Abstract
Zinc transporter LIV-1 (SLC39A6) is estrogen regulated and present in increased amounts in estrogen receptor–positive breast cancer as well as in tumors that spread to the lymph nodes. The LIV-1 subfamily of ZIP zinc transporters consists of nine human sequences that share considerable homology across transmembrane domains. Many of these sequences have been shown to transport zinc and/or other ions across cell membranes. Increasingly, studies have implicated members of the LIV-1 transporter subfamily in a variety of diseases. We review these studies and report our own investigations of the role in breast cancer of the nine LIV-1 zinc transporters. We have documented the response of these transporters to estrogen and antiestrogens, and also their presence in our models of resistance to antiestrogens. Resistance to antiestrogen drugs such as tamoxifen and fulvestrant often occurs in advanced breast cancer. In these models we observed differential expression of individual LIV-1 family members, which may be related to their observed variable tissue expression. We were unable detect ZIP4, which is known to be expressed in the intestine. HKE4/SLC39A7 had elevated expression in both antiestrogen-resistant cell lines, and ZIP8 had elevated expression in fulvestrant-resistant cells. In addition, we investigated the expression of the nine LIV-1 family members in a clinical breast cancer series. Although a number of different LIV-1 family members showed some association with growth factor receptors, LIV-1 was solely associated with estrogen receptor and a variety of growth factors commonly associated with clinical breast cancer. HKE4, however, did show an association with the marker of cell proliferation Ki67 the spread of breast cancer to lymph nodes.
INTRODUCTION TO THE LIV-1 FAMILY OF ZINC TRANSPORTERS
Zinc Transporter Families
Zinc is an essential ion in cells; without it cells cannot sustain life. Zinc is a cofactor for more than 300 enzymes, representing more than 50 different enzyme classes, and is essential for cell growth (1). Zinc is involved in protein, nucleic acid, carbohydrate, and lipid metabolism, as well as in the control of gene transcription, differentiation, development, and growth (2). Zinc deficiency can be detrimental, causing stunted growth and serious metabolic disorders (3), while excess zinc can be toxic to cells (4). Cellular levels of zinc are tightly regulated by specific zinc transporter proteins, of which there are two known families. These two families have opposing action on zinc transport. The ZnT family (SLC30A) (previously termed CDF for cation diffusion facilitator) of zinc transporters (5) transport zinc out of cells or into intracellular compartments from the cytoplasm, whereas the ZIP family (for Zrt-, Irt-like Proteins) (SLC39A) of zinc transporters (6) transport zinc into the cell cytoplasm from either outside the cell or from intracellular compartments. Although these two families are termed zinc transporters, evidence shows that some members are able to transport other divalent cations such as iron, cadmium, copper, and manganese as well as zinc. The exact molecular mechanism for such transport is still unknown, however. The ZnT family contains nine human sequences, and the ZIP family contains 14 human sequences (7). Increasing evidence implicates various members of the SLC39A family of ZIP transporters in disease states and suggests that aberrant expression of zinc transporters leads to uncontrolled growth such that occurring in cancer. Thus any molecules controlling cellular zinc levels are worthy of investigation.
The LIV-1 Family of ZIP Transporters
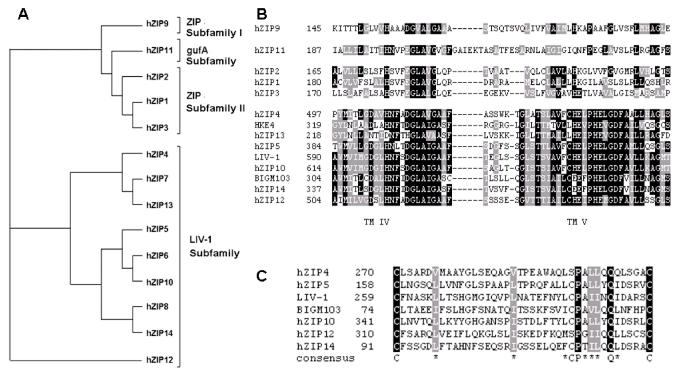
More than 100 SLC39A sequences have been identified, originating from more than 12 species (7,8). The 14 human members of the SLC39A family have been divided into four separate groups (Figure 1A), nine of which are in the LIV-1 subfamily (7). Importantly, the LIV-1 subfamily is a highly conserved group of eight transmembrane domain proteins that are mainly situated on the plasma membrane and transport zinc into cells. At least one SLC39A sequence, however, member HKE4 (SLC39A7/ZIP7), exists on intracellular membranes and transports zinc into the cytosol (9). All of the SLC39A LIV-1 family members contain the consensus sequence present in other ZIP transporters, located in the region of transmembrane domains IV and V, and a histidine-rich motif (of the form HXHXH, where H is histidine and X is any amino acid) thought to be important in zinc transport, located on the long intracellular loop between TM III and IV, immediately preceding TM IV (10). The LIV-1 family sequences also contain a highly conserved potential metalloprotease motif, which closely resembles the active site motif of matrix metalloproteases and is situated in transmembrane domain V (Figure 1B), and considerably increased histidine residues on the N-terminus and extracellular loop between TM II and III (7). For example, ZIP10 has 49 histidines in the extracellular N-terminus, nine on the first extracellular loop between TM II and III and 20 on the intracellular loop between TM III and IV. Interestingly, two of the LIV-1 family sequences, ZIP14 and BIGM103/ZIP8, have the initial H of the HEXXH motif in TM V replaced with an E, which may suggest an ability to transport ions other than zinc. Furthermore, ZIP8 has been demonstrated to transport cadmium in the testis (11), and ZIP14 mediates non–transferrin-bound iron uptake into hepatocytes (12).
Figure 1.
Phylogenetic tree and alignment of the human members of the ZIP superfamily of zinc transporters. (A) This phylogenetic tree was drawn using ClustalW and treeview software. (B) This alignment demonstrating the highly conserved motif in TM V for the LIV-1 family was performed using ClustalW and shaded using Boxshade software. Black shading represents at least 50% identity, gray shading represents at least 50% complementary residues. (C) This alignment demonstrating the CPALLY motif directly upstream of TM I was performed using ClustalW and shaded using Boxshade software. Black shading represents at least 50% identity, gray shading represents at least 50% complementary residues.
The Properties of the LIV-1 Family of ZIP Transporters
Current information about the various members of the LIV-1 subfamily, including clarification of the names of each sequence, is documented in Table 1. Most sequences have widespread tissue distribution, as judged by the EST sequences in the GenBank, whereas ZIP4, ZIP5, and ZIP12 appear to be more tissue specific. Most of the LIV-1 family proteins reside on the plasma membrane of cells, a characteristic consistent with a role as a zinc influx transporter. This role has been demonstrated for ZIP4 (13), ZIP5 (14), LIV-1 (15), and ZIP14 (16,17), but not for HKE4, which has been observed on the endoplasmic reticulum (9) and golgi (18) membranes, or BIGM103/ZIP8, which has been observed on intracellular vesicles such as lysosomes and endosomes (19).
