Abstract
In Saccharomyces cerevisiae, a phosphorelay signal transduction pathway composed of Sln1p, Ypd1p, and Ssk1p, which are homologous to bacterial two-component signal transducers, is involved in the osmosensing mechanism. In response to high osmolarity, the phosphorelay system is inactivated and Ssk1p remains unphosphorylated. Unphosphorylated Ssk1p binds to and activates the Ssk2p mitogen-activated protein (MAP) kinase kinase kinase, which in turn activates the downstream components of the high-osmolarity glycerol response (HOG) MAP kinase cascade. Here, we report a novel inactivation mechanism for Ssk1p involving degradation by the ubiquitin-proteasome system. Degradation is regulated by the phosphotransfer from Ypd1p to Ssk1p, insofar as unphosphorylated Ssk1p is degraded more rapidly than phosphorylated Ssk1p. Ubc7p/Qri8p, an endoplasmic reticulum-associated ubiquitin-conjugating enzyme, is involved in the phosphorelay-regulated degradation of Ssk1p. In ubc7Δ cells in which the degradation is hampered, the dephosphorylation and/or inactivation process of the Hog1p MAP kinase is delayed compared with wild-type cells after the hyperosmotic treatment. Our results indicate that unphosphorylated Ssk1p is selectively degraded by the Ubc7p-dependent ubiquitin-proteasome system and that this mechanism downregulates the HOG pathway after the completion of the osmotic adaptation.
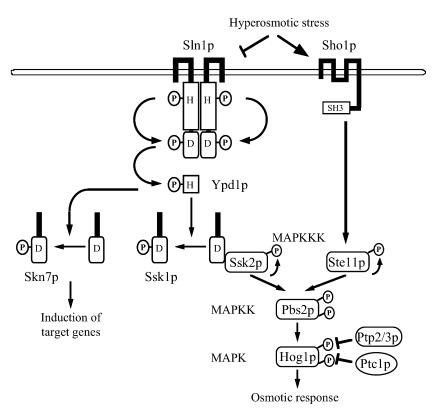
In eukaryotic cells, the mitogen-activated protein kinase (MAPK) cascades are activated by a variety of external stimuli such as growth factors, cytokines, UV irradiation, and osmotic stress. In the yeast Saccharomyces cerevisiae, the high-osmolarity glycerol (HOG) response MAPK pathway is activated by increased external osmolarity (Fig. 1) (6, 18, 20, 49, 54). An upstream His-Asp phosphorelay signaling system, which is homologous to bacterial two-component systems, regulates the downstream HOG MAPK cascade (38, 45, 50). Under normal osmolarity conditions, Sln1p, a membrane-associated histidine kinase, phosphorylates a histidine residue in itself. The phosphate group is in turn transferred to an aspartic acid residue in its receiver domain. Subsequently, the phosphate group is transferred to the Ypd1p phosphotransmitter and then to the receiver domain of the Ssk1p response regulator. High osmolarity inactivates Sln1p and thus lowers the phosphorylation level of Ssk1p (26, 27, 38, 44, 50). Unphosphorylated Ssk1p binds to the Ssk2p, and perhaps the Ssk22p, MAPK kinase kinases (MAPKKKs) through its receiver domain and activates them (48). Activated Ssk2p phosphorylates and activates the Pbs2p MAPK kinase, which in turn phosphorylates and activates the Hog1p MAPK (36, 48, 49). Phosphorylated Hog1p activates the transcription of genes required in the response to high osmolarity (1, 12, 53). Deletion of SLN1 or YPD1 induces constitutive activation of the HOG pathway as the consequence of failure in phosphorylating Ssk1p (38, 50). In addition to Ssk1p, S. cerevisiae has another response regulator, Skn7p, that is also phosphorylated via the Sln1p-Ypd1p phosphorelay (8, 29, 32). Skn7p has a DNA-binding domain homologous to heat shock transcription factors, as well as a receiver domain, and in fact acts as a transcription factor involved in multiple physiological processes, such as maintenance of cell wall integrity, cell cycle regulation, and oxidative stress response (4, 7, 8, 31, 39, 40). Under hypo-osmotic conditions, the receiver domain of Skn7p remains as phosphorylated as that of Ssk1p, and induction of the target genes, including OCH1, is dependent on the phosphorylation (7, 32, 33, 63). In contrast, induction of the target genes for oxidative stress response is independent of the phosphorylation (31). Skn7p is not implicated in the regulation of the HOG pathway except for some marginal genetic interaction (29).
FIG. 1.
The osmosensing signal transduction pathway in yeast. Hyperosmotic stress inhibits the phosphorelay system initiated from the Sln1p membrane-associated histidine kinase, which functions as a homodimer, to cause reduced levels of phosphorylation of Ypd1p and the response regulator Ssk1p. Unphosphorylated Ssk1p activates the Ssk2p MAPKKK, which in turn activates the downstream HOG MAPK cascade. Skn7p is another response regulator phosphorylated by the phosphorelay system, which is a transcriptional factor. Another sensor system that is dependent on the membrane anchor Sho1p also functions, in which Pbs2p is activated by Ste11p (36, 47, 51, 52). Msb2p is also implicated in osmosensing (not shown) (43). MAPKK, MAPK kinase.
In the absence of osmotic stress, or once the adaptation process to high osmolarity is completed, the HOG pathway is negatively regulated by protein phosphatases. Upon activation of Hog1p, a tyrosine residue and a threonine residue in the activation loop are phosphorylated by Pbs2p (6, 25, 37, 38, 64, 66). Hog1p is inactivated by dephosphorylation of the tyrosine residue by the tyrosine phosphatases Ptp2p and Ptp3p (25, 37, 66) or of the threonine residue by the type 2C serine-threonine protein phosphatase Ptc1p (37, 64). The dephosphorylation of constituent kinases by protein phosphatases is a common mechanism for the negative regulation of MAPK cascades. On the other hand, recent reports have shown that the ubiquitin-proteasome system also negatively regulates MAPK cascades. The human extracellular signal-regulated protein kinase (ERK) pathway is downregulated by the degradation of the ERK MAPKs (35), and the yeast pheromone-induced MAPK cascade is downregulated by the degradation of the Ste11p MAPKKK (14).
Here, we report that the yeast osmosensing pathway is also downregulated by the ubiquitin-proteasome system. In this case, the Ssk1p response regulator is the target for degradation. Unphosphorylated Ssk1p is selectively degraded, and Ubc7p/Qri8p, a ubiquitin-conjugating enzyme (E2) implicated in endoplasmic reticulum (ER)-associated degradation (ERAD) (3, 10, 15, 21), is involved in the degradation. This mechanism downregulates the HOG pathway after the completion of the osmotic adaptation. We propose that phosphorelay-regulated degradation of response regulators can be a general regulatory mechanism of eukaryotic His-Asp phosphorelay signaling systems.
