Abstract
Upon chromosomal damage, cells activate a checkpoint response that includes cell cycle arrest and a stimulation of DNA repair. The checkpoint protein Rad24 is key to the survival of a single, repairable double-strand break (DSB). However, the low survival of rad24 cells is not due to their inability to arrest cell cycle progression. In rad24 mutants, processing of the broken ends is delayed and protracted, resulting in extended kinetics of DSB repair and in cell death. The limited resection of rad24 mutants also affects recombination partner choice by a mechanism dependent on the length of the interacting homologous donor sequences. Unexpectedly, rad24 cells with a DSB eventually accumulate and die at the G2/M phase of the cell cycle. This arrest depends on the spindle checkpoint protein Mad2.
Chromosomes may break as a consequence of ionizing radiation, mechanical stress, endonucleases, or errors during replication. In addition, double-strand breaks (DSBs) are instigators of recombination in mitotic and meiotic cells. Repair of DSBs is essential to maintain the integrity of the genome.
Cells have evolved multiple strategies for responding to genomic damage. In eukaryotes, repair of DSBs occurs mainly by two processes: nonhomologous end joining and homologous recombination (31). Recombination can occur between sequences at the same location on homologous chromosomes (allelic) or between dispersed homologous sequences (ectopic). In Saccharomyces cerevisiae, ectopic and allelic recombination occur at comparable levels (13, 18). Current models of recombinational repair of DSBs propose that the broken chromosome ends are resected to generate protruding 3′ single-stranded DNA (ssDNA) that can invade homologous sequences and prime DNA synthesis. Reannealing of the newly synthesized DNA to the opposite broken arm may result in gene conversion; formation of a Holliday junction can also lead to crossing-over (reviewed in references 2 and 31).
Checkpoint proteins delay cell cycle transitions in response to DNA damage. Cell cycle arrest was initially presumed to provide time for the cells to complete DNA repair before mitosis, preventing the segregation of broken chromosomes (40). Subsequent studies showed that checkpoint proteins play additional important roles (1, 3, 19). The DNA damage checkpoint is a signal transduction network consisting of sensors, transducers, and effectors. A group of four conserved sensor proteins are responsible for detecting aberrant DNA structures and initiating the signaling response. Ddc1p, Rad17p, and Mec3p (called in S. cerevisiae the 1-17-3 complex) (10, 21) form a ring resembling the replication factor PCNA (38). The fourth protein, Rad24p, might function as the initial damage sensor, acting as a “clamp loader” to recruit the 1-17-3 sliding clamp (10). A second branch of the DNA damage-sensing group of proteins includes Mec1p, an ATM-like kinase, and Ddc2p. These two proteins form a chromatin-bound complex in response to DNA damage in a manner that is largely independent of Rad24p (27, 30, 33, 42).
The sensor proteins activate signal transducers, such as the Rad53p, Chk1p, and Dun1p protein kinases. This signal transduction eventually leads to cell cycle arrest (5, 42) and induction of repair genes (1, 3) and may play a more direct role in DNA repair (24).
In this paper, we analyze the response of wild-type and rad24 mutants to the creation of a repairable DSB. We demonstrate that the Rad24 checkpoint protein is important for proper DNA processing, recombination partner choice, and survival of the cells after repair. In contrast to what is observed during meiosis (12, 36), deletion of RAD24 impairs ectopic recombination.
MATERIALS AND METHODS
Strains.
All the yeast strains used in this study are isogenic with strain MK205 (MATa-inc ura3-HOcs ade3::GALHO ade2-1 leu2-3,112 his3-11,15 trp1-1 can1-100) (2), a derivative of W303. The ura3-HOcs allele on chromosome V was created by inserting a 39-bp oligonucleotide at the NcoI site of the URA3 gene. Homologous URA3 fragments of different lengths were inserted at an HpaI site within LYS2 sequences as described (17) to create MK203 (1.2 kb), MK202 (5.6 kb), and MK301 (12.8 kb).
Deletion of RAD24 (and other genes, unless indicated otherwise) was obtained by transformation of MK203 with a PCR product produced on the appropriate strain from the Saccharomyces Genome Deletion Project array. For G1 synchronization, strains carrying bar1 deletions were used. MAD2 deletions were generously provided by Jasper Rine and crossed to create MK203mad2. RAD53 and MEC1 deletions were generously provided by Rodney Rothstein and crossed to create MK203rad53 and MK203mec1, respectively. The lethality of rad53Δ and mec1Δ was suppressed by a deletion of the SML1 gene. All chromosomal configurations were verified by Southern blot analysis after transformation. Disomal strains were generated by crosses with kar1-1 strains and selection for transfer of a marked chromosome V.
Media and growth conditions.
S. cerevisiae strains were grown at 30°C except for pds1, which was grown at 25°C. Standard YEP medium (1% yeast extract, 2% Bacto Peptone) supplemented with 3% glycerol (YEPGly), 2% galactose (YEPGal), or 2% dextrose (YEPD) was used for nonselective growth. Bacto Agar (1.8%) was added for solid media. A concentration of 0.04 μM α-factor and 15 μg of nocodazole per ml were used to arrest cells at G1 and G2/M, respectively.
Repair efficiency measurement.
Each strain was streaked on YEPGly plates. Individual colonies were resuspended in water, appropriately diluted, and plated on YEPD and YEPGal plates. Colonies were counted after 3 days of incubation at 30°C.