Table 1.
Nomenclature and Known Function of LIV-1 Family Members.
| Human gene name | Protein name | Other names | Cell location | Tissue distribution | Transport function | Disease link | Chromosome | Accession number |
|---|---|---|---|---|---|---|---|---|
| SLC39A4 | ZIP4 | LZT-Hs5 | PM | Small intestine, stomach, colon, cecum, kidney | Zinc uptake | Acrodermatitis enteropathica (35–37) | 8q24.3 | NM_017767 |
| SLC39A5 | ZIP5 | LZT-Hs7 | Basolateral surface of polarized cells | Kidney, liver, spleen, colon, stomach, pancreas | Zinc removal | 12q13.13 | NM_173596 | |
| SLC39A6 | LIV-1, ZIP6 | LZT-Hs3 | PM | Widespread | Zinc influx | Breast cancer (21–22), | 18q12.1 | NM_012319 |
| SLC39A7 | HKE4, ZIP7 | LZT-Hs1 | ER and Golgi | widespread | Zinc and manganese bidirectional? (20) | Tamoxifen resistance in breast cancer | 6p21.3 | NM_006979 |
| SLC39A8 | BIGM103, ZIP8 | LZT-Hs6 | Vesicles | Widespread | Cadmium influx (11), Zinc influx | Faslodex resistance in breast cancer | 4q22-q24 | NM_022154 |
| SLC39A10 | ZIP10 | LZT-Hs2 | PM | Widespread | Zinc influx | 2q33.1 | NM_020342 | |
| SLC39A12 | ZIP12 | LZT-Hs8 | Brain, lung, testis, retina | Asthma (32) | 10p12.33 | NM_152725 | ||
| SLC39A13 | ZIP13 | LZT-Hs9 | Widespread | 11p11.12 | NM_152264 | |||
| SLC39A14 | ZIP14 | LZT-Hs4 | PM | Widespread | Zinc influx, non-transferrin-bound iron uptake (12) | Asthma (32), inflammation mediated by IL-6 (31) | 8p21.2 | NM_015359 |
PM, plasma membrane.
Because of membrane topology, location on intracellular membranes is presumed to enable ion transport from intracellular compartments to the cytoplasm. However, the yeast equivalent of HKE4, Yke4p, has recently been shown to balance the zinc level between the cytosol and the secretory pathway by transporting zinc in either direction across membranes, depending on the zinc status of the cells (20). This observation is interesting and may explain the presence in the LIV-1 family sequences of additional histidine-rich regions positioned on the side of the membrane opposing that of the other ZIP transporters.
We have previously observed a conserved motif, which we termed CPALLY, immediately preceding the first TM domain (Figure 1C). This motif does not occur in HKE4, which is not present on the plasma membrane, or ZIP13, which is grouped with ZIP7 in the phylogenetic tree and thus may also reside on intracellular membranes. This CPALLY motif contains three conserved cysteines, which usually form disulfide bonds with other cysteine residues. Furthermore, all of the human LIV-1 family members, except HKE4 and ZIP13, have a conserved cysteine immediately preceding their HEXXH motif in TM V. This motif may be involved in binding to this additional cysteine, thus closing the pore and regulating the movement of zinc across the membrane.
Normal Tissue Expression of LIV-1 Family Members
The normal tissue expression of the different members of the LIV-1 family shows considerable variation, which may indicate their diverse roles in different tissues (Table 1). ZIP4 is present in the intestine and kidney (21), particularly the duodenum and jejunum, which are crucial sites of zinc absorption. ZIP5 is expressed in kidney, liver, pancreas, and intestine, particularly the basolateral membrane of the adult and developing mouse intestine (22). LIV-1 has widespread distribution but is not appreciably present in heart and intestine and is primarily increased in hormonally controlled tissues (15). HKE4 (9), ZIP14 (17), and ZIP13 are ubiquitously expressed, appearing in many tissues. ZIP12 is expressed in brain, lung, testis, and retina, and ZIP10 is predominantly expressed in brain and spinal cord (data obtained from expression array in the HUGO database).
The LIV-1 Family of ZIP Transporters and Disease States
The first member of the LIV-1 family to be linked to disease was LIV-1 itself, which was shown to be estrogen regulated and present in increased amounts in estrogen-receptor–positive breast cancers that spread to the lymph nodes (23,24). More recently, this association of LIV-1 with estrogen receptors has been substantiated by larger scale analysis of breast cancer specimens. These studies have shown that LIV-1 is such a reliable marker of estrogen-receptor–positive cancers (25,26) that it is one of the genes used routinely to distinguish the luminal A type of clinical breast cancer (27,28). Furthermore, in zebrafish embryos, LIV-1 was shown to be the downstream target of the transcription factor STAT3, which has a proven role in the development of cancer (29). This work also found that LIV-1 was essential for the nuclear localization of the transcription factor Snail, which plays a major role in the epithelial-to-mesenchymal switch because of its ability to down regulate the expression of genes associated with cell adhesion. This finding suggests that LIV-1 could form a link between cancer and normal development (30) and raises the question of whether any other LIV-1 family members have a similar role. Interestingly, the expression of two other LIV-1 family sequences, ZIP4 and ZIP5, has been observed in the developing mouse intestine (22) and the Drosophila LIV-1 family member called fear of intimacy (FOI) has been shown to be essential for gonad development (31). Another LIV-1 family member, ZIP14, is substantially increased during the zinc-dependant differentiation of adipocytes (16) and is regulated by IL-6, a mechanism that requires STAT3 signaling (32).
Additionally, the LIV-1 family of zinc transporters has been implicated in other disease states. For example, increases in expression of the zinc transporters LIV-1, ZIP12, and ZIP14 have been observed during acute inflammation in the airway and asthma and have been suggested to promote an increase in zinc uptake, which can reduce inflammation (33). The zinc content in brains of individuals with schizophrenia is lower than that of individuals with other cerebral diseases (34), and a role for the ZIP12 gene has been demonstrated by observation of mutations in ZIP12 in a small group of schizophrenic patients. Significantly, brain zinc content was maintained in rats fed a zinc-deficient diet by a compensatory rise in LIV-1 expression in the brain (35). The ZIP4 gene has been demonstrated to be mutated in the zinc deficiency disorder acrodermatitis enteropathica (21,36), and the observed mutations were shown to disrupt the molecule in areas thought important for zinc transport (37).