MATERIALS AND METHODS
Bacterial and yeast methods.
Standard Escherichia coli and yeast manipulations were performed as described previously (2). E. coli strain XL10-Gold (Stratagene, San Diego, Calif.) was used for plasmid propagation. SGal and SRaf media are modified synthetic complete media with dextrose (SD media) in which glucose is replaced by galactose and raffinose, respectively.
Yeast strains.
The yeast strains used in this study are listed in Table 1. UBC7 was disrupted by transformation with pRH1186 (17), SKN7 was disrupted by transformation with pNS396 (see below), and RPN9 was disrupted by transformation with pJUN180 (61).
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| TM141 | MATaleu2-Δ1 his3-Δ200 trp1-Δ63 ura3-52 | 38 |
| NS318 | Same as TM141 except ssk1::HIS3 hog1::kanMX6 | This study |
| NS320 | Same as TM141 except ypd1::LEU2 ssk1::HIS3 hog1::kanMX6 | This study |
| NS325 | Same as TM141 except skn7::TRP1 ssk1::HIS3 hog1::kanMX6 | This study |
| NS327 | Same as TM141 except ubc7::LEU2 | This study |
| NS329 | Same as TM141 except ubc7::LEU2 ssk1::HIS3 hog1::kanMX6 | This study |
| NS333 | Same as TM141 except ubc7::LEU2 ypd1::URA3 ssk1::HIS3 hog1::kanMX6 | This study |
| NS336 | Same as TM141 except rpn9::LEU2 ypd1::URA3 ssk1::HIS3 hog1::kanMX6 | This study |
| NS337 | Same as TM141 except rpn9::LEU2 ssk1::HIS3 hog1::kanMX6 | This study |
| NS354 | Same as TM141 except ssk1::HIS3 ssk2::TRP1 hog1::kanMX6 | This study |
| NS355 | Same as TM141 except ypd1::LEU2 ssk1::HIS3 ssk2::TRP1 hog1::kanMX6 | This study |
| TM107 | MATaleu2-Δ1 trp1-Δ63 ura3-52 ptp2::LEU2 | 37 |
| NS358 | Same as TM107 except ubc7::TRP1 | This study |
| NS362 | Same as TM107 except ubc7::TRP1 hog1::kanMX6 | This study |
| W303-1A | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade2-101 can1-100 | 61 |
| J43 | Same as W303-1A except rpn9::LEU2 | 61 |
| YK109 | Same as W303-1A except rpn12-1 | 30 |
Plasmids.
The plasmids used in this study are listed in Table 2.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| p416GAL1 | PGAL1URA3 CEN | 41 |
| p424GAL1 | PGAL1TRP1 2μm | 41 |
| p426TEF | PTEFURA3 2μm | 42 |
| p426GPD | PGPDURA3 2μm | 42 |
| pRS414 | TRP1 CEN | 58 |
| pRS416 | URA3 CEN | 58 |
| pRS424 | TRP1 2μm | 11 |
| pFA6a-3HA-6His-His3MX6 | 3HA-6His-TADHHis3MX6 | T. Maeda, unpublished |
| pFA6a-TRP1 | TRP1 | 34 |
| pRH1186 | pBluescript KSII ubc7::LEU2 | 17 |
| pJUN180 | pBluescript KS(−) rpn9::LEU2 | 61 |
| pRS315-SKN7 | SKN7 LEU2 CEN | 8 |
| pNS396 | pBluescript KS(+) skn7::TRP1 | This study |
| pNS252 | p424GAL1 SSK1-3HA-6His | This study |
| pNS114 | pRS414 PGAL1-SSK1-3HA-6His | This study |
| pNS476 | pRS416 YPD1 | This study |
| pFP57 | pRS416 ypd1 (H64Q) | 50 |
| pcDNA3.1/N-3×FLAG | 3×FLAG | T. Maeda, unpublished |
| pTB422 | pcDNA3.1 3×FLAG-SSK1 | This study |
| pTB426 | p416GAL1 3×FLAG-SSK1 | This study |
| pNS462 | pRS416 PGAL1-SSK1-3HA-6His | This study |
| pcDNA3-FLAG | FLAG | Gift from M. Kawabata |
| pNS454 | p426TEF SSK1-FLAG | This study |
| pNS456 | p426GPD SSK1ΔUBL-FLAG | This study |
| pNS473 | pRS416 PGAL1-SSK1 | This study |
| pHG11 | pRS424 HOG1 | 38 |
PGAL1, PTEF, and PGPD are, respectively, GAL1, TEF, and GPD promoters. TADH is an alcohol dehydrogenase terminator.
To induce the C-terminally epitope-tagged SSK1 from the galactose-inducible GAL1 promoter, pNS252, pNS114, or pNS462 was used. A three-hemagglutinin (HA)-six-His composite epitope tag was fused to the C terminus of Ssk1p as follows. A SalI site was introduced just before the termination codon of SSK1 by PCR. The 2.2-kb ClaI-SalI fragment covering the SSK1 open reading frame (ORF) and the 0.2-kb SalI-AscI (blunted) fragment containing a three-HA-six-His composite epitope tag derived from pFA6a-3HA-6His-His3MX6 (T. Maeda, unpublished data), which has the three-HA-six-His tag in the common pFA6a-His3MX6 backbone (34), were cloned into the ClaI and HincII sites of p424GAL1 (41) to produce pNS252. The 0.7-kb SalI-SmaI fragment containing the GAL1 promoter from pGSS1 (38), the 2.2-kb ClaI (blunted)-SalI fragment described above, and the 0.4-kb SalI-SacI fragment containing the three-HA-six-His tag and the alcohol dehydrogenase terminator derived from pFA6a-3HA-6His-His3MX6 were cloned into the SalI and SacI sites of pRS414 (58) to produce pNS114. pNS462 is essentially the same as pNS114 except that the parental vector is pRS416 (58). pNS476 is essentially the same as pFP57 (50) except that the insert is wild-type YPD1.
To induce N-terminally three-FLAG-tagged SSK1 from the GAL1 promoter, pTB426 was constructed as follows. An EcoRI site was introduced just in front of the initiation codon of SSK1 by PCR. The 2.6-kb EcoRI-BstXI segment covering the entire ORF and the 3′ flanking region of SSK1, together with a vector-derived linker sequence adjacent to the BstXI site, was isolated as an EcoRI fragment. The EcoRI fragment was cloned into the EcoRI site of pcDNA3.1/N-3×FLAG (T. Maeda, unpublished), which has a three-FLAG tag in the common pcDNA3.1(+) backbone (Invitrogen Corp., Carlsbad, Calif.), to produce pTB422. The 2.7-kb HindIII-XhoI fragment, containing the N-terminally three-FLAG-tagged ORF and the 3′ flanking region of SSK1, was cloned into the HindIII and XhoI sites of p416GAL1 (41) to produce pTB426.
