Abstract
An emerging family of cell surface inhibitory receptors is characterized by the presence of intracytoplasmic immunoreceptor tyrosine-based inhibition motifs (ITIM). These ITIM-bearing inhibitory receptors, which are typically paired with activating isoforms, associate with Src homology domain 2-containing phosphatases following ITIM tyrosine phosphorylation. Two categories of phosphatases are recruited by the ITIM-bearing receptors: the protein-tyrosine phosphatases, SHP-1 and SHP-2, and the polyphosphate inositol 5-phosphatase, SHIP. The dynamic equilibrium of B cell activation is partially controlled by two well known ITIM-bearing receptors, CD22 and FcγRIIB, a low affinity receptor for IgG. We describe here that a murine ITIM-bearing molecule, PIR-B, can also negatively regulate B cell activation. Tyrosine-phosphorylated ITIMs allow PIR-B to associate with SHP-1 but not with SHIP. Engagement of PIR-B thereby initiates a SHP-1-dependent inhibitory pathway that may play an important role in regulating B lymphocyte activation.
The regulation of cell activation involves a dynamic equilibrium between activating and inhibitory signals. Intracytoplasmic immunoreceptor tyrosine-based inhibition motif (ITIM)-bearing receptors that are defined by one or more consensus ITIM (I/L/V/S)XYXX(L/V) in their intracytoplasmic domain mediate an inhibitory function through recruitment of Src homology domain 2 (SH2)-containing intracytoplasmic phosphatases (1, 2). The phosphatases recruited by the tyrosine-phosphorylated form of ITIMs belong to one of two classes: the SHP-1 and SHP-2 protein-tyrosine phosphatases or the polyphosphate inositol 5-phosphatase SHIP (3). ITIM-bearing receptors belong to a large family of molecules with more than 30 distinct members (1, 4). Except for the IgG receptor, FcγRIIB, and the type C lectins, human NKG2A/B and mouse Ly-49s, all ITIM-bearing molecules are encoded by Ig superfamily genes present on human chromosome 19q13 and its syntenic region on mouse chromosome 7. The genes for the Ig-like ITIM-bearing molecules share a conserved organization in which ITIMs are encoded by a single exon preceded by a type 0 intron (5). ITIM-bearing receptors are characteristically paired with activating receptors, which share highly similar extracellular domains but harbor a shorter cytoplasmic domain devoid of ITIM. The activating receptor isoforms are not alternative spliced products of the ITIM-bearing receptor genes but are instead encoded by separate genes. The activating receptor isoforms have a charged amino acid residue in their transmembrane domain, which may allow their association with signal-transducing proteins, as demonstrated recently for killer cell-activating receptors (6, 7).
The paired Ig-like receptors (PIR) recently identified on murine B cells and myeloid-lineage cells include PIR-A molecules, with a short cytoplasmic domain, and PIR-B molecules, which bear a long cytoplasmic domain (178 amino acids) that contains three potential ITIMs centered on tyrosine residues Tyr-713 (SLYASV), Tyr-794 (VTYAQL), and Tyr-824 (SVYATL) (8). The six Ig-like domains in the extracellular region of mouse PIR-B share sequence homology with the two to four Ig-like domain-containing molecules, ILT-2/LIR-1, LIR-2, LIR-3, ILT-3/LIR-5, and LIR-8 (9–11), which may represent their human homologues. In this report, we characterize the inhibitory features of the PIR-B molecules.
MATERIALS AND METHODS
Antibodies.
Rat monoclonal antibodies (mAbs), 10.4 and 6C1, specific for PIR-A and PIR-B molecules (H.K., C.C.C., and M.D.C., unpublished work) were digested with pepsin before passage over a protein-G column to separate the F(ab′)2 fragments, which were then biotinylated. Other mAbs included a mouse anti-2,4,6-trinitrophenyl IgE mAb (2682–1), a rat anti-2,4,6-trinitrophenyl IgE mAb (LO-DNP 30) (12), a mouse anti-rat FcɛRIα mAb (BC4, γ1κ; the gift of R. Siraganian, National Institutes of Health, Bethesda, MD), a rat anti-mouse FcγRII/III mAb (2.4G2; γ2bκ), and a murine anti-phosphotyrosine mAb (4G10; Upstate Biotechnology, Lake Placid, NY). Polyclonal antibodies included F(ab′)2 fragments of goat anti-mouse Ig antibody (GAM), F(ab′)2 fragments of donkey anti-mouse Ig antibodies lacking cross-reactivity to rat Ig (DAM), F(ab′)2 fragments of donkey anti-rat Ig antibodies lacking cross-reactivity to mouse Ig (DAR) (Immunotech, Luminy, France), and biotin-labeled F(ab′)2 fragments of goat anti-mouse μ-chain antibodies (Southern Biotechnology Associates).
Peptides.
Mouse PIR-B and FcγRIIBI peptides containing an N-terminal-biotin were synthesized in phosphorylated and nonphosphorylated forms (Neosystem, Strasbourg, France): CD3ɛ, KEPPPVPNPDYEPIRK; FcγRIIBI, KTEAENTITYSLLK; PIR-B Tyr-713, AAATQEESLYASVED; PIR-B Tyr-742, PEEDPQGETYAQVKP; PIR-B Tyr-770, MSREQLNTEYEQAEE; PIR-B Tyr-794, ESGESQDVTYAQL; PIR-B Tyr-824, GEAPEEPSVYATLAA.
cDNA Constructs.