INVESTIGATION OF THE LIV-1 FAMILY OF ZIP TRANSPORTERS IN BREAST CANCER
The increase of scientific literature implicating ZIP transporters in a variety of diseases has led to our recent investigation of the relevance of the expression of the nine family members of the LIV-1 family of ZIP transporters in breast cancer. Because LIV-1 itself is known to be regulated by estrogen, we initially investigated the response of these nine LIV-1 family members to short-term treatment with estrogen and antiestrogens. We then followed up with investigation of these nine LIV-1 family members in our cell-line models of longer-term treatment, which have developed resistance to antiestrogens, as well as expression in breast cancer samples. These results are detailed below.
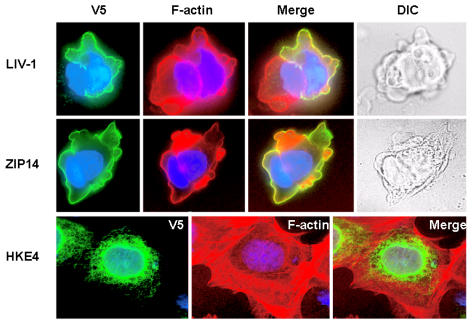
In addition, we transiently transfected MCF-7 cells with constructs for three LIV-1 family members with a C-terminal V5 tag to confirm the same cellular location as previously observed in Chinese hamster ovary cells (9,15,16). Figure 2 shows plasma membrane staining for both LIV-1 and ZIP14 (green) in unpermeabilized MCF-7 cells as determined by colocalization with F-actin filaments (red) and association with the outside of the cell, observed by DIC imaging. In contrast, HKE4 stains intracellular compartments in permeabilized MCF-7 cells, such as the endoplasmic reticulum (green), as evidenced by perinuclear staining and an equal meshlike appearance throughout the cell interior, yet clearly does not reach the cell extremities, which were stained by F-actin (red).
Figure 2.
Cellular location of three human LIV-1 family members in MCF-7 cells. MCF-7 cells expressing recombinant HKE4, LIV-1, or ZIP14 were imaged using a mouse anti-V5 antibody (Invitrogen) conjugated to Alexa Fluor 488 (green) and assembled onto slides using Vectorshield with DAPI (Vector Laboratories). All cells were incubated with Texas red phalloidin to stain F-Actin filaments red. Coverslips were viewed on a Leica RPE automatic microscope using a 63x oil immersion lens. The fluorescent superimposed images were acquired using a multiple bandpass filter set appropriate for DAPI, fluorescein, and Texas Red as well as bright field for differential interference contrast imaging.
The Response of the LIV-1 Family of ZIP Transporters to Antiestrogens
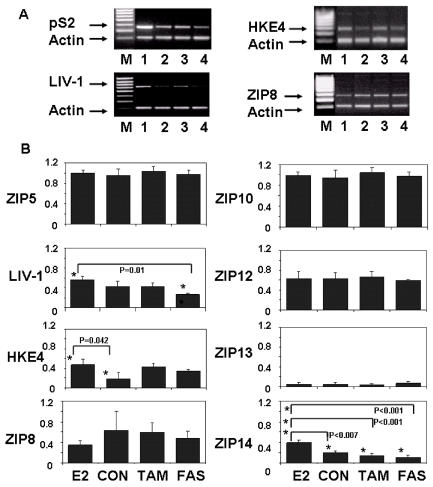
Because of the similarity between the sequences of the nine LIV-1 family members, we investigated whether any of the other LIV-1 family members were also estrogen-regulated and/or expressed in breast cancer cells. To achieve this we compared the RNA expression of all nine members of the LIV-1 family of ZIP transporters in the estrogen-receptor–positive MCF-7 breast cancer cell line that had been exposed to estrogen, the partial antiestrogen tamoxifen, or the pure antiestrogen fulvestrant for 10 days. Cells were harvested, RNA was prepared, RT-PCR was performed, and DNA for individual genes was amplified by PCR in the presence of actin, to allow normalization of the results (Figure 3). To verify the profile of the samples, we first investigated the expression of the estrogen-regulated gene pS2, which was characterized by elevation in response to estrogen, little response to tamoxifen, and reduced expression in response to fulvestrant (see Figure 3A). We observed differential expression of all nine LIV-1 family members, both in the amount of RNA present and in the response to different treatments (see Figure 3B). We show representative gels of the three LIV-1 family members with the largest response (see Figure 3A). ZIP4 was undetectable in all samples, a finding that may reflect its known expression in the intestine and kidney (21). The levels of ZIP5, 10, 12, and 13 did not change across the treatments, and all were expressed at relatively low levels, at which ZIP5, ZIP10, and ZIP12 required an increased number of PCR cycles compared with the other LIV-1 family members. LIV-1 and ZIP14 appeared to be estrogen regulated, showing an increased response to estrogen and reduced response to tamoxifen or fulvestrant, which was statistically significant (see Figure 3). Although HKE4 was also significantly increased in response to estrogen treatment, it was also increased, to a lesser extent, in response to both tamoxifen and fulvestrant. ZIP8 showed a slight decrease in response to estrogen, which was not significant presumably due to the large error bars.
Figure 3.
Comparison of the effect of antihormones on the RNA expression of the nine human LIV-1 family members. MCF-7 cells were treated with 4-hydroxytamoxifen (TAM), fulvestrant (FAS), or oestradiol (E2) for seven days in serum growth factor–free DCCM medium before total RNA was extracted, reverse transcribed to cDNA, and PCR performed for individual ZIPs. All data were normalized to individual β-actin levels. The upper panel shows representative agarose gels of pS2, LIV-1, HKE4, and ZIP8 in MCF-7 treated with oestradiol (lane 1), untreated (lane 2), treated with Tamoxifen (lane 3), or treated with Fulvestrant (lane 4). M represents size markers. Lower panel shows mean ± SEM densitometric values of 8 genes, comparing their expression levels in MCF-7 cells untreated (CON), or treated with oestradiol (E2), Tamoxifen (TAM), or Fulvestrant (FAS). Statistically significant results are indicated with asterisks, and the relevant P value is given. ZIP4 was undetectable.
The observed changes in the SLC39A-family mRNA levels reported here are anticipated to be reflected in the production of the individual proteins, but this possibility has not been examined across the SLC39A family because of the absence of available antibodies. However, recent results in our laboratory with two new antibodies to endogenous LIV-1 and HKE4 suggest that this relationship exists, because both LIV-1 and HKE4 proteins were increased by estrogen treatment, and LIV-1 was decreased with both tamoxifen and faslodex treatments whereas HKE4 was unchanged, mirroring the results that we observed at the mRNA level.