To express C-terminally FLAG-tagged SSK1, pNS454 was constructed as follows. The 2.2-kb ClaI-SmaI fragment containing the SSK1 ORF from pNS252 and the 0.1-kb BamHI (blunted)-ApaI (blunted) fragment containing a single FLAG epitope from pcDNA3-FLAG were cloned into the ClaI and HincII sites of p426TEF (42) to produce pNS454. The ubiquitin-like (UBL) domain of Ssk1p was deleted as follows. An EcoRV restriction site was introduced at nucleotides 709 to 714 of the SSK1 ORF (mutated from GAA TCA to GAT ATC) by PCR. The 0.7-kb ClaI-EcoRV fragment containing the 5′ portion of SSK1, the 1.3-kb EcoRV-SmaI fragment containing the 3′ portion of SSK1 from pNS358, and the 0.1-kb BamHI (blunted)-ApaI (blunted) fragment described above were cloned into the ClaI and HincII sites of p426GPD (42) to produce pNS456 containing SSK1ΔUBL (amino acids 243 to 290 were deleted).
pNS396, the plasmid used for the deletion of SKN7, was constructed as follows. The 3.5-kb SmaI-XbaI fragment containing the SKN7 locus from pRS315-SKN7 (8) was cloned into the HincII and XbaI sites of pBluescript KS(+). The 1.5-kb PstI-HincII fragment within the SKN7 ORF was then replaced with the 1.0-kb PstI-PmeI fragment containing the TRP1 cassette from pFA6a-TRP1 (34).
Detection of epitope-tagged Ssk1p.
Cells expressing tagged SSK1 were cultured in SD medium and then transferred to SRaf medium. Cells were grown to an optical density at 600 nm of ∼0.8, and galactose was added to the medium to a final concentration of 2%. After 3 h, cells were transferred to SD medium containing cycloheximide (0.5 mg/ml). Aliquots were collected at the indicated times and centrifuged. Cell pellets were suspended in the Laemmli sample buffer and boiled for 5 min. Proteins were resolved by sodium dodecyl sulfate-7 to 10% polyacrylamide gel electrophoresis. Proteins were transferred to a polyvinylidene difluoride membrane, and the blot was incubated with the mouse anti-HA monoclonal antibody 12CA5, peroxidase-conjugated sheep anti-mouse immunoglobulin (Amersham Biosciences, Piscataway, N.J.), and then enhanced chemiluminescence detection reagent (Amersham Biosciences). Actin was detected with the mouse anti-actin monoclonal antibody C4 (ICN Biomedicals, Costa Mesa, Calif.). In some experiments (see Fig. 3B), the primary antibody was replaced with the mouse anti-FLAG monoclonal antibody M2 (Sigma, St. Louis, Mo.).
FIG. 3.
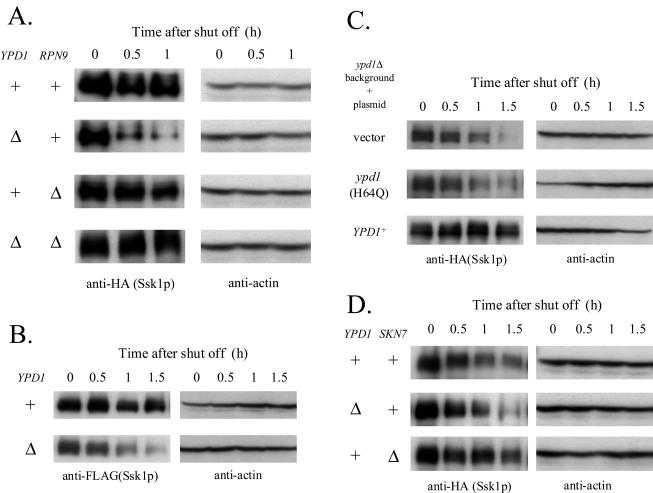
The stability of Ssk1p is dependent on both the phosphotransfer and the proteasome system but not on the SKN7 pathway. (A) The stability of Ssk1p is dependent on the phosphotransfer from Ypd1p and the proteasome system. C-terminally tagged Ssk1p was expressed from the low-copy-number plasmid pNS114 in the YPD1+ ssk1Δ hog1Δ RPN9+ (NS318), ypd1Δ ssk1Δ hog1Δ RPN9+ (NS320), YPD1+ ssk1Δ hog1Δ rpn9Δ (NS337), and ypd1Δ ssk1Δ hog1Δ rpn9Δ (NS336) strains, and the Ssk1p content was monitored by Western blotting. (B) N-terminally three- FLAG-tagged Ssk1p is also degraded in a phosphorelay-regulated manner. N-terminally three-FLAG-tagged Ssk1p was expressed from the low-copy-number plasmid pTB426 in NS318 and NS320 strains, and the Ssk1p content was monitored. (C) Ypd1p-Ssk1p interaction does not affect the stability of Ssk1p. NS320 strains with pNS114 were transformed with pRS416 (vector), pFP57 (ypd1 [H64Q]), or pNS476 (YPD1+), and the Ssk1p content was monitored. (D) Deletion of SKN7 does not reduce the stability of Ssk1p. Tagged Ssk1p was expressed by pNS462 in NS318, NS320, and skn7Δ ssk1Δ hog1Δ (NS325) strains, and the Ssk1p content was monitored. +, present; Δ, deleted.
Coimmunoprecipitation of Ssk1p with the proteasome.
NS318 and NS320 strains expressing SSK1 (wild type or ΔUBL)-FLAG were grown to an optical density at 600 nm of ∼1.0. Cells were collected by centrifugation, suspended in lysis buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 3 mM phenylmethylsulfonyl fluoride, 40 μg of aprotinin/ml, 10 μg of pepstatin A/ml, 20 μg of leupeptin/ml, 0.1 mM MG-132), and mixed with an equal volume of glass beads (Sigma). After being vortexed 10 times with 30-s pulses, samples were centrifuged at 17,000 × g for 15 min at 4°C. The supernatant was then incubated with anti-FLAG M2-agarose beads (Sigma) for 4 h at 4°C. Beads were collected and washed twice with lysis buffer and then suspended in Laemmli sample buffer and boiled for 5 min. Western blotting was performed essentially as described above, with 12CA5, M2, and rabbit anti-20S proteasome core subunit polyclonal antibody (62).
