleotides) and DraI restriction sites. (E) A strain expressing Mtw1-GFP with mutant CF/CEN6(Δ31) (YMB4225 containing pOKMTW1-GFP) or Bub3-GFP with mutant CF/CEN6(Δ31) (YMB4105) was grown to logarithmic phase in synthetic minimal medium lacking uracil at 30°C, and chromatin was prepared. Total chromatin (T), chromatin precipitated with anti-GFP antibodies (GFP), or mock-precipitated chromatin (M) was used in PCR amplifications. Shown are PCR amplifications with centromere-specific primer pairs for CEN6, CEN1, and CEN16 and a non-CEN-related primer pair for actin (ACT1). Control experiments involving immunoprecipitation of the samples with anti-HA antibody did not show PCR amplification products (data not shown). (F) Anti-GFP immunoprecipitates from panel E (Mtw1-GFP and Bub3-GFP) were PCR amplified with CEN6 primers and analyzed for the presence of wild-type CEN6 and CEN6(Δ31) amplification products. This process was accomplished by analysis of individual wild-type CEN6 and CEN6(Δ31) PCR products after separation on 5% polyacrylamide gels, SYBR green staining, and quantitation with a phosphorimager. Error bars indicate averages from two independent experiments.
We decided to directly test the observation that Bub3-GFP may associate with altered kinetochores, such as those of defective CF/CEN6(Δ31). Hence, we used the ChIP technique to determine whether Bub3-GFP could associate with CF/CEN6(Δ31) sequences. Chromatin prepared from strains containing either wild-type CF or defective CF [CF/CEN6(Δ31)] was cross-linked, and Bub3-GFP complexes were immunoprecipitated with anti-GFP antibodies as described in the legend to Fig. 3C. We were able to show specific enrichment of a PCR product derived with only CEN6 primers in the CF/CEN6(Δ31)-containing strain and little or none in the wild-type (control) strain (Fig. 6B). To verify its identity, the CEN6-derived PCR product was cloned and analyzed by restriction enzyme digestion with XhoI (Fig. 6C) and DraI (data not shown) as well as by sequencing (Fig. 6D). We determined that Bub3-GFP ChIP assays contained both CF/CEN6(Δ31) and wild-type CEN6 sequences (data not shown).
Our sequencing data derived from the ChIP-PCR products, even though qualitative, showed that in vivo Bub3-GFP may associate with normal and defective kinetochores. Therefore, we examined whether Bub3-GFP associates with defective [CF/CEN6(Δ31)] kinetochores in comparison with another kinetochore protein (Mtw1-GFP). For this experiment, strains containing both endogenous wild-type CEN6 and CF/CEN6(Δ31) and expressing either Bub3-GFP or Mtw1-GFP were analyzed by ChIP with anti-GFP antibodies as described in the legend to Fig. 3C. Our ChIP results confirmed that Mtw1-GFP associates with CEN DNA (22) and shows an association with CEN6, CEN1, and CEN16 but not noncentromeric ACT1 sequences (Fig. 6E). Under identical experimental conditions, Bub3-GFP associates with only CEN6 (Fig. 6E) and shows little or no association with CEN1, CEN16, and ACT1 sequences. An additional restriction enzyme analysis of CEN6-derived PCR products from Mtw1-GFP- and Bub3-GFP-expressing strains showed the presence of both wild-type and CF/CEN6(Δ31) sequences (data not shown). Thus, even though Mtw1-GFP associates with all CEN sequences, including wild-type and defective CF/CEN6(Δ31), Bub3-GFP associates only with CEN6-derived sequences.
We also analyzed the level of association of Bub3-GFP with CEN6 (wild type) or CF/CEN6(Δ31) and compared it to that observed for Mtw1-GFP. This was done by comparing amplification products of ChIP samples from the Mtw1-GFP- and Bub3-GFP-expressing strains that contain CEN6 (wild type) or CEN6(Δ31) (see Materials and Methods). Our analysis revealed that Bub3-GFP preferentially associated with defective CEN6 [CEN6(Δ31), ∼68.0%; wild type, ∼39.0%]. Mtw1-GFP, on the other hand, preferentially associated with wild-type CEN6 [CEN6(Δ31), ∼24.0%; wild type, ∼80.0%] (Fig. 6F). Both chromatin samples (total starting material) contained equivalent levels of CEN6(Δ31) and CEN6 (wild type) (data not shown). Using the ChIP technique, we have determined for the first time that a checkpoint protein can preferentially associate with defective CEN sequences in vivo. We suggest that the recruitment of Bub3p to a single defective CEN sequence may play a role in the cell cycle delay observed in a CF/CEN6(Δ31)-containing strain (47).