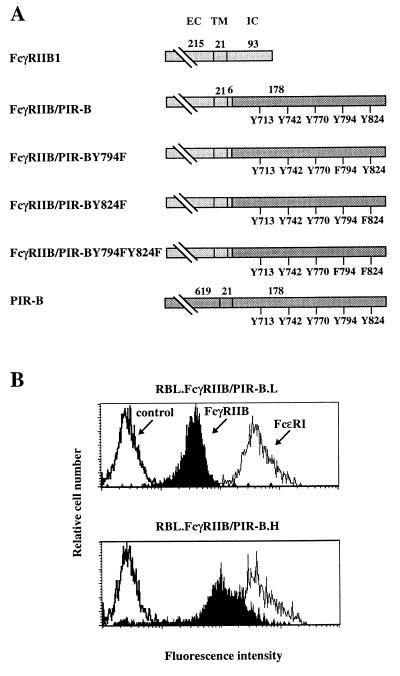
The cDNA constructs were generated from a pNT-Neo plasmid including a chimeric cDNA encoding the extracellular, transmembrane, and the first six amino acids (KKKQVP) of the intracellular region of murine FcγRIIB1 fused with the intracellular region of mouse PIR-B (FcγRIIB/PIR-B) obtained by PCR amplification of the pBluescript-7M5 plasmid (8). The primers generated 5′ KpnI and 3′ SacI sites, allowing in frame ligation with the FcγRIIB1 sequence in pNT-Neo plasmid. Point mutation constructs were generated by PCR-directed mutagenesis of the FcγRIIB/PIR-B construct. Each point mutation involved a tyrosine replacement by phenylalanine. The tyrosine at position 794 in PIR-B was mutated in the FcγRIIB/PIR-BY794F mutant, the tyrosine at position 824 in the FcγRIIB/PIR-BY824F mutant, and both tyrosines in the FcγRIIB/PIR-BY794F,Y824F mutant (Fig. 2A). Fidelity of the constructs was verified by sequencing (Genome Express, Grenoble, France).
Figure 2.
Expression of FcγRIIB/PIR-B chimeric molecules by transfected rat basophil leukemia (RBL) cells. (A) Schematic representation of chimeric molecules. The FcγRIIB1 sequence is indicated in light gray, and the PIR-B sequences in dark gray. Numbers over the bars indicate amino acids included in each region of the protein (EC, extracellular; TM, transmembrane; and IC, intracellular). Tyrosines in the cytoplasmic regions of the chimeric molecules are numbered according to the wild-type PIR-B sequence. Tyrosine (Y) to phenylalanine (F) mutations are indicated for each point mutant construction. (B) Cell surface expression of chimeric molecules by RBL transfectants was assayed by indirect immunofluorescence staining with the 2.4G2 anti-FcγRII/III mAb (filled profile). FcɛRI expression was revealed by staining with the BC4 anti-FcɛR mAb (thin line), and irrelevant isotype-matched mAbs served as negative controls (bold lines). Flow cytometric analysis of the FcγRIIB/PIR-B.L (2A10, Upper) and FcγRIIB/PIR-B.H (2F7, Lower) clones indicates expression of the FcγRIIB/PIR-B chimeric molecule at relatively low and high densities, respectively.
Cell Transfection.
RBL-2H3 cells cultured in DMEM supplemented with 10% fetal calf serum, 100 international units/ml penicillin, and 100 μg/ml streptomycin (complete medium) were transfected by electroporation (12). Stable transfectants were established by culture in the presence of G418 (1 mg/ml), and representative clones were selected for further investigation.
Calcium Mobilization Analysis.
Splenocytes from BALB/c mice were incubated for 30 min at 37°C in HBSS (5 × 106 cells per ml) containing 5% fetal calf serum and 1 μg/ml Indo-1/AM (Calbiochem). Cells held at room temperature for 5 min were resuspended either in HBSS containing 5% fetal calf serum or calcium-free HBSS containing 2 mM EGTA (106 cells per ml). The 395/520 nm fluorescence ratio for viable cells preloaded with Indo-1/AM dye was determined with a FACStar Plus flow cytometer equipped with an argon ion laser operated in UV mode (Becton Dickinson). After establishing the baseline, 10 μg of biotin-labeled F(ab′)2 goat anti-mouse μ and/or F(ab′)2 anti-PIR mAbs were added to the dye-loaded cells, followed by the addition of Neutralite avidin 10 μg (SBA) as a cross-linker. Data represent ≈80,000 events collected over 5–8 min.
Serotonin Release.
RBL-2H3 cell transfectants were examined for serotonin release as described (12). Briefly, cells were incubated at 37°C for 1 h with 2 mCi/ml [3H]serotonin (Amersham), washed, incubated for another hour at 37°C, washed again, and distributed in 96-well microculture plates at 2 × 105 cells/well. Cells were then incubated for 1 h with IgE and the 2.4G2 anti-FcγR antibody in a final volume of 50 μl, washed, and then challenged for 30 min at 37°C with 25 μl of prewarmed GAM, DAM, or DAR F(ab′)2 fragments. Reactions were stopped by adding 50 μl of ice-cold medium and by placing plates on ice. [3H]Serotonin content in the supernatants was assayed as described (12).
Fusion Protein Analysis by Surface Plasmon Resonance.