Detection of phosphorylated Hog1p.
TM141 and NS327 strains expressing multicopy HOG1 from pHG11 (38) were grown overnight in SD medium without sorbitol. Cells were transferred to yeast extract-peptone-dextrose medium without sorbitol and grown for 2.5 h to an optical density at 600 nm of ∼0.8. Cells were then resuspended in yeast extract-peptone-dextrose medium containing 1.5 M sorbitol and collected at the indicated times. Cell pellets were suspended in Laemmli sample buffer containing 1 mM sodium vanadate and boiled for 5 min. Cell extract was prepared with glass beads as described above. Western blotting was performed essentially as described above, with rabbit polyclonal anti-phospho-p38 antibody (New England Biolabs, Beverly, Mass.) and the goat polyclonal anti-Hog1p antibody yC-20 (Santa Cruz Biotechnology, Santa Cruz, Calif.).
RESULTS
Ssk1p is stabilized in proteasome mutants.
A recent paper on the high-throughput mass spectrometric protein complex identification applied to S. cerevisiae (22) reported that Ssk1p binds to the Ssk2p MAPKKK, Ubi4p (ubiquitin), and Ubc7p. Ubc7p is a ubiquitin-conjugating enzyme (E2) responsible for ERAD, which involves the ubiquitination of misfolded proteins retrotranslocated from the ER to the cytoplasm (15, 21, 46). Ubc7p also functions in the regulatory degradation of the following proteins: Hmg2p, a yeast isozyme of the cholesterol-biosynthesis enzyme HMG-coenzyme A reductase (3, 17, 19); Matα2p, a transcriptional repressor (10, 60); and Ole1p, an ER-bound Δ-9 fatty-acid desaturase (5).
This discovery prompted us to investigate the relationship between Ssk1p and the ubiquitin-proteasome system. We examined the stability of Ssk1p in rpn9Δ (61) and rpn12-1 (30) mutants, which are defective in a 19S regulatory subunit of the proteasome. C-terminally epitope-tagged Ssk1p, which had been shown to complement the deletion of SSK1 (data not shown), was expressed under the GAL1 promoter in wild-type (W303-1A), rpn9Δ, and rpn12-1 strains cultured in SGal medium at 30°C, the permissive temperature. Then the GAL1 promoter was shut off in SD medium, and novel protein synthesis was inhibited by the addition of cycloheximide to the medium. Simultaneously, cells were shifted to 37°C, the nonpermissive temperature, and sampled every hour. The Ssk1p content was monitored by Western blotting. In wild-type cells, Ssk1p content constantly decreased over time, whereas Ssk1p was more stable in rpn9Δ and in rpn12-1 cells than in wild-type cells (Fig. 2). These results indicate that Ssk1p is degraded by the activity of the proteasome system.
FIG. 2.
Ssk1p is stabilized in proteasome mutants. HA-tagged Ssk1p was expressed from the high-copy-number plasmid pNS252 in the wild-type (WT) (W303-1A), rpn9Δ (J43), and rpn12-1 (YK109) strains cultured in SGal medium at 30°C. Transcription and translation were shut off in SD medium containing cycloheximide at 37°C. Cells were collected at the indicated times, the proteins were extracted, and the Ssk1p content was monitored by Western blotting with anti-HA antibody. Actin was detected as the loading control.
The stability of Ssk1p is dependent on both the phosphotransfer from Ypd1p and the proteasome system.
In the absence of hyperosmotic stress, Ypd1p phosphorylates and inactivates Ssk1p. Deletion of YPD1 keeps Ssk1p constitutively unphosphorylated (50). To determine whether the phosphorylation state of Ssk1p affects its stability, we examined the stability of Ssk1p in strains in which YPD1 was intact or deleted. To circumvent the lethality of ypd1Δ, the experiments were performed on a hog1Δ background. C-terminally tagged Ssk1p was expressed in both YPD1+ and ypd1Δ cells, and Ssk1p content was monitored by Western blotting as described above, except that SSK1 was expressed from a low-copy-number plasmid and the cells were kept at 30°C throughout the experiment. In ypd1Δ cells, in which Ssk1p was unphosphorylated, the Ssk1p content decreased more rapidly than in YPD1+ cells, in which Ssk1p was kept phosphorylated (Fig. 3A). These results indicate that unphosphorylated Ssk1p is less stable than phosphorylated Ssk1p. The instability of Ssk1p in ypd1Δ cells was almost negated by the deletion of RPN9 (Fig. 3A). This observation suggests that the instability of unphosphorylated Ssk1p is caused by its preferred degradation by the proteasome system. Alternatively tagged Ssk1p, with a different epitope tag at the other terminus, was also degraded in a phosphorelay-regulated manner as was the C-terminally tagged Ssk1p used in the experiments thus far, which indicates that the degradation was unlikely to be affected by the epitope tagging (Fig. 3B). Thus, we used the C-terminally tagged version in the following experiments.
The possibility was tested that the instability of Ssk1p in ypd1Δ cells was caused by phosphorelay-independent effects of Ypd1p deficiency, such as the loss of a physical and stabilizing complex between Ypd1p and Ssk1p. In ypd1Δ cells, expression of mutant Ypd1H64Qp, which is incapable of phosphotransfer but expected to be able to interact with Ssk1p (26, 50), did not significantly stabilize Ssk1p, while wild-type Ypd1p did (Fig. 3C). These results suggest that the phosphorylation state of Ssk1p itself is the determinant of the stability.
Instability of unphosphorylated Ssk1p is independent of the SKN7 pathway.
In ypd1Δ cells, not only is Ssk1p unphosphorylated and/or activated but Skn7p is also unphosphorylated and/or inactivated (8, 29, 32) (Fig. 1). To test the possible involvement of the SKN7 pathway in Ssk1p stability, we examined the stability of Ssk1p in strains with intact or deleted SKN7. The deletion of SKN7 did not decrease the stability of Ssk1p in YPD1+ cells (Fig. 3D), indicating that Ssk1p instability in ypd1Δ cells is not caused by the inactivation of the SKN7 pathway. Conversely, Ssk1p was marginally, but reproducibly, more stable in skn7Δ cells than in SKN7+ cells. This may be due to increased phosphorylation of Ssk1p caused by the absence of Skn7p, which potentially competes with Ssk1p for phosphotransfer from Ypd1p.
Ubc7p is involved in the degradation of unphosphorylated Ssk1p.