DISCUSSION
In this report, we present the first comprehensive analysis of the functional dynamics of S. cerevisiae Bub3p, an evolutionarily conserved spindle checkpoint protein. First, in live-cell studies, we find that Bub3p is a nucleus-localized protein which preferentially associates with and forms foci in a subset of kinetochores. Bub3-GFP may be enriched on certain kinetochores due to the lack or modification of certain kinetochore proteins or detachment from the MT spindle and/or the main kinetochore clusters. Second, we show that overexpression of the checkpoint protein kinase Mps1p leads to the ubiquitous association of Bub3-GFP with kinetochores. Third, we provide the first ChIP-based analysis for the association of a checkpoint protein with both wild-type and mutant CEN DNAs in S. cerevisiae. Fourth, using a unique genetically engineered CF, we show that Bub3-GFP preferentially associates with a single defective centromere, thus providing a powerful tool for studying checkpoint protein assembly and function. Our data are consistent with a model in which alterations or defects in kinetochore or spindle integrity may signal the enrichment of Bub3p at these sites (Fig. 7).
FIG. 7.
Model for in vivo association of Bub3p with altered kinetochores in S. cerevisiae. Bub3-GFP is a spindle checkpoint protein that localizes diffusely in the nucleus and/or localizes in distinct intranuclear foci. The model depicts yeast nuclei (large ovals) with different levels of nucleus-localized Bub3p (dark green, as in panel 1, or light green, as in panels 2, 3, and 4). Inside the nuclei, kinetochores of chromatids are tethered through MTs to the SPB in the nuclear envelope (the black line delineating the nucleus). Kinetochores with Bub3p foci (small green spheres) or without Bub3p foci (small blue spheres) on individual chromatids are depicted. The localization of Bub3-GFP suggests a preferential association with defective kinetochore or spindle structures under conditions that lead to activation of the spindle checkpoint and a delay in G2/M. These events can be induced by treatment of wild-type cells with nocodazole or an MT-depolymerizing agent, overexpression of the checkpoint kinase Mps1p, or the presence of a CF with defective CEN. The diagrams show the observed localization of Bub3-GFP in wild-type strains not treated (1), treated with nocodazole (2), overexpressing GAL-MPS1 (3), or expressing mutated CEN [CF/CEN6/(Δ31)] (4). (1) Metaphase nuclei in untreated wild-type cells exhibit only diffuse nuclear staining of Bub3-GFP without recruitment to kinetochore clusters. The latter is consistent with “double dots” of kinetochores in mitotic cells (2). (2) Nuclei of nocodazole-treated cells with depolymerized MTs show aggregation of bipartite kinetochore clusters due to the absence of spindle tension. Nuclei with one kinetochore cluster rarely, if ever, show Bub3-GFP foci. However, some kinetochores that are not part of the main kinetochore cluster are subject to enrichment of Bub3-GFP foci. (3) Nuclei of cells overexpressing GAL-MPS1 with intact MTs contain bipartite kinetochore clusters due to MT (spindle) tension on kinetochores of cohesive chromatids. The kinetochores of these checkpoint-activated cells contain Bub3-GFP. (4) Nuclei of cells containing a CF with mutated CEN [CF/CEN6/(Δ31)] exhibit a delay in G2/M, with large budded cells and bipartite kinetochore clusters due to MT (spindle) tension on kinetochores of cohesive chromatids. The small, distinctly staining Bub3-GFP foci which do not overlap the main kinetochore cluster may represent mislocalized CF/CEN6/(Δ31). In scenarios 2, 3, and 4, the enrichment of Bub3-GFP in kinetochores may lead to a decrease in the diffuse nuclear localization signal (light green). Our model and the supporting data suggest that arrest or delay in G2/M is mediated by the recruitment of checkpoint protein complexes containing Bub3p to sites of perturbed kinetochore or spindle interactions. The enrichment of a checkpoint protein(s) at kinetochores may provide the signal to halt anaphase.
Fluorescence microscopy of live cells expressing GFP- and CFP-tagged proteins revealed diffusely staining nucleoplasmic Bub3p and/or distinct Bub3 foci that overlapped a subset of Mtw1p-labeled kinetochores. These two Bub3-GFP pools may represent a “snapshot” of Bub3p distribution as it cycles on and off the kinetochores. Bub3-GFP foci in unperturbed cells may mark newly replicated centromeres that have not yet assembled a functional kinetochore. This interaction requires at least some kinetochore structure, as judged from the absence of Bub3-GFP foci in an ndc10-1 kinetochore mutant at the nonpermissive temperature. Interestingly, the absence of checkpoint protein kinase Bub1p may also affect the distribution of Bub3p and is in agreement with previous results (2). These results show that the subcellular distribution of Bub3p and additional checkpoint proteins may provide important clues about their functional interactions.