Surface plasmon resonance measurements were made with a BIAcore apparatus (BIAcore) using GST-SHP 1.SH2(NC), GST-SHP 2.SH2(NC), and GST-SHIP.SH2 fusion proteins generated from murine phosphatase cDNAs and purified from DH5α lysates as previously described (13). Before assay, fusion proteins were dialyzed in pH 7.4 HBS buffer (10 mM Hepes/150 mM NaCl/3.4 mM EDTA). Protein purity was assessed by SDS-12.5% PAGE and Coomassie Blue staining. HBS buffer supplemented with 0.05% surfactant P20 was used as the injection medium in BIAcore experiments employing biotinylated peptides immobilized on streptavidin microchips (13).
Metabolic Cell Labeling and Assay for Peptide-Binding Proteins.
IIA.1.6 murine B cells (2.5 × 108) were labeled using 3 mCi of Tran35S-label ([35S]methionine and [35S]cysteine) and 1 mCi of [35S]cysteine (ICN) and incubated overnight at 37°C as described (14). Briefly, after lysis, samples were precleared with control peptide (nonphosphorylated CD3ɛ ITAM peptide) immobilized on streptavidin-agarose beads (Sigma) prior to analysis in the cell lysate adsorption assay. Indicated biotinylated peptides (5 μg) were coupled to 50-μl streptavidin-agarose slurry beads for 1 hr at 4°C prior to bead saturation with D-biotin (1 mg/ml) for 1 h at 4°C. Cell lysates were incubated with peptide-coated beads for 2 h at 4°C, and after three washes in lysis buffer, samples were fractionated on SDS-8% PAGE under reducing conditions.
Immunoprecipitation and Immunobloting.
FcγRIIB/PIR-B.H transfectants of RBL-2H3 cells were stimulated with mouse IgE (1/10 final dilution of hybridoma supernatant) in complete medium in the presence or absence of F(ab′)2 fragments of the 2.4G2 mAb (3 μg/ml) before washing and stimulation for 2 min at 37°C with 20 μg/ml F(ab′)2 fragments of GAM. Immunoprecipitation and immunoblotting of proteins in the cell lysates were performed as described in ref. 15.
RESULTS
PIR-Mediated Inhibition of B Cell Activation.
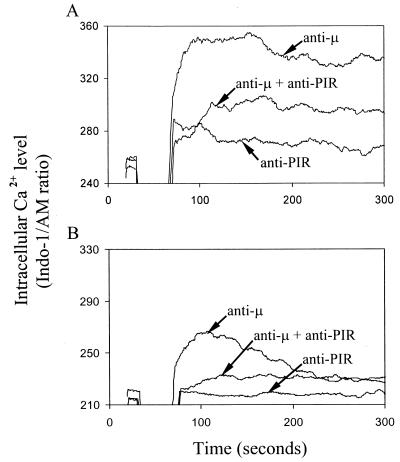
Splenic B cells express both PIR-A and PIR-B molecules. To determine the effects of PIR-A and PIR-B cross-linkage on B cell responses that are initiated by B cell receptor (BCR) ligation, we employed F(ab′)2 fragments of anti-μ and anti-PIR mAbs that react with epitopes common to both the putatively activating PIR-A and inhibitory PIR-B molecules. Whereas ligation of the PIR-A and PIR-B molecules alone had no demonstrable effect on intracellular Ca2+ levels, co-ligation of the PIR molecules with the BCR effectively inhibited the BCR-mediated Ca2+ mobilization response (Fig. 1A). When this experiment was repeated in calcium-free media, efficient inhibition of the BCR-triggered release of intracellular Ca2+ was again observed when the BCR and PIR molecules were co-ligated (Fig. 1B). These results suggest a dominant inhibitory role for the PIR-B molecule that may be mediated via phosphorylation of its candidate ITIM motifs.
Figure 1.
Inhibition of BCR-induced Ca2+ mobilization by coligation with PIR molecules. Mouse splenic B cells loaded with INDO-1 dye were analyzed by flow cytometry for intracellular Ca2+ levels in the presence (A) or absence (B) of extracellular Ca2+. Cells were stimulated with biotinylated F(ab′)2 fragments of anti-μ and anti-PIR mAbs either alone or in combination as indicated by arrows. Biotin (10 μg) was used as a cross-linker. Independent ligation with the anti-PIR mAb had no inhibitory effect on the anti-μ induced Ca2+ response (not shown).
Engagement of an FcγRIIB/PIR-B Chimeric Molecule Extinguishes RBL-2H3 Cell Activation Induced by FcɛRI Cross-Linking.