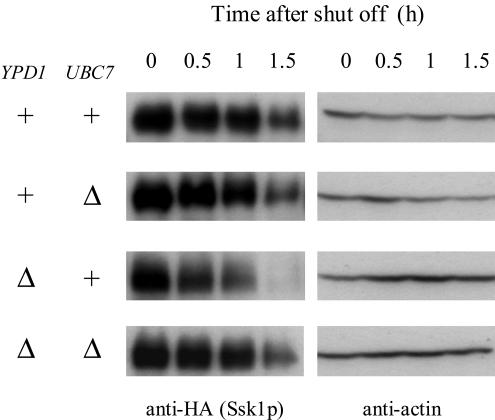
In light of the results of high-throughput mass spectrometric protein complex identification applied to S. cerevisiae as mentioned above (22), Ubc7p was expected to be involved in the degradation of Ssk1p. To test the possibility, we examined the stability of Ssk1p in ubc7Δ cells. In YPD1+ cells, deletion of UBC7 did not significantly affect the stability of Ssk1p. In ypd1Δ cells, on the other hand, disruption of UBC7 stabilized Ssk1p cells in comparison to UBC7+ cells (Fig. 4). These results indicate that unphosphorylated Ssk1p is selectively degraded by the Ubc7p-dependent degradation pathway.
FIG. 4.
Ubc7p-dependent degradation of unphosphorylated Ssk1p. Ubc7p is involved in the degradation of Ssk1p. Tagged Ssk1p was expressed from pNS114 in the YPD1+ ssk1Δ hog1Δ UBC7+ (NS318), YPD1+ ssk1Δ hog1Δ ubc7Δ (NS329), ypd1Δ ssk1Δ hog1Δ UBC7+ (NS320), and ypd1Δ ssk1Δ hog1Δ ubc7Δ (NS333) strains, and the Ssk1p content was monitored as described in the legend to Fig. 3. +, present; Δ, deleted.
Ssk2p-independent degradation of Ssk1p.
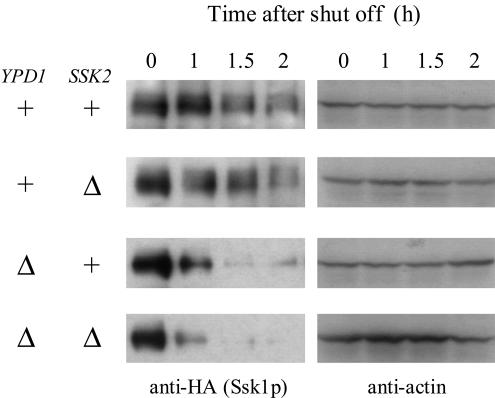
To examine the possible involvement of the Ssk1p-Ssk2p interaction in the regulation of the Ssk1p stability, the level of Ssk1p was monitored in ssk2Δ cells. Both in YPD1+ and ypd1Δ cells, deletion of SSK2 did not significantly affect the stability of Ssk1p (Fig. 5). These results indicate that the Ssk1p-Ssk2p interaction is not required for the degradation of Ssk1p.
FIG. 5.
Ssk2p-independent degradation of Ssk1p. Tagged Ssk1p was expressed from pNS462 in the YPD1+ ssk1Δ hog1Δ SSK2+ (NS318), ypd1Δ ssk1Δ hog1Δ SSK2+ (NS320), YPD1+ ssk1Δ hog1Δ ssk2Δ (NS354), and ypd1Δ ssk1Δ hog1Δ ssk2Δ (NS355) strains, and the Ssk1p content was monitored. +, present; Δ, deleted.
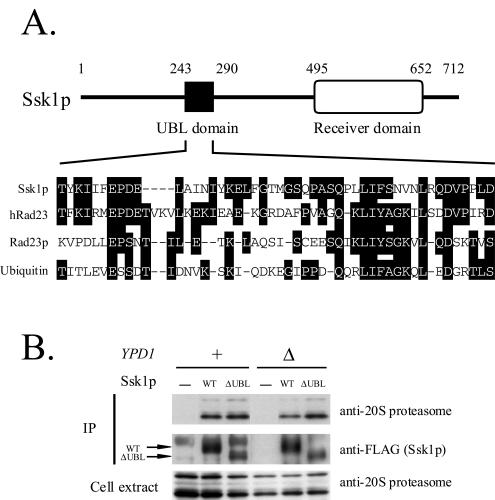
The UBL domain of Ssk1p is not required for the interaction with the proteasome.
We found a UBL domain in the N-terminal region (amino acids 243 to 290) of Ssk1p (Fig. 6A). Recent reports showed that the UBL domains of Rad23p and Dsk2p bind to Rpn1p, a component of the base complex of the 26S proteasome (13, 16, 56, 65). Thus, we examined the interaction between the UBL domain of Ssk1p and the proteasome. Wild-type or UBL domain-deleted Ssk1p (Ssk1ΔUBLp) fused to a FLAG tag was expressed in both YPD1+ and ypd1Δ cells. When expressed from SSK1's own promoter, the expression level of Ssk1ΔUBLp was much lower than that of wild-type Ssk1p, probably because of the gross structural perturbation of this mutant protein (data not shown). To minimize the difference between the expression levels in this experiment, Ssk1ΔUBLp was overproduced from the strong GPD promoter while wild-type Ssk1p was overproduced from the weaker TEF promoter. Ssk1p was immunoprecipitated with anti-FLAG antibody, and proteasomes bound to Ssk1p were detected by Western blotting with anti-20S proteasome antibody. Even when the UBL domain was deleted, Ssk1p coprecipitated with the proteasome (Fig. 6B). These results indicate that the UBL domain of Ssk1p is atypical in that it is not required for interaction with the proteasome.
FIG. 6.
The UBL domain of Ssk1p is not required for the interaction with the proteasome. (A) UBL domain of Ssk1p. In the middle of the molecule (amino acids 243 to 290), Ssk1p contains a small domain homologous to ubiquitin and the UBL domains of human and yeast Rad23p. The UBL domain of Ssk1p shares amino acid identities of 42.6% with human Rad23p, 25.5% with yeast Rad23p, and 25.5% with ubiquitin. (B) Coimmunoprecipitation of Ssk1p with the proteasome. SSK1 (wild type [WT] or ΔUBL)-FLAG was expressed from pNS454 or pNS456, respectively, in the YPD1+ ssk1Δ hog1Δ (NS318) and ypd1Δ ssk1Δ hog1Δ (NS320) strains. Ssk1p (WT or ΔUBL)-FLAG was immunoprecipitated with anti-FLAG M2 beads, and proteasomes bound to Ssk1p were detected by Western blotting. +, present; Δ, deleted; −, not present.
The Hog1p MAPK is ectopically activated in ubc7Δ cells.