During nocodazole-induced G2/M arrest, only a subset of kinetochores stained for Mtw1-CFP foci also stained for Bub3-GFP foci. Therefore, it is possible that one or several of the kinetochores that fail to associate with other clustered kinetochores upon release of MT tension cause the accumulation of Bub3-GFP at sufficient (focus-forming) levels and become visually detectable. In accordance with the report by Goshima and Yanagida (22), we observed that the majority of double dots of Mtw1-CFP (∼80%) in large budded cells collapse to form a single kinetochore cluster after nocodazole treatment. We find that nocodazole-arrested cells with a single kinetochore cluster rarely, if ever, show visible Bub3-GFP foci. We noticed, however, that when cells were synchronized with α-factor and subsequently treated with nocodazole, we observed both single and double dots of Mtw1-CFP-stained kinetochore clusters. This result was probably not due to incomplete nocodazole arrest, as judged by flow cytometry and visual analysis of the treated cells (Fig. 2C). We speculate that the observed Mtw1p unclustering phenotype may be due to the assembly of new kinetochores (on replicated centromeres) in the absence of MT spindles. While we cannot completely explain the presence of two Mtw1-GFP foci after α-factor treatment and nocodazole arrest, we took advantage of this observation to further analyze the association of Bub3p with kinetochores. Using the above-described procedure for a strain coexpressing Bub3-GFP and Mtw1-CFP, we were able to show that Bub3-GFP foci colocalize with a subset of kinetochores labeled with Mtw1-CFP (Fig. 3B). Furthermore, under these conditions, Bub3-GFP associates with all centromeres tested in ChIP experiments (Fig. 3C). The weak association of Bub3-GFP with CEN6 in logarithmically grown cells (Fig. 3C) most likely represents a background amplification product, as we failed to observe an enrichment of Bub3-GFP in ChIP experiments with CEN3 (Fig. 3C) or CEN1 and CEN16 (Fig. 6B).
Unlike Bub3-GFP foci in unperturbed and nocodazole-arrested cells, the overexpression of MPS1 results in the ubiquitous association of Bub3-GFP with all Mtw1-CFP-labeled kinetochores. The overexpression of Mps1p leads to Bub1p-, Bub3p-, and Mad2p-dependent activation of the spindle checkpoint, leading to arrest in G2/M phase of the cell cycle (23). Our inability to detect a biochemical association of Bub3-GFP with CEN DNA in the absence of Mps1p overexpression or in untreated cells may indicate a weak or transient association of Bub3-GFP with these centromeres. Alternatively, the small number of cells that show kinetochores may be below the limit of detection in our ChIP analysis. Therefore, the enrichment of Bub3-GFP foci may be physiologically relevant, serving to mark kinetochores in response to Mps1p activity. Even though the precise mechanism of checkpoint activation by Mps1p is not clearly understood, based on our data, we propose that the overexpression of MPS1 may increase the affinity of Bub3-GFP (or a protein complex containing Bub3-GFP) for the kinetochore or modify a kinetochore protein to increase its affinity for Bub3-GFP binding.
Under physiological conditions, cells may rarely encounter a situation wherein all of the chromosomes have a defect in kinetochore or spindle integrity. It seems more likely that during mitosis, the interaction of a single kinetochore or spindle is compromised, thereby triggering spindle checkpoint activation. How can we probe the interaction of a checkpoint protein like Bub3p with a single defective kinetochore? In a novel approach with genetically engineered defective CEN [CF/CEN6(Δ31)], we determined that Bub3-GFP associates with a single defective centromere. Using strains containing both CEN6 and CF/CEN6(Δ31), we compared Bub3-GFP ChIP assay results to those obtained with the GFP-tagged kinetochore protein Mtw1p. Unlike the ubiquitous CEN association of Mtw1-GFP, Bub3-GFP only showed amplification products with CEN6-specific primers. We cannot exclude a low level of association of Bub3-GFP with other centromeres. Sequence analysis of the PCR products revealed the presence of both CF/CEN6(Δ31) and wild-type CEN6 sequences with CEN6 primers. It is possible that the apparent association of Bub3-GFP with both forms of CEN6 is due to the pairing of analogous chromosomes that has been observed in premeitoic G1 and mitotic dividing budding yeast cells (6). Alternatively, these results could represent an in vitro artifact of the ChIP technique. We suggest that our ChIP assays with strains containing CF/CEN6(Δ31) provide a novel way to examine the unique checkpoint-specific association of Bub3-GFP (and potentially other checkpoint proteins) with altered kinetochores due to a single defective centromere. We have taken the first step to demonstrate that the protein compositions of defective and functional kinetochores may be different. For example, we have shown that Bub3p can preferentially associate with defective CF/CEN6(Δ31), unlike kinetochore protein Mtw1p.