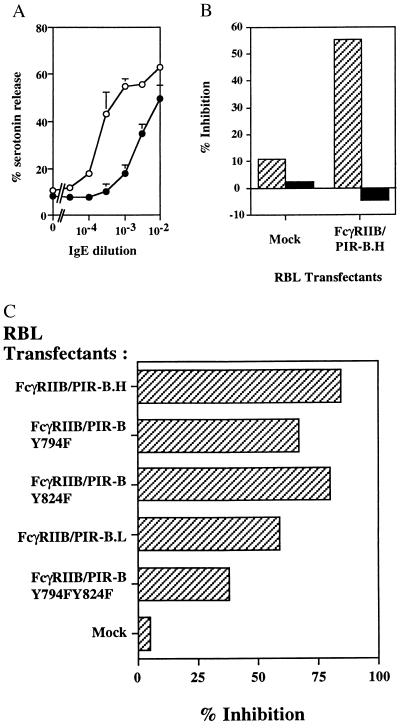
To dissect the structural basis for the PIR-B inhibitory function, we generated chimeric molecules composed of the extracellular and transmembrane domains of murine FcγRIIB1 fused to the entire intracytoplasmic domain of PIR-B (Fig. 2A). Stable transfectants of the rat basophilic leukemia cell line, RBL-2H3, were generated using this chimeric construct, and cell surface immunofluorescence analysis indicated that transfected RBL cells express FcγRIIB/PIR-B chimeric molecules at different levels, whereas expression of the endogenous FcɛRI complex remains at the same level. Two clones, RBL FcγRIIB/PIR-B.H and RBL FcγRIIB/PIR-B.L, which express the FcγRIIB/PIR-B chimeric molecule at relatively high and low densities, respectively (Fig. 2B), were analyzed in a standard serotonin release assay, involving stimulation via the endogenous rat FcɛRI complex by IgE and engagement of the FcγRIIB/PIR-B chimeric molecule by the mouse 2.4G2 anti-FcγRIIB mAb. Consistent with the inhibitory function of endogenous PIR molecules on splenic B cells (Fig. 1), co-aggregation of FcγRIIB/PIR-B with the FcɛRI complex led to an inhibition of IgE-induced serotonin release for both RBL FcγRIIB/PIR-B.H and RBL FcγRIIB/PIR-B.L cells, whereas no effect was observed for mock-transfected RBL cells (Fig. 3). The inhibitory function mediated by the PIR-B cytoplasmic domain was dependent upon the dosage of IgE used to induce FcɛRI cross-linkage (Fig. 3A). The inhibitory function of the PIR-B fusion protein was profound at suboptimal concentrations of IgE, being 56–77% between 10−3 and 10−4 IgE dilutions, and was reduced at optimal and supraoptimal IgE concentrations, being 21–38% between 10−2 and 3 × 10−3 IgE dilutions. Using mouse and rat IgE, both of which bind to the FcɛRI complex on RBL-2H3 cells, we investigated whether co-aggregation of FcɛRI and FcγRIIB/PIR-B molecules is required for the inhibition. As shown in Fig. 3B, the inhibitory function was observed only when FcɛRI and the FcγRIIB/PIR-B fusion protein were brought into close proximity; no inhibition was observed when FcɛRI and FcγRIIB/PIR-B were independently aggregated. The inhibitory function of FcγRIIB/PIR-B thus implicates the cytoplasmic domain of PIR-B in the inhibition of immunoreceptor tyrosine-based activation motif (ITAM)-dependent signals initiated via the FcɛRI complex on basophilic cells and via the BCR complex on B cells.
Figure 3.
Inhibition of FcɛRI-dependent cell activation by co-engagement of FcγRIIB/PIR-B chimeric molecules. (A) Assay of serotinin release by FcγRIIB/PIR-B.H cells (•) and mock transfectant cells (○). Cells were stimulated with varying dilutions of mouse IgE-producing hybridoma supernatants and the 2.4G2 anti-FcγRII/III mAb (20 μg/ml). Wild-type RBL cells yielded similar results to the mock RBL transfectant (not shown). Results expressed as mean ± SD of three independent experiments. (B) Analysis of serotonin release after co-aggregation (striped bars) versus independent aggregation (black bars) of FcγRIIB/PIR-B chimeric molecules and FcɛRI. Co-aggregation was induced by incubation of the cells with the 2.4G2 anti-FcγRII/III mAb (20 μg/ml), mouse IgE (10−3 dilution), and F(ab′)2 GAM (50 μg/ml). Independent aggregation was induced by incubation of the cells with the 2.4G2 mAb (20 μg/ml) plus F(ab′)2 DAM (100 μg/ml) and rat IgE plus F(ab′)2 DAR (100 μg/ml). Percentage inhibition was defined as the serotonin release (in cpm) induced by mouse IgE stimulation and 2.4G2 minus the background release divided by the serotonin stimulation induced by mouse IgE stimulation minus the background release. Results are representative of three independent experiments. (C) Comparative analysis of the inhibitory functions of FcγRIIB/PIR-B mutants versus wild-type chimeras. Inhibition percentages were calculated according to the above formula. FcγRIIB/PIR-B.H, FcγRIIB/PIR-B.Y794F, and FcγRIIB/PIR-B.Y824F transfected RBL cells expressed comparative levels of chimeric molecule; FACS analysis indicated mean fluorescence intensities for 2.4G2 anti-FcγR staining of 138, 165, and 89 for FcγRIIB/PIR-B.H, FcγRIIB/PIR-B.Y794F, and FcγRIIB/PIR-B.Y824F, respectively. FcγRIIB/PIR-B.L and FcγRIIB/PIR-BY794F,Y824F have similar but lower 2.4G2 fluorescence staining intensities, being 59 and 44, respectively, for FcγRIIB/PIR-B.L and FcγRIIB/PIR-B.Y794F,Y824F. The difference in inhibition exerted by FcγRIIB/PIR-B versus the double mutant was confirmed in two additional experiments (P < 0.01).
Definition of PIR-B as an ITIM-Bearing Molecule That Recruits SHP-1 and SHP-2 Protein-Tyrosine Phosphatases.