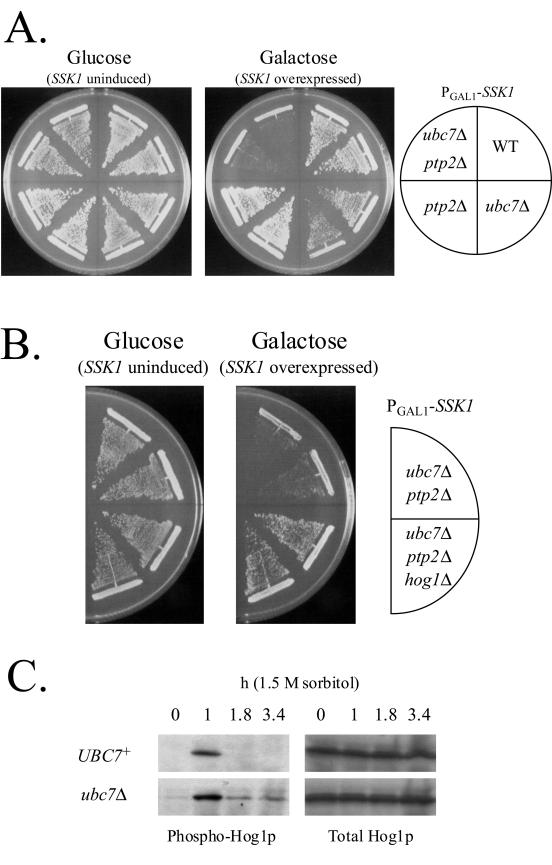
In ubc7Δ cells, the delay of the Ssk1p degradation was expected to cause ectopic activation of the Hog1p MAPK. To examine the possibility, we overexpressed SSK1 under the strong GAL1 promoter in wild-type, ubc7Δ, ptp2Δ, and ubc7Δ ptp2Δ cells on SGal medium. Overexpression of SSK1 was slightly toxic to ubc7Δ cells compared to wild-type cells, as observed by poorer growth of ubc7Δ cells than of wild-type cells. More remarkably, in ptp2Δ cells, in which the inactivation process of Hog1p is partially defective (66), the overexpression of SSK1 was lethal when UBC7 was deleted (Fig. 7A). This lethality was suppressed by deletion of HOG1 (Fig. 7B). These results suggest that the Ssk1p degradation by the Ubc7p-dependent mechanism represses the activity of the HOG pathway.
FIG.7.
Ectopic activation of the HOG pathway in ubc7Δ cells. (A) Overexpression of SSK1 is toxic to ubc7Δ cells. Wild-type (TM141), ubc7Δ (NS327), ptp2Δ (TM107), and ubc7Δ ptp2Δ (NS358) strains were transformed with pNS473. Transformants were grown on SD and SGal plates. SSK1 was overexpressed under the GAL1 promoter on an SGal plate. Each plate was incubated at 30°C for 4 days. (B) Deletion of HOG1 suppresses the lethality caused by SSK1 overexpression. SSK1 was overexpressed from pNS473 in the NS358 and ubc7Δ ptp2Δ hog1Δ (NS362) strains. Each plate was incubated at 30°C for 5 days. (C) Prolonged phosphorylation of the Hog1p MAPK in ubc7Δ cells after the hyperosmotic treatment. TM141 and NS327 strains expressing HOG1 from pHG11 were treated with or without 1.5 M sorbitol, and phospho-Hog1p was detected by Western blotting.
Next, we monitored the phosphorylation level of Hog1p in wild-type and ubc7Δ cells. To improve the sensitivity of our assay, HOG1 was expressed from a high-copy-number plasmid. Cells were treated with or without 1.5 M sorbitol then sampled at the indicated times, and phosphorylated Hog1p was detected by Western blotting with anti-phospho-p38 antibody. After the hyperosmotic treatment, the dephosphorylation and/or inactivation process of Hog1p was delayed in ubc7Δ cells compared with wild-type cells. On the other hand, in the absence of osmotic stress, no significant increase in the basal activity of Hog1p was observed in ubc7Δ cells compared with wild-type cells (Fig. 7C). These results indicate that the Ubc7p-dependent degradation mechanism, by degrading an unphosphorylated pool of Ssk1p, downregulates the HOG pathway after the completion of the osmotic adaptation, in cooperation with the protein phosphatases that inactivate the pathway (25, 37, 64, 66).
DISCUSSION
Activation of the yeast HOG MAPK cascade is essential in the organism's adaptation to high extracellular osmolarity (6, 36, 38, 48). On the other hand, proper inactivation of the HOG cascade is also essential for growth because hyperactivation of this cascade is toxic to cells (36, 37, 38, 66). Therefore, the HOG cascade must remain inactive unless cells are exposed to high osmolarity and must be inactivated promptly once the adaptation process to high osmolarity is completed. Dephosphorylation of a phosphotyrosine or a phosphothreonine residue in active Hog1p by protein phosphatases is a well-characterized inactivation mechanism (25, 37, 38, 64, 66). Our results demonstrate an additional downregulation mechanism in the pathway: unphosphorylated Ssk1p response regulator is degraded by the ubiquitin-proteasome system.
Two distinct physiological roles are conceivable for this downregulation mechanism. One is to maintain the low basal activity of the HOG pathway in the absence of osmotic stress. This aids cells in avoiding ectopic induction of osmotic responses under normal osmotic conditions, which is not only unnecessary but also deleterious. The other is to downregulate the HOG pathway after a sufficient signal is transmitted to complete the adaptation process. This ensures that adequate adaptation is achieved and that the immediate resumption of growth occurs once the adaptation is complete. Here, we show that the latter is the main role for the degradation mechanism because, as shown in Fig. 7C, the deletion of UBC7 caused the prolonged activation of the HOG pathway after the hyperosmotic treatment but did not significantly increase the basal activity of Hog1p in the absence of osmotic stress.
The reaction steps in the ubiquitination of substrates to be degraded require three kinds of enzymes: ubiquitin-activating enzyme (E1), E2, and ubiquitin ligase (E3). E1 activates ubiquitin by forming a thiol-ester bond between ubiquitin and E1. Activated ubiquitin is then transferred to E2. E3 interacts with E2 and the substrate and facilitates ubiquitination. The E3 involved in Ssk1p ubiquitination has yet to be identified. In the case of the human ERK pathway, MEKK1 (MAPKKK), which contains a RING finger-like PHD domain, functions as E3 for the ERK1/2 MAPKs and downregulates the pathway (35). However, our observation that deletion of SSK2 does not affect the stability of Ssk1p excludes the possibility that the Ssk2p MAPKKK acts as E3 for Ssk1p. Consistently, Ssk2p does not appear to have any of the motifs frequently found in E3s, such as a RING, HECT, or U-box domain. The yeast Ste11p MAPKKK is activated by both mating pheromone and osmotic stress (18, 20, 47, 49). When activated by pheromone, Ste11p is degraded by the ubiquitin-dependent system (14). However, E3 for Ste11p has also yet to be identified. In the case of ERAD, Hrd1p (3, 19) and Doa10p (60) have been defined as ER-associated E3s that act together with Ubc7p, whereas degradation of Ole1p is independent of these E3s (5). Whether Hrd1p and/or Doa10p is also an E3 in the ubiquitination of Ssk1p awaits further investigation.