In summary, our data are consistent with models for the molecular roles of checkpoint proteins previously based solely on genetic and in vitro analyses of biochemical complexes in S. cerevisiae. Future studies aimed at identifying kinetochore and checkpoint proteins required for the kinetochore association of Bub3p will further the understanding on the temporal and spatial requirements for the assembly of spindle checkpoint complexes in yeast and other systems. These studies are particularly important, as mutations in checkpoint genes lead to chromosome instability in yeasts (52). Furthermore, for humans it has been shown that some cancers displaying a chromosomal instability (CIN) phenotype show a loss of function of Bub1p which is found in a complex with Bub3p (7). Hence, understanding the molecular role of checkpoint proteins in the context of their biological functions will contribute greatly to the study of aneuploidy, cancers, and developmental catastrophes.
Acknowledgments
We thank past and present members of the laboratory of M. A. Basrai for many fruitful discussions. Special thanks are also due to C. Babic, C. Carter, C. Dunbar, J. Kastenmayer, M. Lee, M. Nau, T. Rice, R. Shroff, F. Spencer, and B. Todd for expert help and intellectual contributions. We also acknowledge D. Burke, T. Davis, S. Jaspersen, K. Hardwick, P. Hieter, M. Lichten, F. Spencer, and M. Winey for providing reagents and/or advice.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Basu, J., E. Logarinho, S. Herrmann, H. Bousbaa, Z. Li, G. K. Chan, T. J. Yen, C. E. Sunkel, and M. L. Goldberg. 1998. Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma 107:376-385. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, P., K. Hardwick, and J. P. Javerzat. 1998. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143:1775-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady, D. M., and K. G. Hardwick. 2000. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 10:675-678. [DOI] [PubMed] [Google Scholar]
- 5.Breeden, L. L. 1997. Alpha-factor synchronization of budding yeast. Methods Enzymol. 283:332-341. [DOI] [PubMed] [Google Scholar]
- 6.Burgess, S., and N. Kleckner. 1999. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes Dev. 13:1871-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahill, D. P., et al. 1998. Mutations of mitotic checkpoint genes in human cancers. Nature 392:300-303. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, M. S., G. K. Chan, and T. J. Yen. 2001. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114:953-963. [DOI] [PubMed] [Google Scholar]
- 9.Castillo, A. R., J. B. Meehl, G. Morgan, A. Schutz-Geschwender, and M. Winey. 2002. The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p. J. Cell Biol. 156:453-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheeseman, I. M., D. G. Drubin, and G. Barnes. 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, R. H., A. Shevchenko, M. Mann, and A. W. Murray. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, R. H., J. C. Waters, E. D. Salmon, and A. W. Murray. 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 274:242-246. [DOI] [PubMed] [Google Scholar]
- 13.Chen, R. H., D. M. Brady, D. Smith, A. W. Murray, and K. G. Hardwick. 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell 10:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchenko, M. Mann, and K. Nasmyth. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93:1067-1076. [DOI] [PubMed] [Google Scholar]
- 15.Daum, J. R., N. Gomez-Ospina, M. Winey, and D. J. Burke. 2000. The spindle checkpoint of Saccharomyces cerevisiae responds to separable microtubule-dependent events. Curr. Biol. 10:1375-1378. [DOI] [PubMed] [Google Scholar]
- 16.Elliott, S., M. Knop, G. Schlenstedt, and E. Schiebel. 1999. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc. Natl. Acad. Sci. USA 96:6205-6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farr, K. A., and M. A. Hoyt. 1998. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol. Cell. Biol. 18:2738-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraschini, R., E. Formenti, G. Lucchini, and S. Piatti. 1999. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 145:979-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraschini, R., A. Beretta, L. Sironim, A. Musacchio, G. Lucchini, and S. Piatti. 2001. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 20:6648-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraschini, R., A. Beretta, G. Lucchini, and S. Piatti. 2001. Role of the kinetochore protein Ndc10 in mitotic checkpoint activation in Saccharomyces cerevisiae. Mol. Genet. Genomics 266:115-125. [DOI] [PubMed] [Google Scholar]
- 21.Goh, P. Y., and J. V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100:619-633. [DOI] [PubMed] [Google Scholar]