Induction experiments.
Single colonies were resuspended in rich YEPGly medium, grown to logarithmic phase, centrifuged, and resuspended in YEPGal. At timely intervals, samples were plated on YEPD plates to score viability, and DNA was extracted with HMW-DNA isolation kits (Gentra Systems). At least three independent experiments were carried out for each strain analyzed.
Survival.
Survival was assayed by plating samples of cells grown on liquid YEPGal onto YEPD plates at different times during a DSB induction experiment.
Southern blot analysis.
Southern blot analysis was carried out as previously described (17) and quantified with a PhosphorImager.
Nondenaturing slot blots.
DNA was either directly spotted on nylon Hybond+ filters or denatured first by boiling for 5 min. Hybridization and exposure were carried out as in Southern blot analysis. The results obtained for the denatured samples were used to calibrate those obtained without denaturation and quantified with a PhosphorImager.
PCR assays.
We amplified 5 ng of genomic DNA in each sample. For quantitative measurements of intact chromosome V and PCR standards, samples were removed at cycle 18. Otherwise, reactions were allowed to proceed to cycle 35. The composition of individual primers is available upon request.
Quantitation of results.
Southern blot phosphoimages and ethidium bromide-stained agarose gels were quantified with the TINA (version 2.1) and NIHimage (version 1.62) computer programs.
FACS analysis.
For fluorescence-activated cell sorter (FACS) analysis, samples of 107 cells were harvested, fixed in 1 ml of 80% ethanol, and stored at 4°C. All samples were processed together and analyzed by standard procedures.
Allelic/ectopic recombination assays.
MK235 is a diploid isogenic derivative of MK203 in which the DSB can be repaired by recombination with either allelic or ectopic sequences. These strains possess a single URA3 gene (chromosome V) in addition to one allelic ura3-HOcs (chromosome V) and one ectopic ura3-HOinc (chromosome II). Recombination with the allelic URA3 template creates two copies of the URA3 gene, whereas ectopic recombination with the ura3-HOinc template renders cells heterozygous for URA3. In both cases the resulting cells are unable to grow on plates containing the uracil analogue 5-fluoroorotic acid. However, mitotic recombination or mutation can give rise to 5-fluoroorotic acid-resistant papillae in the heterozygotes. Thus, 5-fluoroorotic acid-resistant papillation is indicative that the broken chromosome was repaired by ectopic recombination. This was confirmed by PCR, followed by restriction analysis with NcoI, EcoRI, and BamHI. Strain MK219 is a diploid carrying ura3-HOcs alleles on both copies of chromosome V. One copy of the 1.2-kb ura3-HO-inc is located on each chromosome II, allowing only ectopic recombination to occur.
RESULTS
Experimental system.
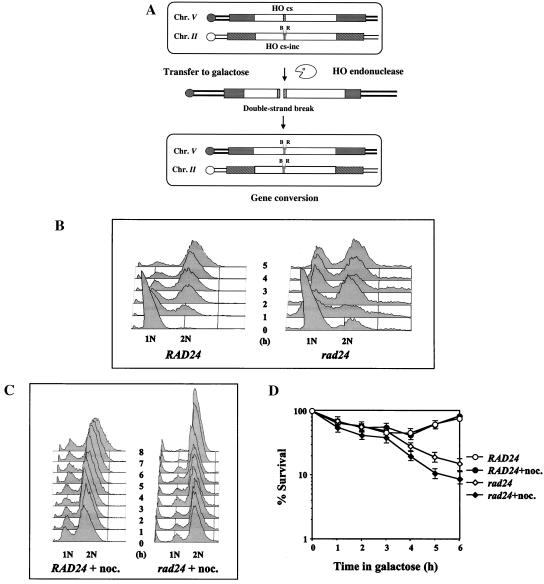
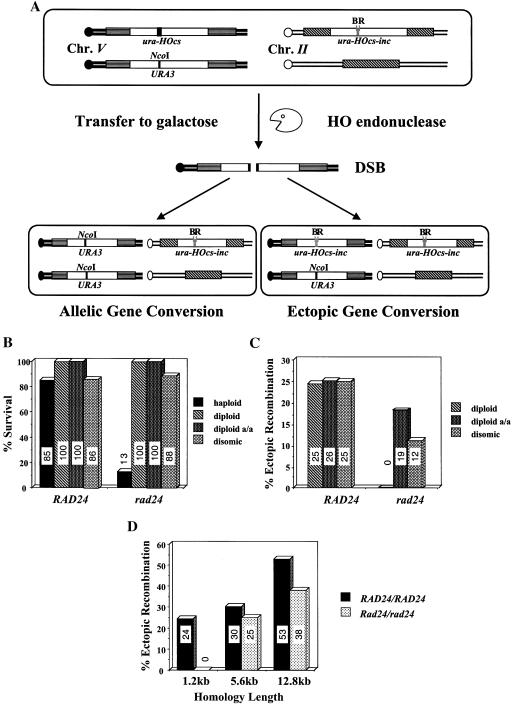
In order to study the effect of a single, defined chromosomal break in the genome, we generated (2) haploid strains of the yeast S. cerevisiae that bear two copies of the URA3 gene (Fig. 1A). One copy, located on chromosome V, carries the recognition site for the yeast HO site-specific endonuclease inserted as a short oligonucleotide (ura3-HOcs). The second copy, located on chromosome II, carries a similar site containing a single-base-pair mutation that prevents recognition by the endonuclease (ura3-HOcs-inc). In addition, the ura3 alleles differ at two restriction sites, located to the left (BamHI) and to the right (EcoRI) of the HOcs-inc insertion. These polymorphisms are used to follow the transfer of information between the chromosomes. In these strains, the HO gene is under the transcriptional control of the GAL1 promoter. Upon transfer of the cells to galactose-containing medium, the HO endonuclease is produced at high levels. The enzyme creates a single DSB in each cell of the population. The broken chromosomes are then repaired by a mechanism that copies the HOcs-inc information together with the flanking markers, resulting in a gene conversion event (Fig. 1A). During the repair, the donor chromosome remains unchanged. Survival remains high in wild-type cells, and there is no need for genetic selection of recombinational products; instead, repair can be monitored in the entire cell population (2, 16, 17).