The domains in Ssk1p required for its interaction with E2, E3, or proteasomes, if any, have yet to be determined. We found a UBL domain in the N-terminal region (amino acids 243 to 290) of Ssk1p (Fig. 6A). Recent reports showed that the UBL domains of Rad23p and Dsk2p bind to Rpn1p, a component of the base complex of the 26S proteasome (13, 16, 56, 65). In contrast, the UBL domain of Ssk1p is dispensable for its interaction to proteasomes. In accord with the difference in function, there are some structural differences between the UBL domains of Ssk1p and Rad23p/Dsk2p. The UBL domain of Ssk1p is shorter than those of Rad23p and Dsk2p, and its location is in the center of the molecule, whereas those of Rad23p and Dsk2p are near the N termini. The UBL domain of Ssk1p may bind to degradation-related proteins other than components of the 26S proteasome, such as E2 and/or E3.
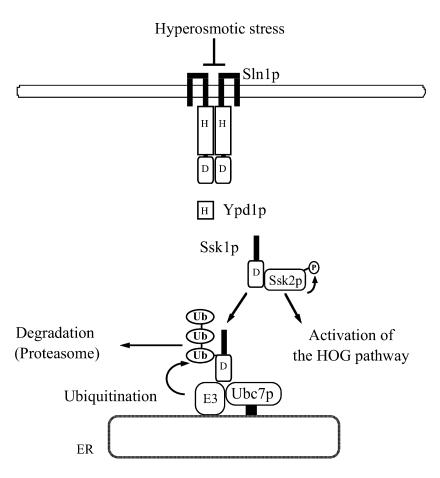
A model for the functional connection between Ssk1p and the ubiquitin-proteasome system is shown in Fig. 8. Once the osmotic adaptation is completed, unphosphorylated Ssk1p, which potentially activates Ssk2p, is ubiquitinated by the Ubc7p E2 and an unidentified E3 on the surface of the ER. Then unphosphorylated and ubiquitinated Ssk1p is targeted to the 26S proteasome and degraded, which ensures the timely inactivation of the HOG pathway after the completion of the osmotic adaptation.
FIG. 8.
Model for the functional connection between Ssk1p and the ubiquitin-proteasome system. See Discussion for explanation.
Phosphorelay-regulated degradation of response regulators by the ubiquitin-proteasome system may be a general regulatory mechanism in eukaryotic His-Asp phosphorelay signaling systems. Higher plants make use of His-Asp phosphorelay signaling systems in the signal transduction of plant hormones ethylene and cytokinin (9, 23, 24, 28, 55, 57, 67). Recently, a mutant of an Rpn12p homolog in Arabidopsis was reported to show altered responses to cytokinin (59). Although response regulators for cytokinin signaling are still to be identified, impaired degradation of them may cause the altered responses to the hormone in this proteasome mutant.
Acknowledgments
We thank Randolph Hampton, Mark Longtine, Haruo Saito, Howard Bussey, and Masahiro Kawabata for plasmids. We also thank Eugene Futai, Michio Hayashi, Hiroyuki Sorimachi, Koichi Suzuki, and all the members of the Maeda laboratory for help, advice, and discussion.
This work was supported by a grant-in-aid for scientific research on priority areas (KAKENHI 14086203) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), a grant (no. 0253) from the Salt Science Research Foundation, and a grant from the Japan Foundation for Applied Enzymology (to T.M.).
REFERENCES
- 1.Albertyn, J., S. Hohmann, J. M. Thevelein, and B. A. Prior. 1994. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14:4135-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. Wiley, New York, N.Y.
- 3.Bays, N. W., R. G. Gardner, L. P. Seelig, C. A. Joazeiro, and R. Y. Hampton. 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3:24-29. [DOI] [PubMed] [Google Scholar]
- 4.Bouquin, N., A. L. Johnson, B. A. Morgan, and L. H. Johnston. 1999. Association of the cell cycle transcription factor Mbp1 with the Skn7 response regulator in budding yeast. Mol. Biol. Cell 10:3389-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, S., K. Matuschewski, M. Rape, S. Thoms, and S. Jentsch. 2002. Role of the ubiquitin-selective CDC48UFD1/NPL4 chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760-1763. [DOI] [PubMed] [Google Scholar]
- 7.Brown, J. L., H. Bussey, and R. C. Stewart. 1994. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 13:5186-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, J. L., S. North, and H. Bussey. 1993. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall β-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 175:6908-6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, C., S. F. Kwok, A. B. Bleecker, and E. M. Meyerowitz. 1993. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262:539-544. [DOI] [PubMed] [Google Scholar]
- 10.Chen, P., P. Johnson, T. Sommer, S. Jentsch, and M. Hochstrasser. 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell 74:357-369. [DOI] [PubMed] [Google Scholar]
- 11.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 12.de Nadal, E., P. M. Alepuz, and F. Posas. 2002. Dealing with osmostress through MAP kinase activation. EMBO Rep. 3:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsasser, S., R. R. Gali, M. Schwickart, C. N. Larsen, D. S. Leggett, B. Müller, M. T. Feng, F. Tübing, G. A. G. Dittmar, and D. Finley. 2002. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4:725-730. [DOI] [PubMed] [Google Scholar]
- 14.Esch, R. K., and B. Errede. 2002. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc. Natl. Acad. Sci. USA 99:9160-9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedlander, R., E. Jarosch, J. Urban, C. Volkwein, and T. Sommer. 2000. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2:379-384. [DOI] [PubMed] [Google Scholar]
- 16.Funakoshi, M., T. Sasaki, T. Nishimoto, and H. Kobayashi. 2002. Budding yeast Dsk2 is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 99:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner, R. G., A. G. Shearer, and R. Y. Hampton. 2001. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol. Cell. Biol. 21:4276-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampton, R. Y. 2002. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14:476-482. [DOI] [PubMed] [Google Scholar]
- 20.Herskowitz, I. 1995. MAP kinase pathways in yeast: for mating and more. Cell 80:187-197. [DOI] [PubMed] [Google Scholar]
- 21.Hiller, M. M., A. Finger, M. Schweiger, and D. H. Wolf. 1996. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273:1725-1728. [DOI] [PubMed] [Google Scholar]
- 22.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S.-L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sørensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. V. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 23.Hua, J., C. Chang, Q. Sun, and E. M. Meyerowitz. 1995. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269:1712-1714. [DOI] [PubMed] [Google Scholar]
- 24.Inoue, T., M. Higuchi, Y. Hashimoto, M. Seki, M. Kobayashi, T. Kato, S. Tabata, K. Shinozaki, and T. Kakimoto. 2001. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409:1060-1063. [DOI] [PubMed] [Google Scholar]
- 25.Jacoby, T., H. Flanagan, A. Faykin, A. G. Seto, C. Mattison, and I. Ota. 1997. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J. Biol. Chem. 272:17749-17755. [DOI] [PubMed] [Google Scholar]