The LIV-1 Family of ZIP Transporters in Antiestrogen Resistance
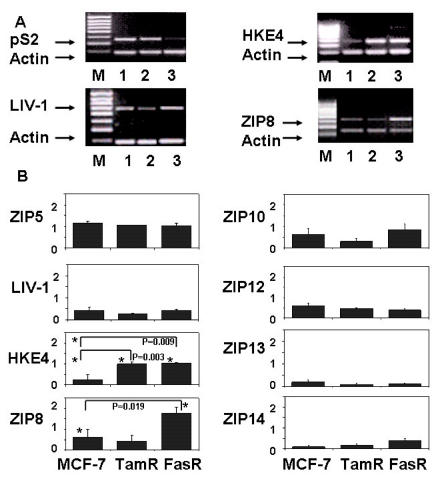
Although estrogen-receptor–positive breast cancers are routinely treated clinically with antihormones such as tamoxifen or fulvestrant, with time the tumors develop resistance to these agents, leading to subsequent regrowth, usually with an altered and more aggressive phenotype (38). To better understand the mechanisms underlying the occurrence of resistance, we have developed a unique panel of antiestrogen responsive and resistant cell lines derived from the estrogen-receptor–positive human breast cancer cell line MCF-7 (39,40). We have investigated the expression of all nine human LIV-1 family members in both our tamoxifen (TamR)- and fulvestrant (FasR)-resistant cell lines and compared them to the wild-type MCF-7 cells (Figure 4) in an effort to examine any potential role that individual LIV-1 family members may play in the development of antihormone-resistant breast cancer. These antihormone-resistant cell lines are able to grow in the presence of antiestrogens by efficiently utilizing signaling pathways such as epidermal growth factor receptor (EGFR) (39,40), Src (41), Insulin-like growth factor receptor 1 (IGF1-R) (42), and c-Met (43), which allows them to exhibit a more aggressive phenotype (44). Cells were harvested, RNA prepared, RT-PCR performed, and DNA for individual genes was amplified by PCR in the presence of actin, to allow normalization of the results. The cell samples were first characterized for pS2 levels, and showed the expected decrease in the FasR cells (45). Representative gels are given for the three LIV-1 family members that appeared most altered, LIV-1, HKE4, and ZIP8. ZIP4 levels were again undetectable, and the levels of ZIP5, ZIP10, ZIP12, and ZIP13 were relatively low, with ZIP5, ZIP10, and ZIP12 again requiring an increased number of PCR cycles. Levels of ZIP5, ZIP12, and ZIP13 did not change, and the levels of LIV-1, ZIP12, and ZIP13 were either unchanged or reduced in the resistant cell lines. ZIP14, although present in low amounts, was increased in the resistant cells, and ZIP10 was decreased in TamR cells. Only HKE4 and ZIP8 produced statistically significant changes, with HKE4 elevated in both resistant states, whereas ZIP8 was considerably elevated in FasR cells. These results suggest that a number of LIV-1 family members (ZIP4, ZIP5, LIV-1, ZIP10, ZIP12, and ZIP13) appear unaltered or decreased by the acquisition of endocrine resistance. However, both HKE4 and ZIP8 were elevated in one or both antihormone-resistant cell lines, suggesting a possible role in the development of resistance. These results are summarized in Table 2.
Figure 4.
Comparison of RNA expression of the nine human LIV-1 family members in anti-hormone-resistant MCF-7 cells. The expression of different ZIPs in tamoxifen (TamR)- and fulvestrant (FasR)-resistant MCF-7 cells was compared with wild-type MCF-7 cells. RNA was extracted and reverse transcribed to cDNA. The upper panel shows representative gels of pS2, LIV-1, HKE4, and ZIP8 in MCF-7 (lane 1), TamR (lane 2) and FasR (lane 3) cell lines. M represents size markers. Lower panel shows mean ± SEM densitometric values of 8 genes comparing their expression levels in MCF-7, TamR, and FasR cells. Statistically significant results are indicated with asterisks, and the relevant P value is given. ZIP4 was undetectable.
Table 2.
Summary of LIV-1 Family Expression Response to Treatments or Antihormone Resistance
| Cells | ZIP4 | ZIP5 | LIV-1 | HKE4 | ZIP8 | ZIP10 | ZIP12 | ZIP13 | ZIP14 |
|---|---|---|---|---|---|---|---|---|---|
| MCF-7 + E2 | ND | — | Up | Up | Down | — | — | — | Up |
| MCF-7 + TAM | ND | — | — | — | — | — | — | — | Down |
| MCF-7 + FAS | ND | — | Down | — | — | — | — | — | Down |
| Tamoxifen-resistant cells | ND | — | — | Up | — | — | — | — | — |
| Fulvestrant-resistant cells | ND | — | — | Up | Up | — | — | — | — |
ND, not determined; —, no change.
We have previously demonstrated that our TamR cells have increased intracellular zinc levels (46; Taylor et al, unpublished results), increased EGFR (39), Src (41), and IGF1-R (42) signaling as well as increased growth and invasion (41). Interestingly, treatment of these cells with 20 μM zinc can activate EGFR, Src, and IGF1-R signaling as well as growth and invasion (46; Taylor et al, unpublished results). Furthermore, we have demonstrated a role for HKE4 in the development of the aggressive phenotype in TamR cells by using siRNA for HKE4 which prevented the observed zinc-induced activation of signaling pathways (Taylor et al, unpublished results). Whether ZIP8 has a similar role in the FasR cells needs to be investigated. We have, however, not examined whether these transporters have the ability to transport metals other than zinc or whether these pathways can be stimulated by other metals.
LIV-1 FAMILY OF ZIP TRANSPORTERS IN BREAST CANCER SAMPLES
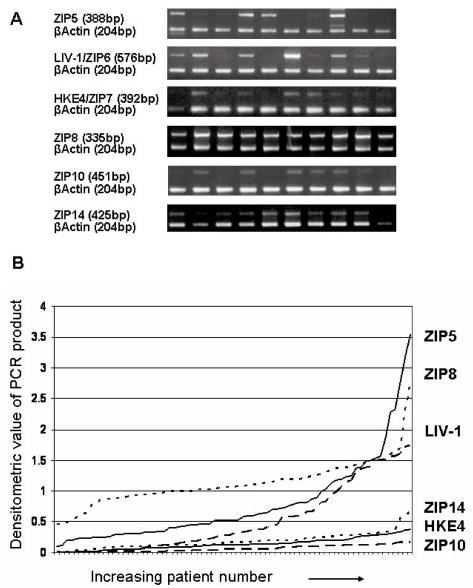
LIV-1 is an estrogen-regulated gene that has been implicated in estrogen-receptor–positive breast cancer and the subsequent spread to the regional lymph nodes (23–26), and more recently it has been used as a reliable marker of luminal A type clinical breast cancer (27,28). We therefore investigated whether any other LIV-1 family members also had a positive association with breast cancer. For this analysis we used a series of tumor samples from 74 patients presenting with primary breast cancer to the Breast Cancer Unit, City Hospital, Nottingham, between 1987 and 1989. For sample analysis we used the same PCR conditions described in Figure 3. The expression of three LIV-1 family members, ZIP4, ZIP12, and ZIP13, was undetectable in these samples, and examples of the expression levels of the remaining six LIV-1 family members are documented in Figure 5 for 10 patients. The graph depicting the variation of each LIV-1 family member with increasing patient number (see Figure 5) demonstrates that LIV-1 and ZIP5 show the most heterogeneity, with samples varying from low to medium and high values. However, it is noteworthy that ZIP5 required an increased number of PCR cycles, suggesting low levels in the breast cancer samples. In contrast, the distribution of HKE4, ZIP8, ZIP10, and ZIP14 across the samples showed little variation.