FIG. 1.
(A) Schematic representation of our experimental system. Open rectangles represent the ura3 alleles on chromosomes II and V. A stippled box represents the HOcs; a grey box depicts the inactive HOcs-inc flanked by the BamHI (designated B) and EcoRI (designated R) restriction sites. Transfer of the cells to galactose-containing medium results in a DSB that is repaired by gene conversion. (B) FACS analysis of strains MK203 and MK203rad24. Cells were synchronized at G1 with α-factor and released into medium containing galactose at 0 h. (C) Exponentially growing MK203 and MK203rad24 cells were artificially arrested at G2/M with nocodazole, transferred to galactose with nocodazole, and subjected to FACS analysis to confirm complete G2/M arrest. (D) Aliquots from the same experiment were also plated on glucose at different times after HO induction. Survival was determined as relative number of CFU.
Rad24p is required to survive a single, repairable DSB.
In order to assess the efficiency with which wild-type and rad24 cells can repair and survive a single DSB, we compared the colony-forming ability of cells on galactose-containing medium (constitutive HO induction) to that observed on medium containing glucose (no HO induction). In wild-type cells, recombinational repair is quite efficient even when homology is limited. For example, strain MK203 (bearing ectopically located ura3 alleles that share 1.2 kb of homology) exhibited 85% efficiency of repair (2).
In contrast, in isogenic strains deleted for the RAD24 gene, only 12.6% of the cells formed colonies on galactose-containing plates. This indicates that most rad24 cells are unable to form colonies after a single chromosomal DSB despite possessing potential ectopic donor sequences.
rad24 cells do not die due to lack of cell cycle arrest.
Plating of timely aliquots from a DSB induction experiment onto glucose-containing medium reveals the proportion of viable cells over time. Despite a transient dip of survival, the viability of wild-type cells remained high throughout the experiments. Survival of rad24 cells was initially comparable to that of wild-type cells. However, starting 3 h after transfer to galactose, the survival of rad24 cells steadily decreased to approximately 10%. By 3 h after transfer, wild-type cells arrested at the G2/M phase of the cell cycle, while most of the rad24 cells had passed through mitosis and entered an additional cell cycle (Fig. 1B and D).
Conceivably, the low survival of rad24 cells is due to their inability to prevent mitotic progression despite the presence of a broken chromosome. Given additional time in G2/M, rad24 cells may be able to complete repair and survive. In order to test this hypothesis, cells were prevented from progressing into mitosis by treatment with the spindle toxin nocodazole. Nocodazole causes cells to arrest at the G2/M phase of the cell cycle (Fig. 1C) through a mechanism independent of the DNA repair checkpoint. Survival of wild-type cells with a broken chromosome was unaffected by nocodazole-imposed arrest. Moreover, arrest of rad24 mutants at G2/M did not prevent cell death (Fig. 1D), demonstrating that the low survival of rad24 mutants due to DSBs is not due to their inability to arrest the cell cycle in response to DNA damage.
rad24 cells exhibit protracted kinetics of DSB repair.
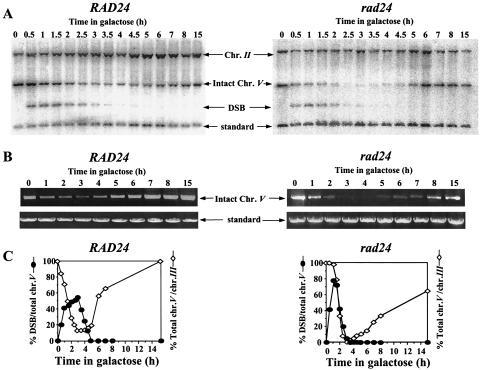
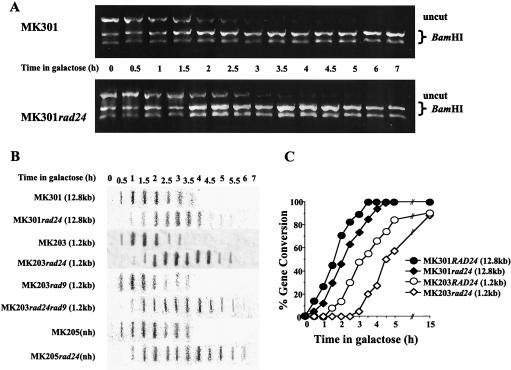
In order to understand the nature of the defective response to a DSB in rad24 mutants, we followed the fate of a single broken chromosome in these cells (2). In both wild-type and rad24 cells, broken chromosome arms were detected in Southern blots within 30 min, and by 1.5 h after induction, DSB formation was maximal. The relative amount of intact chromosome V diminished as the parental chromosome was broken and subsequently increased as the break was repaired. In wild-type cells, repair commenced rapidly, and therefore repaired chromosome V was detected starting from 2.5 h after DSB induction. In rad24 cells, there was a delay in recovery of intact chromosome V, causing a dramatic decrease in its levels between 2 and 4 h (Fig. 2A and C). Since repair intermediates are not detected by Southern blot analysis (2), the decrease observed is symptomatic of the prolonged kinetics of repair in rad24 cells. Intact chromosome V was also measured by quantitative PCR (Fig. 2B). The results from PCR analysis fully corroborated those obtained with Southern blots (Fig. 2C).