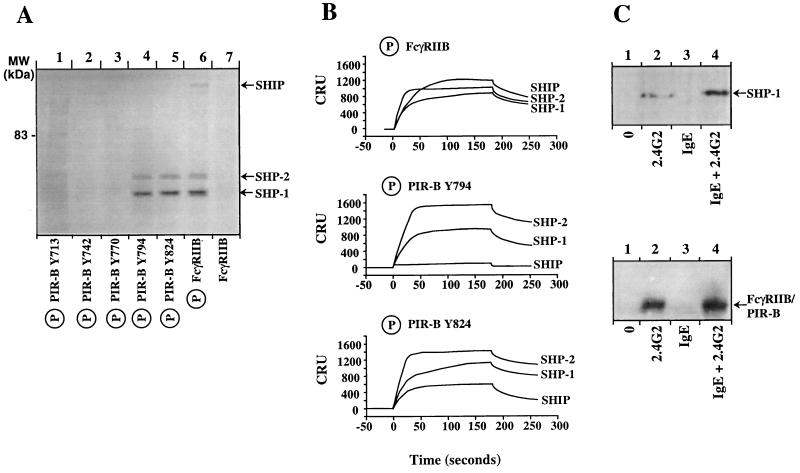
Among the five tyrosine residues in the PIR-B cytoplasmic domain (Tyr-713, Tyr-742, Tyr-770, Tyr-794, and Tyr-824), only Tyr-713, Tyr-794, and Tyr-824 conform to the ITIM (I/L/V/S)XYXX(L/V) consensus motif (1, 2). ITIMs may associate with either the protein-tyrosine phosphatases, SHP-1 and SHP-2, or the polyphosphate inositol 5-phosphatase, SHIP. When synthetic phosphopeptides corresponding to 15-amino acid stretches (including each of the PIR-B intracytoplasmic tyrosine residues) were used to adsorb lysates prepared from 35S metabolically labeled B cells (Fig. 4A), two prominent protein bands of 64 and 68 kDa were found to bind selectively to the synthetic peptides that included the phosphorylated form of Tyr-794 and Tyr-824. No consistent association was detected with nonphosphorylated peptides or phosphorylated forms of the Tyr-742 and Tyr-770 peptides, and only minimal binding of 68 and 85 kDa proteins were detected for the phosphorylated peptide corresponding to Tyr-713. The 64- and 68-kDa proteins comigrated with SHP-1 and SHP-2 bands previously shown to associate with the phosphorylated FcγRIIB1 ITIM peptide (16, 17). No protein association was observed for the nonphosphorylated forms of FcγRIIB (Fig. 4A) or recombinant PIR-B (data not shown). Importantly, none of the phosphorylated PIR-B peptides could be shown to associate with SHIP (≈145 kDa), a finding that contrasts with the binding of SHIP to the phosphorylated form of FcγRIIB (Fig. 4A).
Figure 4.
Analysis of phosphatase binding of candidate ITIMs in the PIR-B cytoplasmic domain (A) PIR-B tyrosine-containing peptides were incubated with lysates prepared from metabolically 35S-labeled IIA1.6 B cells, and adsorbed molecules were separated under reducing conditions on an 8% SDS-PAGE. (B) Binding potential of phosphorylated PIR-B peptides to the GST-SHP 1.SH2(NC), GST-SHP 2.SH2(NC), and GST-SHIP.SH2 GST fusion proteins (150 nM) was analyzed with a BIAcore apparatus. The flow rate was constant at 10 μl/min. In this experiment, 80 resonance units of phosphorylated peptides were immobilized on streptavidin-coated sensorchips. The regeneration was performed using HBS buffer supplemented with 0.02% SDS. Results expressed as corrected resonance units (CRU) corresponding to raw values after subtraction of background RU values due to the injection medium. (C) Analysis of tyrosine phosphorylation induced by co-aggregation and independent aggregation of FcɛRI and the FcγRIIB/PIR-B chimera. Cell lysates of transfected RBL cells untreated (lane 1) or treated with the 2.4G2 anti-FcγR mAb (lane 2), mouse IgE (lane 3), or both 2.4G2 and mouse IgE (lane 4) followed by F(ab′)2 GAM were subjected to immunoprecipitation with protein-G beads. The bound materials were either separated on an SDS-8% PAGE, transferred onto membranes, and immunoblotted with anti-SHP-1 (Upper, 45 × 106 FcγRIIB/PIR-B.H cells per lane) or separated on an SDS-10% PAGE, transferred onto nitrocellulose membranes, and immunobotted with anti-phosphotyrosine antibodies (Lower, 5 × 106 FcγRIIB/PIR-B.H cells per lane). The experiment shown is representative of four independent experiments.
Surface plasmon resonance was used to examine the direct binding of the PIR-B phosphopeptides to the soluble recombinant SH2 domains of SHP-1, SHP-2, and SHIP. These studies employed the phosphorylated FcγRIIB ITIM peptide as a positive control, as it can associate with all three phosphatases in vitro. Phosphopeptides corresponding to the PIR-B Tyr-794 and Tyr-824 motifs both bound to the tandem SH2 domains of SHP-1 and SHP-2, and a weaker but detectable association was observed between the SHIP SH2 domain and the Tyr-824 phosphopeptide. Comparable binding of the phosphatase SH2 domains was not detected with phosphopeptides corresponding to the PIR-B Tyr-742 or Tyr-770 motifs; only minimal binding with SHP-2 was observed for the PIR-B Tyr-713 phosphopeptide (data not shown). These results indicate that the phosphorylated forms of PIR-B Tyr-794 and Tyr-824 motifs can bind to the SH2 domain of SHP-1 and SHP-2 phosphatases in vitro.