Figure 5.
Comparison of the expression of LIV-1 family members in a series of samples from breast cancer patients. (A) representative results from 10 patient tumor samples amplified with gene specific primers for ZIP5, HKE4, ZIP10, LIV-1, ZIP8, ZIP14, and β-actin. (B) distribution of the densitometric values for ZIP5, HKE4, ZIP10, LIV-1, ZIP8, and ZIP14 in the patient samples, with no correction for the number of PCR cycles.
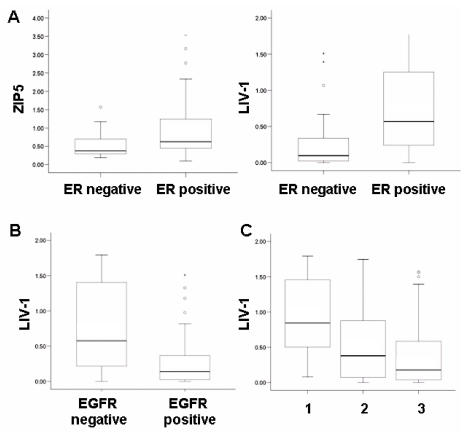
Comparative analysis of densitometric data was performed using the SPSS (version 10) statistical analysis package using either a two-sided Mann-Whitney U test with the previously described cut off values or a Spearman rank correlation test where indicated (Table 3). Statistical significance was assumed if P < 0.05 and is shown for a number of common indicators of breast cancer progression and grade. LIV-1 has previously been identified as an estrogen-regulated gene and a prognostic marker of endocrine response (23,24). Here we confirm this association (P < 0.001, Figure 6A) as well as an inverse relationship to EGFR (P < 0.001, Figure 6B) and a positive association with two other erbB receptor tyrosine kinase members, erbB3 (P = .001, r = 0.414) and erbB4 (P = .028), and another growth factor receptor, IGF1-R (P = .02). No other LIV-1 family member exhibits the same profile as LIV-1; however, ZIP5 (P = .009, see Figure 6A) and ZIP10 (P = .033) both show a positive association with estrogen receptor (ER), although the distribution of ZIP10 in Figure 5B suggests that ZIP10 may be of less clinical relevance. No other LIV-1 family member has an association with EGFR or ErbB4; however, HKE4 (P = .001) and ZIP10 (P < 0.001) both show a positive association with ErbB3. ZIP8 behaves differently from other LIV-1 family members and shows a negative association with both ER (P = .04) and ErbB2 (P = .027). This negative association with ER was also suggested in response of MCF-7 cells to estrogen treatment (see Figure 3) but was not statistically significant. These results together suggest that the different LIV-1 family members may transport zinc, but their activation may be regulated differently, allowing them variation of function in different cell types.
Table 3.
Statistical Analysis of LIV-1 Family Members with Indicators of Breast Cancer Progression
| ZIP5 | LIV-1 | HKE4 | ZIP8 | ZIP10 | ZIP14 | |
|---|---|---|---|---|---|---|
| ER | 0.009 (+) | < 0.001 (+) | NS | 0.04 (−) | 0.033 (+) | NS |
| EGFR | NS | < 0.001 (−) | NS | NS | NS | NS |
| ErbB2a | NS | NS | NS | 0.027 (−) | NS | NS |
| ErbB3a | NS | < 0.001 (+) | 0.001 (+) | NS | < 0.001 (+) | NS |
| ErbB4 | NS | 0.028 (+) | NS | NS | NS | NS |
| IGF1-R | NS | 0.02 (+) | NS | NS | NS | NS |
| STAT3a | NS | 0.007(+) | 0.031(+) | NS | < 0.001(+) | 0.004 (+) |
| Ki 67 | NS | NS | 0.026 (+) | NS | NS | NS |
| Grade | NS | 0.007 (−) | NS | NS | NS | NS |
Comparative analysis of densitometric data assumed statistical significance was assumed if P < 0.05. NS, not significant.
Spearman’s correlation test.
Figure 6.
Comparison of the expression of LIV-1 and/or ZIP5 with ER, EGFR and grade in a series of breast cancer samples. (A) shows the positive correlation of both LIV-1 and ZIP5 with estrogen receptor (ER). (B) shows the reverse relationship of LIV-1 with EGFR. (C) shows the decreasing levels of LIV-1 with worsening grade.
Recently, LIV-1 has been shown to be the downstream target of STAT3 in zebrafish embryos (29), and ZIP14 is regulated by IL-6 (32), a mechanism that uses STAT3 signaling. We have observed a positive correlation in these breast cancer samples between STAT3 and LIV-1 (P = .007), ZIP14 (P = .004), HKE4 (P = .031), and ZIP10 (P = .001). This observation is interesting, especially because the level of STAT3 has been well documented to be associated with breast cancer progression (47).
We next investigated the association of LIV-1 family members with histological grade. LIV-1 was the only family member associated with grade and was highly expressed in low histological grade tumors (P = .007, see Figure 6C). The only other observed relationship between any other LIV-1 family members was a positive association of HKE4 with the well-known proliferation marker Ki67 (P = .026) and those cancers with increased lymph node involvement (P = .036). Although this patient group was small, this latter result was an interesting observation which had previously been documented for LIV-1 (23) and may support a role of HKE4 in proliferating and metastatic tumors.