FIG. 2.
(A) Southern blot analysis of DNA extracted from MK203 and MK203rad24 cells at intervals after HO induction. The DNA was digested with ClaI and probed with the URA3 gene. A probe for the LEU2 sequence served as a loading standard. (B) Quantitative PCR of the relative amounts of intact chromosome V in MK203 and MK203rad24 cells. PCR was quantified relative to the unrelated gene PRP8 in the same PCR. (C) Quantitation of Southern blot and PCR analysis. “Total chr. V” refers to the sum of broken and intact chromosome V sequences detected in the Southern blot.
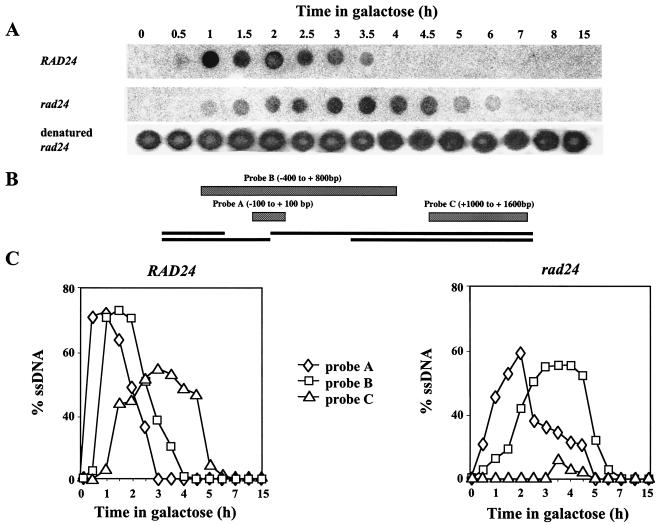
Current models of recombination (reviewed in reference 31) propose that there is a 5′-to-3′ resection of DNA flanking the broken chromosome ends. The resulting ssDNA is a pivotal intermediate in all homologous recombination pathways. We used nondenaturing dot blot analysis to track and quantify ssDNA intermediates (Fig. 3). In wild-type cells, resection following a DSB is highly synchronous. ssDNA immediately flanking the DSB was exposed as early as 30 min after transfer to galactose. The hybridization signal of this probe (probe A) peaked 1 h after DSB induction and was no longer detected by 3 h postinduction. ssDNA resection in a 600-bp region adjacent to the DSB was maximal 1.5 h after DSB induction in wild-type cells. A majority of wild-type cells underwent resection of approximately 1,400 bp by 3 h after DSB induction.
FIG. 3.
Dot blot assays. (A) A sample dot blot representing the differential accumulation of ssDNA intermediates in MK203 and MK203rad24 cells. Samples were hybridized with probe B. (B) Schematic representation of probes used in dot blot assays. Distance from HOcs is shown. (C) Quantitation of dot blots. The percentage of ssDNA was determined relative to the amount of denatured DNA hybridizing to the same probe in each sample.
In the rad24 strain, even initial resection was protracted compared to that of wild-type cells. Subsequent exposure of a single-stranded filament was markedly delayed and reduced (Fig. 3). Two hours after DSB induction, only 100 bp were processed in most rad24 cells. ssDNA complementary to probe B remained exposed and unprocessed for a prolonged period after the chromosome had been broken. In contrast to wild-type cells, in rad24 mutants hybridization levels of ssDNA reached a plateau instead of peaking, indicating that the processing of DSB ends was asynchronous. Most rad24 cells did not successfully resect sequences located 1 kb away from the DSB. We conclude that processing of DSB ends in rad24 cells is reduced, asynchronous, and protracted compared to that in the wild type.
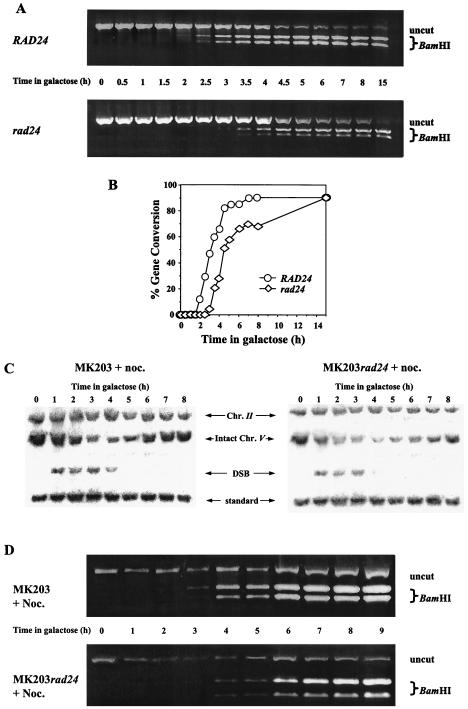
The inefficient processing of the DSB ends in rad24 cells causes loss of synchronization and delay of subsequent steps in the repair process. The final stage of repair, involving religation of the broken chromosome ends, can be visualized in our strains by the transfer of two polymorphic restriction enzyme sites on either side of the HOcs from chromosome II to chromosome V. We followed the extent and kinetics of this gene conversion event by measuring the relative amount of intact chromosome V carrying these restriction sites (Fig. 4A and B).