- 26.Janiak-Spens, F., D. P. Sparling, and A. H. West. 2000. Novel role for HPt domain in stabilizing the phosphorylated state of a response regulator domain. J. Bacteriol. 182:6673-6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janiak-Spens, F., J. M. Sparling, M. Gurfinkel, and A. H. West. 1999. Differential stabilities of phosphorylated response regulator domains reflect functional roles of the yeast osmoregulatory SLN1 and SSK1 proteins. J. Bacteriol. 181:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kakimoto, T. 1996. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274:982-985. [DOI] [PubMed] [Google Scholar]
- 29.Ketela, T., J. L. Brown, R. C. Stewart, and H. Bussey. 1998. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol. Gen. Genet. 259:372-378. [DOI] [PubMed] [Google Scholar]
- 30.Kominami, K., G. N. DeMartino, C. R. Moomaw, C. A. Slaughter, N. Shimbara, M. Fujimuro, H. Yokosawa, H. Hisamatsu, N. Tanahashi, Y. Shimizu, K. Tanaka, and A. Toh-e. 1995. Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 14:3105-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krems, B., C. Charizanis, and K. D. Entian. 1996. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29:327-334. [DOI] [PubMed] [Google Scholar]
- 32.Li, S., A. Ault, C. L. Malone, D. Raitt, S. Dean, L. H. Johnston, R. J. Deschenes, and J. S. Fassler. 1998. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 17:6952-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, S., S. Dean, Z. Li, J. Horecka, R. J. Deschenes, and J. S. Fassler. 2002. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol. Biol. Cell 13:412-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 35.Lu, Z., S. Xu, C. Joazeiro, M. H. Cobb, and T. Hunter. 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9:945-956. [DOI] [PubMed] [Google Scholar]
- 36.Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554-558. [DOI] [PubMed] [Google Scholar]
- 37.Maeda, T., A. Y. M. Tsai, and H. Saito. 1993. Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTC1) cause a synthetic growth defect in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5408-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeda, T., S. M. Wurgler-Murphy, and H. Saito. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242-245. [DOI] [PubMed] [Google Scholar]
- 39.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan, B. A., N. Bouquin, G. F. Merrill, and L. H. Johnston. 1995. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 14:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mumberg, D., R. Müller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mumberg, D., R. Müller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 43.O'Rourke, S. M., and I. Herskowitz. 2002. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 22:4739-4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostrander, D. B., and J. A. Gorman. 1999. The extracellular domain of Saccharomyces cerevisiae Sln1p membrane osmolarity sensor is necessary for kinase activity. J. Bacteriol. 181:2527-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ota, I. M., and A. Varshavsky. 1993. A yeast protein similar to bacterial two-component regulators. Science 262:566-569. [DOI] [PubMed] [Google Scholar]
- 46.Plemper, R. K., and D. H. Wolf. 1999. Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem. Sci. 24:266-270. [DOI] [PubMed] [Google Scholar]
- 47.Posas, F., and H. Saito. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702-1705. [DOI] [PubMed] [Google Scholar]
- 48.Posas, F., and H. Saito. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17:1385-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Posas, F., M. Takekawa, and H. Saito. 1998. Signal transduction by MAP kinase cascades in budding yeast. Curr. Opin. Microbiol. 1:175-182. [DOI] [PubMed] [Google Scholar]
- 50.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 51.Raitt, D. C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiser, V., S. M. Salah, and G. Ammerer. 2000. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2:620-627. [DOI] [PubMed] [Google Scholar]
- 53.Rep, M., V. Reiser, U. Gartner, J. M. Thevelein, S. Hohmann, G. Ammerer, and H. Ruis. 1999. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol. 19:5474-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 55.Sakai, H., T. Honma, T. Aoyama, S. Sato, T. Kato, S. Tabata, and A. Oka. 2001. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294:1519-1521. [DOI] [PubMed] [Google Scholar]
- 56.Schauber, C., L. Chen, P. Tongaonkar, I. Vega, D. Lambertson, W. Potts, and K. Madura. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391:715-718. [DOI] [PubMed] [Google Scholar]
- 57.Sheen, J. 2002. Phosphorelay and transcriptional control in cytokinin signal transduction. Science 296:1650-1652. [DOI] [PubMed] [Google Scholar]
- 58.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smalle, J., J. Kurepa, P. Yang, E. Babiychuk, S. Kushnir, A. Durski, and R. D. Vierstra. 2002. Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14:17-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swanson, R., M. Locher, and M. Hochstrasser. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 15:2660-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeuchi, J., M. Fujimuro, H. Yokosawa, K. Tanaka, and A. Toh-e. 1999. Rpn9 is required for efficient assembly of the yeast 26S proteasome. Mol. Cell. Biol. 19:6575-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka, K., T. Yoshimura, A. Kumatori, A. Ichihara, A. Ikai, M. Nisihigai, K. Kameyama, and T. Takagi. 1988. Proteasomes (multi-protease complexes) as 20S ring-shaped particles in a variety of eukaryotic cells. J. Biol. Chem. 263:16209-16217. [PubMed] [Google Scholar]
- 63.Tao, W., R. J. Deschenes, and J. S. Fassler. 1999. Intracellular glycerol levels modulate the activity of Sln1p, a Saccharomyces cerevisiae two-component regulator. J. Biol. Chem. 274:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warmka, J., J. Hanneman, J. Lee, D. Amin, and I. Ota. 2001. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 21:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watkins, J. F., P. Sung, L. Prakash, and S. Prakash. 1993. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol. 13:7757-7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wurgler-Murphy, S. M., T. Maeda, E. A. Witten, and H. Saito. 1997. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol. Cell. Biol. 17:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada, H., T. Suzuki, K. Terada, K. Takei, K. Ishikawa, K. Miwa, T. Yamashino, and T. Mizuno. 2001. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42:1017-1023. [DOI] [PubMed] [Google Scholar]