It appears from this small patient series that LIV-1 is the LIV-1 family member most associated with endocrine response in breast cancer, although HKE4 does have some association with factors suggestive of aggressive behavior such as increased proliferation and lymph node involvement. However, ZIP5 and ZIP10 also appear to be regulated by estrogen within the breast cancer samples, but due to their apparent low levels of expression may not have any useful clinical relevance. Some LIV-1 family members show an association with members of the erbB growth factor family as well as STAT3, an association that may be suggestive of their mechanism of action. Clearly, ZIP5 and ZIP8 are not regulated in the same way. The observed diversity of expression within this LIV-1 family may be indicative of the different role that each molecule has in a variety of different tissues. Zinc has a vital role in cellular processes, and deregulated expression of zinc transporters could have dramatic consequences in the regulation of zinc, which in its turn could be pivotal in the initiation or progression of breast cancer. Investigations to decipher such actions and assess zinc transporters as novel therapeutic targets are currently underway.
ACKNOWLEDGMENTS
The authors wish to thank the Tenovus cancer charity for funding and Mrs Lynne Farrow for help with the statistical analysis.
APPENDIX
Materials and Methods
Cell culture
MCF-7 cells were grown in the presence of 4-hydroxytamoxifen (10−7 M), fulvestrant (10−7 M), or estradiol (10−9M) for seven days in serum growth-factor–free DCCM medium [Biosynergy (Europe), Cambridge, UK]. The production of TamR- and FasR-resistant MCF-7 cells has been described previously (40). The two antiestrogen-resistant cell lines were grown for four days before being transferred into phenol-red/steroid-free, serum growth-factor–free DCCM medium for 24 h prior to harvest.
RNA extraction and RT-PCR
Total RNA was extracted and reverse transcribed to cDNA using random hexamers as previously described (48). The following PCR primers (MWG Biotech) were used to detect the ZIP genes: ZIP4 5′-CCCATCACCA TGGCGTCCCTGG-3′ 5′-GGTGCC CTCG GGGTTGCTGA GG-3′ 330 bp, ZIP5 5′-GGGTGACCTG GAA GAGTCAA –3′ 5′-CAGCAA GGGC CGTAGTAGAC-3′ 388 bp, LIV-1 5′-GTC TAACAG C TCTAGGAGGC-3′ 5′-CACCAATTGC TAGGCCATCG-3′ 576bp, HKE4 5′-ATC GCTCTCT ACTTCAGATC-3′ 5′-CTCTTCTGAA CCCCTC TTG-3′ 392 bp, ZIP8 5′-CCCATCACCA TGGCCCCGGG TCGCGCG-3′ 5′-GGGTGAAAGT TCAATT GCTG TAA-3′ 335 bp, ZIP10 5′-TTGGCAGTTC AA-GAGG GAAAG-3′ 5′-CGA TTATGCT CATACTGT-3′ 451 bp, ZIP12 5′-AAACTTGCCT TCCCCAGACT-3′ 5′-TGAGTGAGAG GCCCTTCTGT-3′ 297 bp, ZIP13 5′-CCCATCACCA TGG CGGGCCC AAG-3′ 5′-GGGAATGACA AGCAACGGGA A-3′ 242 bp, ZIP14 5′-TGCTTGGCTT ATGGAGAACC-3′ 5′-GAGATGACGG TCA CACAGAGG-3′ 425 bp, β-Actin (NM_001101) 5′-GGAGCAATGA TCTTGATC TT-3′ 5′-CCTTCC TGGG CATGGAGTCCT-3′. PCR parameters were optimized accordingly from the previously described protocol (48) using 29 cycles for ZIP4, LIV-1, HKE4, ZIP8, ZIP13, and ZIP14; 31 cycles for ZIP10 and ZIP12; and 33 cycles for ZIP5. All data were normalized with respect to individual β-actin levels and statistical analysis (one-way ANOVA with post hoc Dunnett test) was performed.
Fluorescent microscopy
Recombinant proteins for LIV-1 (15), HKE4 (9), and ZIP14 (17) were engineered in vector pcDNA3.1/V5-His-TOPO to provide a C-terminal V5 tag as previously described. MCF-7 cells were seeded on coverslips for 24 h before transfection with lipofectamine 2000 as described previously (15), and after 16–24 h were prepared for fluorescence microscopy by fixing with 4% formaldehyde, permeabilizing with 0.4% saponin if required, and blocking with 10% normal goat serum before incubating with a mouse anti-V5 antibody (1/2000, Invitrogen) conjugated to Alexa Fluor (1/2000) 594 (red) or 488 (green), and then assembled onto slides using Vectorshield with DAPI (Vector Laboratories). All coverslips were viewed on a Leica RPE automatic microscope using a 63x oil immersion lens. The fluorescent superimposed images were acquired using a multiple bandpass filter set appropriate for DAPI, fluorescein, and Texas Red as well as bright field for differential interference contrast (DIC) imaging.
Tumor samples
Tumor samples were from 74 patients presenting with primary breast cancer to the Breast Cancer Unit, City Hospital, Nottingham, between 1987 and 1989. The samples were snap frozen in liquid nitrogen and stored at −70°C. Clinical and pathological data was provided, and that which was used in the study is presented in Table 2. Total RNA was extracted from each breast tumor sample as previously described (48). RNA was extracted and reverse transcribed to cDNA as described above.
Statistical analysis
Comparative analysis of densitometric data from the breast cancer samples was performed using the SPSS (version 10) statistical analysis package using either a two-sided Mann-Whitney U test or a Spearman rank correlation test with the previously described cutoff values. Statistical significance was assumed if P < 0.05. Immunocytochemical assays for ER, EGFR, and Ki67 were performed as previously reported and the positivity cutoff points used for statistical analysis (49–51). The levels of other growth factors were determined by RT-PCR and quantified as described previously (52).