FIG. 4.
Real-time gene conversion assay. (A) DNA from MK203 and MK203rad24 cells was extracted at intervals after HO induction. Equal amounts of PCR products flanking the DSB were digested with BamHI. Only fragments originating from a template repaired by gene conversion can be digested. (B) Quantitation of PCR. Percent gene conversion represents the portion of PCR fragments cut by BamHI. (C and D) Exponentially growing MK203 and MK203rad24 cells were arrested at G2/M with nocodazole (noc.) and transferred to galactose in the presence of nocodazole. Southern blot (C) and PCR (D) analyses were carried out as described above.
In wild-type cells, gene conversion reached detectable levels by 2 h after transfer to galactose, and the population was completely repaired 6 h after induction. Surprisingly, despite their low survival and delayed kinetics, rad24 mutants were fully capable of repairing the broken chromosome. Initial gene conversion products were detected in rad24 mutants with a delay of approximately 1 h compared to wild-type cells, but roughly 90% of the rad24 population eventually underwent gene conversion (Fig. 4A and B). These results were corroborated by Southern blot analysis (Fig. 2A).
Thus, in the absence of Rad24p, cells are able to repair a broken chromosome, albeit with delayed kinetics. Despite the fact that the total level of repair is remarkably similar to that observed in the wild type, most rad24 cells were unable to survive a single genomic DSB. G2/M arrest imposed with nocodazole did not alter the kinetics of DSB repair in rad24 cells (Fig. 4C and D). In fact, the vast majority of the cells were unable to form viable colonies even when released from arrest and plated at times when most of the repair had been completed (Fig. 1C and 4C and D). This indicates that the low viability of rad24 cells on galactose is due neither to lack of repair of the broken chromosome nor to lack of cell cycle arrest.
Rad24p is important for recombination partner choice.
To test whether the low survival of rad24 cells following a DSB is particular to a requirement to utilize ectopic donors, we generated the isogenic diploid strains MK235 and MK235rad24 (Fig. 5A). In these strains, a single DSB is created by the HO endonuclease on one of the two copies of chromosome V. This broken chromosome can be repaired with either allelic or ectopic donor sequences.
FIG. 5.
(A) A schematic representation of the diploid strain used to assay ectopic and allelic recombination. MK235 is an isogenic derivative of MK203 in which the DSB (chromosome V) can be repaired by recombination with either URA3 sequences (chromosome V) or ura-Hocs-inc sequences (chromosome II). A stippled box represents the HOcs; a light grey box depicts the inactive HOcs-inc flanked by the BamHI (designated B) and EcoRI (designated R) restriction sites; a dark grey line depicts the NcoI site in URA3. (B) Survival of wild-type and rad24 strains on YEPGal (constitutive expression of the HO endonuclease) compared to YEPD (no HO expression) plates. (C) Proportion of colonies that utilized an ectopic recombination donor in order to repair a single DSB. (D) Proportion of diploid colonies that utilized an ectopic recombination donor in order to repair a single DSB. Homology length indicates the extent of homologous sequences on chromosome II flanking the ura-HOcs-inc.
Wild-type MK235 cells exhibited 100% survival following a DSB and utilized ectopic sequences for homologous recombination even when homologous allelic sequences were also available. Surprisingly, rad24 diploid yeast strains with homologous allelic donor sequences also demonstrated 100% survival in response to a DSB (Fig. 5B). In contrast to the wild type, however, rad24/rad24 cells did not perform ectopic recombination when allelic sequences were provided (Fig. 5C). This suggests that, due to limited resection, rad24 mutants may be specifically defective in contending with ectopic recombination. Survival was consistently dramatically decreased in rad24/rad24 strains in which only ectopic donors were available for recombination. This phenotype of rad24/rad24 mutants is the opposite of that seen in meiosis, where deletion of RAD24 increased the levels of ectopic recombination (12, 36). Nevertheless, if the extent of ectopic homology was increased (≥5.6 kb), rad24/rad24 mutants were able to locate and exploit ectopic homologous sequences for DSB repair almost as efficiently as wild-type cells (Fig. 5D).
To further study this point, we generated wild-type and rad24 haploids carrying longer stretches of ectopic homology. When homology length was increased, the survival of rad24 mutants increased accordingly (13%, 28%, and 35% survival for 1.2 kb, 5.6 kb, and 12.8 kb, respectively). Although longer homology of the donor did not appear to affect the kinetics of ssDNA resection (Fig. 6B), it nevertheless hastened the repair kinetics and increased the survival of rad24 strains. rad24 strains consistently exhibited a delay in the processing and repair of DSBs compared to the wild type (Fig. 6).
FIG. 6.