Because discrepancy between the in vitro and in vivo binding capacities of FcγRIIB phosphopeptides has been reported (15, 18), we performed in vivo experiments to examine the association of PIR-B with SHP-1, SHP-2, and SHIP. In these experiments, FcγRIIB/PIR-B.H cells were either untreated or stimulated via the FcγRIIB/PIR-B chimeric molecule, the endogenous FcɛRI complex, or coligated FcγRIIB/PIR-B and FcɛRI. This analysis indicated that co-aggregation between FcγRIIB/PIR-B and FcɛRI leads to the recruitment of SHP-1 with PIR-B (Fig. 4C, Upper, lane 4), whereas no association of the PIR-B cytoplasmic domain with SHP-2 or SHIP could be detected (data not shown). Weak constitutive association of the PIR-B cytoplasmic domain with SHP-1 was also observed. Tyrosine phosphorylation of the PIR-B chimera was detected upon FcγRIIB/PIR-B and FcɛRI co-engagement (Fig. 4C, Lower, lane 4), as well as following FcγRIIB/PIR-B stimulation (Fig. 4C, Lower, lane 2). We did not detect any association of FcɛRI with SHP-1, SHP-2, or SHIP in this experimental model (Fig. 4C, Upper, lane 3, and data not shown) in contrast to previous reports (19, 20).
Mutations of Tyr-794 and Tyr-824 Impair the PIR-B Inhibitory Function.
Because the binding assays clearly implicated the two ITIMs centered on Tyr-794 and Tyr-824 in the inhibitory function of PIR-B, Y794F and Y824F point mutants of the FcγRIIB/PIR-B chimeric molecule were generated. Stable RBL-2H3 cell transfectants were obtained, which expressed FcγRIIB/PIR-B, FcγRIIB/PIR-B.Y794F, and FcγRIIB/PIR-B.Y824F at similar cell surface densities. However, RBL-2H3 cell transfectants expressing reduced levels of the chimeric molecule were obtained when both Tyr-794 and Tyr-824 were mutated. No significant alteration of PIR-B inhibitory function was detected in the serotonin release assay when the endogenous FcɛRI receptor was co-aggregated with the Y794F or Y824F single mutants (Fig. 3C), suggesting that the PIR-B ITIMs may be redundant. Simultaneous mutation of both Tyr-794 and Tyr-824 led to significant impairment of the PIR-B inhibitory function (Fig. 3C), although the FcγRIIB/PIR-B.Y794F,Y824F double mutant molecule was still capable of partial inhibition of the FcɛRI-induced activation.
DISCUSSION
The results presented in this report establish an inhibitory function for PIR-B on murine B cells and show that its tyrosine-phosphorylated ITIMs can recruit the protein-tyrosine phosphatase SHP-1. These observations raise several issues relative to the function and mechanisms of action of the PIR-B molecules.
The two ITIMs centered on Tyr-794 (VTYAQL) and Tyr-824 (SVYATL) in the PIR-B cytoplasmic domain are capable of associating with SHP-1 in vitro and are highly homologous to those present in the human ILT-2/LIR-1 (VTY614AQL and SIY644ATL) and ILT-3/LIR-5 (VTY412ARL and SVY442ATL) molecules. Because PIR-B, ILT-2/LIR-1, and ILT-3/LIR-5 can recruit SHP-1 in vivo and are expressed by B cells and myelomonocytic cells (8, 21, 22), our results support the idea that the murine PIR-B and human ILT/LIR molecules are homologues. The recruitment of SHP-1 occurs via direct binding of the SHP-1 SH2 domains to phosphorylated ITIMs, therefore indicating that the tyrosine phosphorylation is a critical step in the inhibitory function of ITIM-bearing molecules. As for the killer cell inhibitory receptors (KIRs) (23), but in contrast to FcγRIIB1 (15), mAb engagement of FcγRIIB/PIR-B chimeric molecules induces PIR-B tyrosine phosphorylation. The identity of the protein-tyrosine kinase associated with PIR-B remains to be elucidated. Nevertheless, the co-aggregation between the FcγRIIB/PIR-B chimera and FcɛRI in RBL-2H3 transfectants led to enhanced PIR-B tyrosine phosphorylation, suggesting that protein-tyrosine kinases associated with activating receptors (e.g., lyn, Syk) may contribute to PIR-B phosphorylation. The inhibitory strategy employed by ITIM-bearing receptors involves the catalytic function of recruited phosphatases, which disrupts the signaling cascade initiated by the engagement of activating receptors (24). As demonstrated for SHP-2, an increase in SHP-1 phosphatase activity presumably depends upon the simultaneous engagement of its N- and C-terminal SH2 domains (25). In this regard, the length of the spacer between the PIR-B Tyr-794 and Tyr-824 ITIMs (29 amino acids) is similar to that found between the KIR (29 amino acids), NKG2A (31 amino acids), and ILTs/LIRs/MIRs (29 amino-acids) ITIMs and is likely to support the simultaneous engagement of the N- and C-terminal SHP-1 SH2 domains. However, our analysis shows that the inhibitory function of PIR-B is minimally affected by single Y794F and Y824F PIR-B mutations. This suggests either that the N- or C-SH2 domains of SHP-1 are capable of interacting in trans with individual ITIMs of co-aggregated PIR-B molecules or that single occupancy of the SHP-1 SH2 domains is sufficient to mediate the PIR-B inhibitory function, thus indicating redundancy of PIR-B ITIMs. Although functionality of the Tyr-794 and Tyr-824 PIR-B ITIMs is indicated by the reduced inhibition of FcɛRI-mediated cell activation observed for the FcγRIIB/PIR-B Y794FY824F double-mutant molecule, this mutant chimera still possessed some inhibitory capacity. Other PIR-B motifs may therefore contribute to its inhibitory function. In this regard, Tyr-713 (SLYASV) might also act as an ITIM, as minimal binding to SHP-2 was detected by lysate adsorption assays and by surface plasmon resonance analysis. In addition, weak association of SHP-2 and the noncanonical motif centered on PIR-B Tyr-742 (ETYAQV) could be detected by surface plasmon resonance. Although association of PIR-B and SHP-2 was not detected in vivo, the Tyr-742 motif might contribute to an association of PIR-B with SHP-2, as the distance between Tyr-713 and Tyr-742 is in the range observed between canonical ITIMs (25–31 amino acids). Other potential ITIMs are also present in ILT-2/LIR-1 (NLY533AAV and VTY562ASV), ILT-3/LIR-5 (VTY360AKV), as well as in SIRPα molecules. The family of ITIM-bearing receptors may therefore include receptors with one (FcγRIIB), two (KIRs), as well as three or four ITIMs (PIR-B, ILTs/LIRs/MIRs, and SIRPα).