Footnotes
Online address: http://www.molmed.org
REFERENCES
- 1.Vallee BL, Auld DS. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–59. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- 2.Vallee BL, Falchuk KH. The biochemical basis of zinc physiology. Physiological Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 3.Truong-Tran AQ, Carter J, Ruffin RE, Zalewski PD. The role of zinc in Caspase activation and apoptotic cell death. Biometals. 2001;14:315–30. doi: 10.1023/a:1012993017026. [DOI] [PubMed] [Google Scholar]
- 4.Koh JY, Suh SW, Gwag BJ, He YY, Hsu CY, Choi DW. The role of zinc in selective neuronal death after transient global cerebral ischemia. Science. 1996;272:1013–6. doi: 10.1126/science.272.5264.1013. [DOI] [PubMed] [Google Scholar]
- 5.Palmiter RD, Huang L. Efflux and compartmentalization of zinc by members of the SLC30 family of solute carriers. Pflugers Arch. 2004;447:744–51. doi: 10.1007/s00424-003-1070-7. [DOI] [PubMed] [Google Scholar]
- 6.Eide DJ. The SLC39 family of metal ion transporters. Pflugers Arch. 2004;447:796–800. doi: 10.1007/s00424-003-1074-3. [DOI] [PubMed] [Google Scholar]
- 7.Taylor KM, Nicholson RI. The LZT proteins; the new LIV-1 subfamily of ZIP zinc transporters. BBA Biomembranes. 2003;1611:16–30. doi: 10.1016/s0005-2736(03)00048-8. [DOI] [PubMed] [Google Scholar]
- 8.Gaither LA, Eide DJ. Eukaryotic zinc transporters and their regulation. Biometals. 2001;14:251–70. doi: 10.1023/a:1012988914300. [DOI] [PubMed] [Google Scholar]
- 9.Taylor KM, Morgan HE, Johnson A, Nicholson RI. Structure-function analysis of HKE4, a member of the new LIV-1 subfamily of zinc transporters. Biochem J. 2003;377:131–9. doi: 10.1042/BJ20031183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerinot ML. The ZIP family of metal transporters. Biochim Biophys Acta. 2000;1465:190–8. doi: 10.1016/s0005-2736(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 11.Dalton TP, He L, Wang B, et al. Identification of mouse SLC39A8 as the transporter responsible for cadmium-induced toxicity in the testis. Proc Natl Acad Sci U S A. 2005;102:3401–6. doi: 10.1073/pnas.0406085102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liuzzi JP, Aydemir F, Nam H, Knutson MD, Cousins RJ. Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc Natl Acad Sci U S A. 2006;103:13612–7. doi: 10.1073/pnas.0606424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim BE, Wang F, Dufner-Beattie J, Andrews GK, Eide DJ, Petris MJ. Zn2+-stimulated endocytosis of the mZIP4 zinc transporter regulates its location at the plasma membrane. J Biol Chem. 2004;279:4523–30. doi: 10.1074/jbc.M310799200. [DOI] [PubMed] [Google Scholar]
- 14.Wang F, Kim BE, Petris MJ, Eide DJ. The mammalian Zip5 protein is a zinc transporter that localizes to the basolateral surface of polarized cells. J Biol Chem. 2004;279:51433–41. doi: 10.1074/jbc.M408361200. [DOI] [PubMed] [Google Scholar]
- 15.Taylor KM, Morgan HE, Johnson A, Nicholson RI. Structure-function analysis of LIV-1, the breast cancer associated protein that belongs to a new subfamily of zinc transporters. Biochem J. 2003;375:51–9. doi: 10.1042/BJ20030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tominaga K, Kagata T, Johmura Y, Hishida T, Nishizuka M, Imagawa M. SLC39A14, a LZT protein, is induced in adipogenesis and transports zinc. FEBS J. 2005;272:1590–9. doi: 10.1111/j.1742-4658.2005.04580.x. [DOI] [PubMed] [Google Scholar]
- 17.Taylor KM, Morgan HE, Johnson A, Nicholson RI. Structure-function analysis of a novel member of the LIV-1 subfamily of zinc transporters, ZIP14. FEBS Letters. 2005;579:427–32. doi: 10.1016/j.febslet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Huang L, Kirschke CP, Zhang Y, Yu YY. The ZIP7 gene (Slc39a7) encodes a zinc transporter involved in zinc homeostasis of the Golgi apparatus. J Biol Chem. 2005;280:15456–63. doi: 10.1074/jbc.M412188200. [DOI] [PubMed] [Google Scholar]
- 19.Begum NA, Kobayashi M, Moriwaki Y, Matsumoto M, Toyoshima K, Seya T. Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics. 2002;80:630–45. doi: 10.1006/geno.2002.7000. [DOI] [PubMed] [Google Scholar]
- 20.Kumanfasrovics A, Poruk KE, Osborn KA, Ward DM, Kaplan J. YKE4 (YIL023C) encodes a bidirectional zinc transporter in the endoplasmic reticulum of Saccharomyces cerevisiae. J Biol Chem. 2006;281:22566–74. doi: 10.1074/jbc.M604730200. [DOI] [PubMed] [Google Scholar]
- 21.Kury S, Dreno B, Bezieau S, Giraudet S, Kharfi M, Kamoun R, Moisan JP. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat Genet. 2002;31:239–40. doi: 10.1038/ng913. [DOI] [PubMed] [Google Scholar]
- 22.Huang ZL, Dufner-Beattie J, Andrews GK. Expression and regulation of SLC39A family zinc transporters in the developing mouse intestine. Dev Biol. 2006;295:571–9. doi: 10.1016/j.ydbio.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 23.Manning DL, Robertson JFR, Ellis IO, et al. Estrogen-regulated genes in breast-cancer: association of pliv1 with lymph-node involvement. Eur J Cancer. 1994;30A:675. doi: 10.1016/0959-8049(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 24.Manning DL, McClelland RA, Knowlden JM, et al. Differential expression of estrogen-regulated genes in breast-cancer. Acta Oncologica. 1995;34:641–6. doi: 10.3109/02841869509094041. [DOI] [PubMed] [Google Scholar]
- 25.Tozlu S, Girault I, Vacher S, et al. Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription-PCR approach. Endocr Relat Cancer. 2006;13:1109–20. doi: 10.1677/erc.1.01120. [DOI] [PubMed] [Google Scholar]
- 26.Schneider J, Ruschhaupt M, Buness A, et al. Identification and meta-analysis of a small gene expression signature for the diagnosis of estrogen receptor status in invasive ductal breast cancer. Int J Cancer. 2006;119:2974–9. doi: 10.1002/ijc.22234. [DOI] [PubMed] [Google Scholar]
- 27.Chung CH, Bernard PS, Perou CM. Molecular portraits and the family tree of cancer. Nat Genet. 2002;32:533–40. doi: 10.1038/ng1038. [DOI] [PubMed] [Google Scholar]
- 28.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita S, Miyagi C, Fukada T, Kagara N, Che YS, Hirano T. Zinc transporter LIVI controls epithelial-mesenchymal transition in zebrafish gastrula organizer. Nature. 2004;429:298–302. doi: 10.1038/nature02545. [DOI] [PubMed] [Google Scholar]