Real-time gene conversion assay. (A) DNA from MK301 and MK301rad24 cells was extracted at intervals after HO induction. Equal amounts of PCR products flanking the DSB were digested with BamHI. In this strain, homology length is 12.8 kb, and therefore the donor URA3 sequences are also detected in the PCR assay. At 0 h, only half the PCR products can be digested with BamHI. Repair of the broken chromosome by gene conversion progressively increases the relative proportion of BamHI-containing fragments. Uncut PCR fragments represent the portion of the population that has not undergone gene conversion. (B) Slot blot assays. Nondenatured DNA was hybridized with probe B (complementary to 1.2-kb URA3, as described previously). (C) Graphic representation of the kinetics of gene conversion. Percent gene conversion represents the relative percentage of PCR fragments digested by BamHI compared to those seen at 0 h.
Following DSB induction, a majority of haploid rad24 cells died, whereas diploid rad24/rad24 cells exhibited 100% survival. Interestingly, whereas all wild-type cells promptly arrested at G2/M, both haploid and diploid rad24 cells, irrespective of their survival levels, exhibited cell cycle arrest defects. These observations substantiated our previous conclusion that cell cycle arrest does not contribute to survival after a DSB.
Diploid cells possess homologous chromosomes containing potential allelic donors. They are also heterozygous at the yeast mating type locus, MAT, and therefore nonmaters, whereas haploid cells have a single copy of each chromosome and retain the ability to mate. To test which diploid attribute might explain the high survival of diploid rad24 cells, we repeated the above experiments in wild-type and rad24 disomic haploid strains carrying an extra, allelic, copy of chromosome V. We also tested diploid rad24/rad24 strains homozygous for the MATa allele. All of these strains are phenotypically a-maters. Even with limited ectopic homology (<5.6 kb), in rad24 disomic and MATa/MATa strains, the frequency of ectopic recombination was dramatically increased (Fig. 5C) and survival was high (Fig. 5B). Consistently, expression of both mating types in haploid rad24 cells (lacking potential allelic donors) dramatically lowered survival (MATa rad24 cells plus MATa, 13% survival, versus MATa rad24 cells plus MATα, 3% survival). This indicates that in addition to homology length, mating type heterozygosity controls recombination partner choice in rad24 cells.
Genetic interactions with other checkpoint genes.
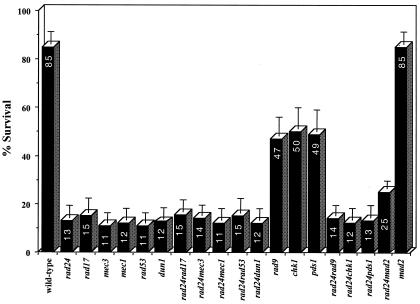
In order to analyze potential genetic interactions with other DNA damage checkpoint genes, we tested the survival of a series of checkpoint mutants alone and in combination with rad24. Strains combining rad24 with rad17, mec3, mec1, rad53, or dun1 deletions exhibited survival similar to that of each of the single mutants, confirming that these gene products act in a common pathway (Fig. 7). Previous work has assigned the function of Rad9p to a separate branch of the checkpoint sensing mechanism that contributes equally to survival (reviewed in reference 23). However, in our system, rad9 mutants showed markedly higher survival after a single DSB than rad24 cells, and processing of DSB ends was at least as fast as that of the wild type (Fig. 6B and 7). The rad24 rad9 double mutant exhibited repair kinetics and survival similar to those of the rad24 mutant (Fig. 6B and 7), consistent with a more prominent role for Rad24p in survival of a DSB. Other checkpoint mutants with DSB repair phenotypes similar to rad9 include chk1 and pds1 (Fig. 7), suggesting that the products of these three genes may act together in a pathway that contributes only partially to cell survival of a broken chromosome.
FIG. 7.
Survival of different strains on YEPGal (constitutive expression of the HO endonuclease) compared to YEPD (no HO expression) plates.
Activation of spindle checkpoint is responsible for residual G2/M arrest of rad24 cells and exacerbates rad24 DSB lethality.
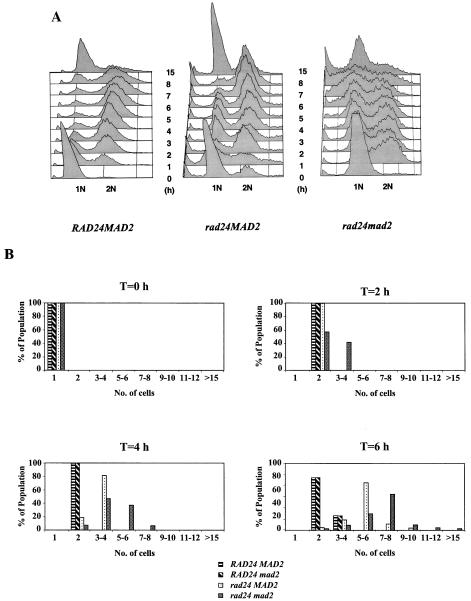
Surprisingly, elimination of the spindle damage checkpoint gene MAD2 in conjunction with a deletion of RAD24 increased cell survival in response to a DSB to 24%, virtually double the survival of a single rad24 mutant (Fig. 7). DSB induction experiments were performed on wild-type, rad24, and rad24 mad2 strains (Fig. 8A). As observed previously, wild-type cells rapidly accumulated at the G2/M phase of the cell cycle in response to a DSB. rad24 mutants initially progressed beyond the G2/M transition and entered mitosis despite possessing an unrepaired chromosome (as previously depicted in Fig. 1B). However, by 6 h after DSB induction, rad24 cells had distinctly accumulated at the G2/M phase of the cell cycle. By 8 h after DSB induction, 90% of the population had arrested with a 2N DNA content. To the best of our knowledge, this delayed but complete G2/M arrest of rad24 mutants has not been reported previously.