The identity of the intracellular molecules that serve as substrates for the phosphatases recruited by the ITIM-bearing receptors is central to the understanding of this inhibitory pathway. In this regard, the differential association of SHIP or SHP-1/SHP-2 with distinct ITIM-bearing molecules indicates that alternative strategies of inhibition are used by this family of receptors. The inhibition of BCR-induced Ca2+ mobilization by PIR-B occurs in the absence of extracellular Ca2+, indicating that PIR-B aborts the initial release of Ca2+ from the intracellular stores. These observations, which parallel those following KIR and ILT-2 engagement (12, 22) and contrast with those following engagement of FcγRIIB, a SHIP-associating molecule (26), support the idea that SHP-1 substrates are upstream signaling molecules in protein-tyrosine kinase-dependent activating pathways. The KIRs have similarly been shown to recruit SHP-1, which in turn dephosphorylates tyrosine-phosphorylated substrates including pp36–38 and ZAP-70 (27, 28). It is likely therefore that the co-engagement of PIR-B and BCR leads to the dephosphorylation of tyrosine-phosphorylated proteins, such as Syk, belonging to the BCR-dependent signaling cascade.
ITIM-bearing receptors on B cells thus include FcγRIIB, CD22, and PIR-B. The SHIP-independent mechanism of action for PIR-B constitutes an alternative inhibitory pathway to that initiated by FcγRIIB and could contribute to significant in vivo differences, as exemplified by the differential regulation of Ca2+ mobilization. The SHP-1-deficient motheaten mouse exhibits severe B cell immunodeficiency and auto-antibody production. In addition, decrease of SHP-1 expression has been demonstrated for germinal center B cells, which have a low threshold for cellular activation (29). Consistent with these observations, SHP-1 is involved in a “censoring” mechanism that determines the threshold of B cell activation through the BCR, thereby affecting the negative selection of B cells (30). The control of BCR signals by SHP-1 might be mediated via ITIM-bearing receptors such as PIR-B and CD22. CD22 may serve as a continuous negative regulator of BCR-induced B cell activation (31) and regulates calcium entry into B cells by a mechanism downstream from or independent of calcium release from intracellular stores (32). PIR-B thus provides an alternative inhibitory pathway for B cells, one that is complementary to those exerted by FcγRIIB and CD22.
Acknowledgments
We thank Marc Daëron, Pierre Brühns, and Odile Malbec (Institut National de la Santé et de la Recherche Médicale U255, Institut Curie, Paris) for valuable advice and reagents, and Pierre Golstein, Peter Burrows, and Louis Justement for their critique of the manuscript. This work was supported by institutional grants from Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Ministère de l’Enseignement Supérieur et de la Recherche, individual grants from Association pour la Recherche contre le Cancer (E.V.), Ligue Nationale contre le Cancer (E.V.), and U.S. Public Health Service Grant AI39816 (M.D.C.). E.V. is a member of the Institut Universitaire de France. M.D.C. is a Howard Hughes Medical Institute Investigator.