- 23.Hardwick, K. G., E. Weiss, F. C. Luca, M. Winey, and A. W. Murray. 1996. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 273:953-956. [DOI] [PubMed] [Google Scholar]
- 24.Hardwick, K. G., R. C. Johnston, D. L. Smith, and A. W. Murray. 2000. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, X., D. R. Rines, C. W. Espelin, and P. K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106:195-206. [DOI] [PubMed] [Google Scholar]
- 26.Hoyt, M. A., L. Totis, and B. T. Roberts. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66:507-517. [DOI] [PubMed] [Google Scholar]
- 27.Hyland, K. M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 145:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikui, A. E., K. Furuya, M. Yanagida, and T. Matsumoto. 2002. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 115:1603-1610. [DOI] [PubMed] [Google Scholar]
- 29.Iouk, T., O. Kerscher, R. J. Scott, M. A. Basrai, and R. W. Wozniak. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint J. Cell Biol. 159:807-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irniger, S. 2002. Cyclin destruction in mitosis: a crucial task of Cdc20. FEBS Lett. 532:7-11. [DOI] [PubMed] [Google Scholar]
- 31.Kitagawa, K., R. Abdulee, P. K. Bansal, G. Cagney, S. Fields, and P. Hieter. 2003. Requirement of Skp1-Bub1 interaction for kinetochore-mediated activation of the spindle checkpoint. Mol. Cell 11:1201-1213. [DOI] [PubMed] [Google Scholar]
- 32.Lauze, E., B. Stoelcker, F. C. Luca, E. Weiss, A. R. Schutz, and M. Winey. 1995. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 14:1655-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, R., and A. W. Murray. 1991. Feedback control of mitosis in budding yeast. Cell 66:519-531. [DOI] [PubMed] [Google Scholar]
- 34.Li, Y., and R. Benezra. 1996. Identification of a human mitotic checkpoint gene: hsMAD2. Science 274:246-248. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Exposito, M. J., K. B. Kaplan, J. Copeland, and P. K. Sorger. 1999. Retention of the BUB3 checkpoint protein on lagging chromosomes. Proc. Natl. Acad. Sci. USA 96:8493-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meluh, P. B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millband, D. N., and K. G. Hardwick. 2002. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 22:2728-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millband, D. N., L. Campbell, and K. G. Hardwick. 2002. The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol. 12:205-209. [DOI] [PubMed] [Google Scholar]
- 39.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell. Biol. 10:731-741. [DOI] [PubMed] [Google Scholar]
- 40.Panzeri, L., L. Landonio, A. Stotz, and P. Philippsen. 1985. Role of conserved sequence elements in yeast centromere DNA. EMBO J. 4:1867-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson, C. G., P. S. Maddox, E. D. Salmon, and K. Bloom. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira, G., T. Hofken, J. Grindlay, C. Manson, and E. Schiebel. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell 6:1-10. [PubMed] [Google Scholar]
- 43.Roberts, B. T., K. A. Farr, and M. A. Hoyt. 1994. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol. 14:8282-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharp-Baker, H., and R. H. Chen. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153:1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Spencer, F., S. L. Gerring, C. Connelly, and P. Hieter. 1990. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124:237-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer, F., and P. Hieter. 1992. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89:8908-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sudakin, V., G. K. Chan, and T. J. Yen. 2001. Checkpoint inhibition of the APC/C HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tavormina, P. A., and D. J. Burke. 1998. Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiae is independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics 148:1701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, S. S., E. Ha, and F. McKeon. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toyoda, Y., K. Furuya, G. Goshima, K. Nagao, K. Takahashi, and M. Yanagida. 2002. Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 12:347-358. [DOI] [PubMed] [Google Scholar]
- 52.Warren, C. D., D. M. Brady, R. C. Johnston, J. S. Hanna, K. G. Hardwick, and F. Spencer. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13:3029-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss, E., and M. Winey. 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132:111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wigge, P. A., O. N. Jensen, S. Holmes, S. Soues, M. Mann, and J. V. Kilmartin. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winey, M., and E. T. O'Toole. 2001. The spindle cycle in budding yeast. Nat. Cell. Biol. 1:E23-E27. [DOI] [PubMed]