FIG. 8.
(A) FACS analysis of MK203, MK203rad24, and MK203rad24 mad2 strains. Cells were synchronized at G1 with α-factor and released into medium containing galactose at 0 h. (B) Single, unbudded MK203, MK203mad2, MK203rad24, and MK203rad24 mad2 cells were manipulated on galactose-containing medium, observed microscopically, and tallied.
In contrast, rad24 mad2 cells did not accumulate at any specific stage of the cell cycle (Fig. 8A). This implies that Mad2p is responsible for the eventual G2/M arrest observed in rad24 mutants. rad24 mad2 cells suffering a DSB are thus expected to experience shorter cell cycles and divide more often than rad24 cells encountering the same damage. We conducted an analysis of the cell cycle progression of individual wild-type, rad24, mad2, and rad24 mad2 cells experiencing a single DSB (Fig. 8B). Single, unbudded cells were micromanipulated onto galactose-containing medium and were monitored and tallied at different times. As early as 2 h after DSB induction, a clear difference was seen between rad24 and rad24 mad2 cells. Whereas the former were almost entirely at the G2/M phase of the cell cycle, more than 35% of the rad24 mad2 cells had already undergone an additional cell division to produce three to four cells. At 6 h after DSB induction, rad24 mutants had progressed to an average of six cells, whereas rad24 mad2 mutants continued dividing rapidly. Throughout this time period, wild-type and mad2 cells remained arrested at G2/M (one large-budded cell) and only recommenced dividing 8 h after DSB induction (Fig. 8B). Even after prolonged incubation, most rad24 microcolonies contained six to eight large cells, whereas rad24 mad2 cells died as nonuniform multicellular microcolonies. These results, combined with the FACS analysis, imply that rad24 cells initially progress through the G2/M transition but eventually arrest and die with a spindle checkpoint-mediated G2/M-arrested phenotype.
DISCUSSION
When a chromosome is broken, cells must sense the presence of damage, activate the course of events that will eventually restore genomic integrity, and then return to normal growth. Sensing mechanisms and repair processes, which potentially compete for the same substrate, must be coordinated. In order to hasten repair, proteins responsible for sensing damaged DNA might also process the broken ends into intermediates easily recognized and handled by the repair machinery. Accordingly, lack of DSB sensor proteins may hamper the proper processing of DSB ends and delay repair of the DNA damage. This is precisely what we observed in our experiments.
End processing is defective in rad24 cells.
We carefully analyzed the repair of a DSB in wild-type and rad24 cells. Although DSBs were formed with similar timing in both strains, the processing and repair of the broken ends in rad24 cells were delayed. The discrepancy in repair kinetics could be observed at the first stage of repair, in which the broken ends are resected. Initiation of ssDNA resection was delayed in rad24 cells. At 2 h after HO induction, the majority of wild-type cells had resected approximately 600 nucleotides; in contrast, rad24 cells had only resected 100 nucleotides, and 1 h later, most wild-type cells had already processed and exposed more than 1 kb of DNA, but in rad24 cells a significant proportion of the population had only resected an average of 600 nucleotides. Whereas wild-type cells resect approximately 3.5 kb of DNA flanking the DSB ends (2), most rad24 cells did not successfully process 1,000 bp of DNA. The net effect observed is that resection of DSB ends in rad24 is delayed, uncoordinated, and impaired compared to resection in the wild type. Rad24p has also been associated with processing of ssDNA at telomeres (4, 24).
Inefficient processing of the DSB ends in rad24 cells causes loss of synchrony and a delay of subsequent steps in the repair process. Fully repaired gene conversion products are detected in rad24 mutants with a delay of approximately 1 h compared to wild-type cells, but roughly 90% of the rad24 cell population eventually undergoes gene conversion. Since the total level of gene conversion is remarkably similar to that of the wild type, it is the proper and rapid processing of DSBs that is essential for survival.
Repair kinetics, not cell cycle progression, are essential for survival.
We have shown that 87% of rad24 cells do not survive a single DSB. Significantly, lethality is not due to insufficient time to repair the broken chromosome before being released into mitosis. Cells kept at G2/M until repair has occurred in most of the population still die (Fig. 1D and 4C and D). Therefore, cell cycle delay is not the essential function of checkpoint activation for survival of a DSB. Although checkpoint proteins were originally defined by their ability to inhibit cell cycle progression when damage occurs (40), current evidence indicates that checkpoint proteins activate multiple cellular processes, only one of which is cell cycle arrest (1, 3, 19). Accordingly, some mutations in the Schizosaccharomyces pombe homolog of RAD24 show radiation sensitivity despite implementing normal checkpoint delays (11).