ABBREVIATIONS
- BCR
B cell receptor
- DAM
donkey anti-mouse Ig antiserum
- DAR
donkey anti-rat Ig antiserum
- GAM
goat anti-mouse Ig antiserum
- GST
glutathione S-transferase
- ITIM
immunoreceptor tyrosine-based inhibition motif
- PIR
pair of Ig-like receptors
- SH2
Src homology domain 2
- KIR
killer cell inhibitory receptor
References
- 1.Vivier E, Daëron M. Immunol Today. 1997;18:286–291. doi: 10.1016/s0167-5699(97)80025-4. [DOI] [PubMed] [Google Scholar]
- 2.Burshtyn D N, Yang W, Yi T, Long E O. J Biol Chem. 1997;272:13066–13072. doi: 10.1074/jbc.272.20.13066. [DOI] [PubMed] [Google Scholar]
- 3.Scharenberg A M, Kinet J-P. Cell. 1996;87:961–964. doi: 10.1016/s0092-8674(00)81790-0. [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama W M. J Exp Med. 1997;186:1803–1808. doi: 10.1084/jem.186.11.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alley, T. L., Cooper, M. D., Chen, M. & Kubagawa, H. (1998) Tissue Antigens 51, in press. [DOI] [PubMed]
- 6.Olcese L, Cambiaggi A, Bottino C, Moretta A, Vivier E. J Immunol. 1997;158:5083–5086. [PubMed] [Google Scholar]
- 7.Vély F, Vivier E. J Immunol. 1997;159:2075–2077. [PubMed] [Google Scholar]
- 8.Kubagawa H, Burrows P D, Cooper M D. Proc Natl Acad Sci USA. 1997;94:5261–5266. doi: 10.1073/pnas.94.10.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samaridis J, Colonna M. Eur J Immunol. 1997;27:660–665. doi: 10.1002/eji.1830270313. [DOI] [PubMed] [Google Scholar]
- 10.Wagtmann N, Rojo S, Eichler E, Mohrenweiser H, Long E O. Curr Biol. 1997;7:615–618. doi: 10.1016/s0960-9822(06)00263-6. [DOI] [PubMed] [Google Scholar]
- 11.Borges L, Hsu M-L, Fanger N, Kubin M, Cosman D. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- 12.Bléry M, Delon J, Trautmann A, Cambiaggi A, Olcese L, Biassoni R, Moretta L, Chavrier P, Moretta A, Daëron M, Vivier E. J Biol Chem. 1997;272:8989–8996. doi: 10.1074/jbc.272.14.8989. [DOI] [PubMed] [Google Scholar]
- 13.Vély F, Olivero S, Olcese L, Moretta A, Damen J E, Liu L, Krystal G, Cambier J C, Daëron M, Vivier E. Eur J Immunol. 1997;27:1994–2000. doi: 10.1002/eji.1830270825. [DOI] [PubMed] [Google Scholar]
- 14.Le Dréan E, Vély F, Olcese L, Cambiaggi A, Guia S, Krystal G, Gervois N, Moretta A, Jotereau F, Vivier E. Eur J Immunol. 1998;28:264–276. doi: 10.1002/(SICI)1521-4141(199801)28:01<264::AID-IMMU264>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 15.Fong D C, Malbec O, Arock M, Cambier J C, Fridman W H, Daëron M. Immunol Lett. 1996;54:83–91. doi: 10.1016/s0165-2478(96)02654-5. [DOI] [PubMed] [Google Scholar]
- 16.D’Ambrosio D, Hippen K L, Minskoff S A, Mellman I, Pani G, Siminovitch K A, Cambier J C. Science. 1995;268:293–296. doi: 10.1126/science.7716523. [DOI] [PubMed] [Google Scholar]
- 17.Olcese L, Lang P, Vély F, Cambiaggi A, Marguet D, Blery M, Hippen K L, Biassoni R, Moretta A, Moretta L, Cambier J C, Vivier E. J Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 18.Ono M, Bolland S, Tempst P, Ravetch J V. Nature (London) 1996;383:263–266. doi: 10.1038/383263a0. [DOI] [PubMed] [Google Scholar]
- 19.Kimura T, Sakamoto H, Appella E, Siraganian R P. J Biol Chem. 1997;272:13991–13996. doi: 10.1074/jbc.272.21.13991. [DOI] [PubMed] [Google Scholar]
- 20.Kimura T, Zhang J, Sagawa K, Sakaguchi K, Appella E, Siraganian R P. J Immunol. 1997;159:4426–4434. [PubMed] [Google Scholar]
- 21.Cella M, Döhring C, Samaridis J, Dessing M, Brockhaus M, Lanzavecchia A, Colonna M. J Exp Med. 1997;185:1743–1751. doi: 10.1084/jem.185.10.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colonna M, Navarro F, Bellon T, Liano M, Garcia P, Samaridis J, Angman L, Cella M, Lopez-Botet M. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bottino C, Vitale M, Olcese L, Sivori S, Morelli L, Augugliaro R, Ciccone E, Moretta L, Moretta A. Eur J Immunol. 1994;24:2527–2534. doi: 10.1002/eji.1830241040. [DOI] [PubMed] [Google Scholar]
- 24.Leibson P J. Immunity. 1997;6:655–661. doi: 10.1016/s1074-7613(00)80441-0. [DOI] [PubMed] [Google Scholar]
- 25.Pei D, Wang J, Walsh C. Proc Natl Acad Sci USA. 1996;93:1141–1145. doi: 10.1073/pnas.93.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono M, Okada H, Bolland S, Yanagi S, Kurosaki T, Ravetch J V. Cell. 1997;90:293–301. doi: 10.1016/s0092-8674(00)80337-2. [DOI] [PubMed] [Google Scholar]
- 27.Binstadt B A, Brumbaugh K M, Dick C J, Scharenberg A M, Williams B L, Colonna M, Lanier L L, Kinet J-P, Abraham R T, Leibson P J. Immunity. 1996;5:629–638. doi: 10.1016/s1074-7613(00)80276-9. [DOI] [PubMed] [Google Scholar]
- 28.Valiante N M, Phillips J H, Lanier L L, Parham P. J Exp Med. 1996;184:2243–2250. doi: 10.1084/jem.184.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delibrias C C, Floettmann J E, Rowe M, Fearon D T. J Exp Med. 1997;186:1575–1583. doi: 10.1084/jem.186.9.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cyster J G, Goodnow C C. Immunity. 1995;2:13–24. doi: 10.1016/1074-7613(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 31.Tedder T F, Tuscano J, Sato S, Kehrl J H. Annu Rev Immunol. 1997;15:481–504. doi: 10.1146/annurev.immunol.15.1.481. [DOI] [PubMed] [Google Scholar]
- 32.Nadler M J S, McLean P A, Neel B G, Wortis H H. J Immunol. 1997;159:4233–4243. [PubMed] [Google Scholar]