- 30.Taylor KM, Hiscox S, Nicholson RI. Zinc transporter LIV-1: a link between cellular development and cancer progression. Trends Endocrinol Metab. 2004;15:461–3. doi: 10.1016/j.tem.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Mathews WR, Ong D, Milutinovich AB, Van Doren M. Zinc transport activity of Fear of Intimacy is essential for proper gonad morphogenesis and DE-cadherin expression. Development. 2006;133:1143–53. doi: 10.1242/dev.02256. [DOI] [PubMed] [Google Scholar]
- 32.Liuzzi JP, Lichten LA, Rivera S, et al. In-terleukin-6 regulates the zinc transporter Zip14 in liver and contributes to the hypozincemia of the acute-phase response. Proc Natl Acad Sci U S A. 2005;102:6843–8. doi: 10.1073/pnas.0502257102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang CJ, Murgia C, Leong M, et al. Anti-inflammatory effects of zinc and alterations in zinc transporter mRNA in mouse models of allergic inflammation. Am J Physiol Lung Cell Mol Physiol. 2006 doi: 10.1152/ajplung.00280.2006. In press. [DOI] [PubMed] [Google Scholar]
- 34.Bly M. Examination of the zinc transporter gene, SLC39A12. Schizophr Res. 2006;81:321–2. doi: 10.1016/j.schres.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 35.Chowanadisai W, Kelleher SL, Lonnerdal B. Zinc deficiency is associated with increased brain zinc import and LIV-1 expression and decreased ZnT-1 expression in neonatal rats. J Nutr. 2005;135:1002–7. doi: 10.1093/jn/135.5.1002. [DOI] [PubMed] [Google Scholar]
- 36.Wang K, Zhou B, Kuo YM, Zemansky J, Gitschier J. A novel member of a zinc transporter family is defective in acrodermatitis enteropathica. Am J Hum Genet. 2002;71:66–73. doi: 10.1086/341125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F, Kim BE, Dufner-Beattie J, Petris MJ, Andrews G, Eide DJ. Acrodermatitis enteropathica mutations affect transport activity, localization and zinc-responsive trafficking of the mouse ZIP4 zinc transporter. Hum Mol Genet. 2004;13:563–71. doi: 10.1093/hmg/ddh049. [DOI] [PubMed] [Google Scholar]
- 38.Nicholson RI, Johnston SR. Endocrine therapy: current benefits and limitations. Breast Cancer Res Treat. 2005;93(Suppl 1):S3–10. doi: 10.1007/s10549-005-9036-4. [DOI] [PubMed] [Google Scholar]
- 39.Knowlden JM, Hutcheson IR, Jones HE, et al. Elevated levels of EGFR/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in Tamoxifen-resistant MCF-7 cells. Endocrinology. 2003;144:1032–44. doi: 10.1210/en.2002-220620. [DOI] [PubMed] [Google Scholar]
- 40.McClelland RA, Barrow D, Madden TA, et al. Enhanced epidermal growth factor receptor signaling in MCF7 breast cancer cells after long-term culture in the presence of the pure antiestrogen ICI 182,780 (Faslodex) Endocrinology. 2001;142:2776–88. doi: 10.1210/endo.142.7.8259. [DOI] [PubMed] [Google Scholar]
- 41.Hiscox S, Morgan L, Green TP, Barrow D, Gee J, Nicholson RI. Elevated Src activity promotes cellular invasion and motility in tamoxifen resistant breast cancer cells. Breast Cancer Res Treat. 2006;97:263–74. doi: 10.1007/s10549-005-9120-9. [DOI] [PubMed] [Google Scholar]
- 42.Knowlden JM, Hutcheson IR, Barrow D, Gee JM, Nicholson RI. Insulin-like growth factor-I receptor signaling in tamoxifen-resistant breast cancer: a supporting role to the epidermal growth factor receptor. Endocrinology. 2005;6:4609–18. doi: 10.1210/en.2005-0247. [DOI] [PubMed] [Google Scholar]
- 43.Hiscox S, Jordan NJ, Jiang W, Harper M, McClelland R, Smith C, Nicholson RI. Chronic exposure to fulvestrant promotes over-expression of the c-Met receptor in breast cancer cells: implications for tumor-stroma interactions. Endocr Relat Cancer. 2006;13:1085–99. doi: 10.1677/erc.1.01270. [DOI] [PubMed] [Google Scholar]
- 44.Hiscox S, Morgan L, Barrow D, Dutkowski C, Wakeling A, Nicholson RI. Tamoxifen resistance in breast cancer cells is accompanied by an enhanced motile and invasive phenotype: inhibition by gefitinib (‘Iressa’, ZD1839) Clin Exp Metastasis. 2004;21:201–12. doi: 10.1023/b:clin.0000037697.76011.1d. [DOI] [PubMed] [Google Scholar]
- 45.Kuske B, Naughton C, Moore K, et al. Endocrine therapy resistance can be associated with high estrogen receptor {alpha} (ER{alpha}) expression and reduced ER{alpha} phosphorylation in breast cancer models. Endocr Relat Cancer. 2006;13:1121–33. doi: 10.1677/erc.1.01257. [DOI] [PubMed] [Google Scholar]
- 46.Taylor KM, Vichova P, Hiscox S, Nicholson RI. Zinc-dependant stimulation of Src, EGFR and IGFR signaling pathways in tamoxifen-resistant breast cancer and the role of zinc transporters. Breast Cancer Res Treat. 2005;94:S162. [Google Scholar]
- 47.Gamero AM, Young HA, Wiltrout RH. Inactivation of Stat3 in tumor cells: releasing a brake on immune responses against cancer? Cancer Cell. 2004;5:111–2. doi: 10.1016/s1535-6108(04)00028-5. [DOI] [PubMed] [Google Scholar]
- 48.Knowlden JM, Gee JM, Bryant S, et al. Use of reverse transcription-polymerase chain reaction methodology to detect estrogen-regulated gene expression in small breast cancer specimens. Clin Cancer Res. 1997;3:2165–72. [PubMed] [Google Scholar]
- 49.Nicholson RI, McClelland RA, Gee JM, et al. Epidermal growth factor receptor expression in breast cancer: association with response to endocrine therapy. Breast Cancer Res Treat. 1994;29:117–25. doi: 10.1007/BF00666187. [DOI] [PubMed] [Google Scholar]
- 50.Snead DR, Bell JA, Dixon AR, Nicholson RI, Elston CW, Blamey RW, Ellis IO. Methodology of immunohistological detection of oestrogen receptor in human breast carcinoma in formalin-fixed, paraffin-embedded tissue: a comparison with frozen section methodology. Histopathology. 1993;23:233–8. doi: 10.1111/j.1365-2559.1993.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 51.van Dierendonck JH, Keijzer R, van de Velde CJ, Cornelisse CJ. Nuclear distribution of the Ki-67 antigen during the cell cycle: comparison with growth fraction in human breast cancer cells. Cancer Res. 1989;49:2999–3006. [PubMed] [Google Scholar]
- 52.Knowlden JM, Gee JM, Seery LT, Farrow L, Gullick WJ, Ellis IO, Blamey RW, Robertson JF, Nicholson RI. c-erbB3 and c-erbB4 expression is a feature of the endocrine responsive phenotype in clinical breast cancer. Oncogene. 1998;17:1949–57. doi: 10.1038/sj.onc.1202107. [DOI] [PubMed] [Google Scholar]