The presence of checkpoint proteins ensures proper kinetics of chromosome repair that are essential for cell survival. The kinetics of DSB repair in rad24 cells are unaffected by cell cycle progression, since nocodazole-arrested cells exhibit the same kinetics of repair as freely cycling cells. (Fig. 4C and D). This implies that the defective DNA processing observed is inherently associated with the lack of Rad24 activity. In addition, the complete repair of nondividing nocodazole-arrested cells confirms that the high levels of DSB repair observed in the rad24 culture are not due to a subpopulation of cells properly repairing the DSB and outgrowing a general population of cells that did not repair the broken chromosome.
One surprising implication of our results is that rad24 cells are able to repair a broken chromosome despite having undergone cell divisions. This indicates that the broken, acentric chromosomal fragment is not lost during mitosis. Evidence that a broken chromosome can segregate without repair has been documented previously (25, 28) even in strains with checkpoint defects (34). Chromatin may tether broken DSB ends together through mitotic divisions, preserving the possibility of repair in the next cell cycle.
Although, as reported (14), rad24 cells fail to arrest in the first cycle following DNA damage, our FACS analysis shows that rad24 cells progressively accumulate with 2N DNA content (Fig. 8A). This cell cycle arrest is due to the triggering of the spindle checkpoint. Whereas rad24 mutants with a single DSB eventually accumulate and die at the G2/M phase of the cell cycle, rad24 mad2 double mutants do not accumulate at any specific stage and continue cycling (Fig. 8A and B). Recently, Garber and Rine (18) showed a role for the spindle checkpoint in mediating cell cycle arrest in response to replication defects and DNA damage (7). Similarly, incomplete DNA replication in Drosophila embryos results in a spindle checkpoint-dependent mitotic arrest (9). Defective replication due to misprocessing of the DSB may activate the spindle checkpoint in rad24 cells. Inactivation of the spindle checkpoint partially increased survival of rad24 strains (Fig. 7), indicating that under some conditions, checkpoint activation may have deleterious effects.
Rad24p affects recombination partner choice.
Our results imply that the definition of “allelic” partners for recombination is based on the extent of homology rather than on physical proximity or somatic pairing. When ectopic homology was increased to 12.8 kb, wild-type cells elevated ectopic recombination to frequencies similar to those of allelic recombination (Fig. 5D). The mechanism of homology search can therefore equally recognize extended homology at any location in the genome. rad24 cells are deficient in this mechanism.
The mating type locus plays an important role in regulating partner choice in the absence of Rad24p. MATa/MATα cells do not carry out ectopic recombination, whereas MATa/MATa diploids or disomic haploids do so at levels comparable to those of wild-type cells (Fig. 5C). The negative effect of heterozygosity of mating type on ectopic recombination is observed only when Rad24p is absent. The mating type locus of S. cerevisiae, MAT, is a master regulator that influences global transcription and developmental decisions. It may be advantageous to MATa/MATα diploids to promote homologous recombination, since, unlike haploids, they carry a potential substrate for homologous recombination at all stages of the cell cycle. Indeed, heterozygosity at the MAT locus affects the choice between alternative repair pathways, favoring homologous recombination (15, 41). For instance, the NEJ1 gene, which plays a role in nonhomologous end joining, is repressed in MATa/MATα cells, channeling repair from end joining to homologous recombination (6, 37). In the absence of Rad24p, we observed a similar control by the mating type locus between two different forms of homologous recombination, allelic (extended homology) and ectopic (limiting homology).
Recombination partner choice as well as survival is influenced by prompt repair of the broken chromosome ends. In rad24 strains with restricted homology, approximately 15% of the cells successfully completed ectopic recombination to create viable colonies (Fig. 5B). When allelic sequences were also provided, the remaining 85% of the population could be rescued by prompt allelic recombination (Fig. 5B and C).
Resected single-stranded filaments sheathed with Rad51p play a central role in homology search (35). Our results suggest that the Rad51 filament is shorter in rad24 cells. However, our results show that donor sequences also affect partner choice: longer stretches of ectopic homology hastened repair kinetics, increased ectopic recombination rates, and improved the survival of rad24 cells. Since longer homology did not markedly affect the kinetics of resection (Fig. 6B), homology length may exert its effect at a later step in the repair process, such as the recognition between the homologous partners. Other Rad24-asociated processes are also important for survival. Although fast repair improves the survival of rad24 cells, it is not sufficient to rescue the entire population.
Interactions between DNA damage checkpoint genes.
Our epistasis analysis shows that RAD24 acts in a common pathway with RAD17, MEC3, RAD53, MEC1, and DUN1 in the survival of a repairable DSB. Lee et al. (22) reported that rad17 mutants showed normal resection of sequences flanking the MAT locus in strains lacking potential homologous donors. Since rad17 and rad24 are in the same epistasis group, this seems to contradict our results. We repeated our assays in rad17 and rad24 rad17 strains carrying or lacking ectopic donor sequences. In all cases, we obtained results identical to those with the rad24 single mutants (Fig. 6B and data not shown). We therefore conclude that the discrepancy between our results and those of Lee et al. must be due to the different experimental systems used.
Mating type switch is unique in several aspects: resection is unidirectional, signal transducers such as Rad53p are not activated, and no cell cycle checkpoint arrest is elicited. Therefore, it is conceivable that resection at the MAT locus may differ from processing of DSBs at other genomic locations. The results we reported here are consistent with the effects of Rad24p in the processing of ssDNA at cdc13-1-induced telomere damage (24). In both systems, mutations in RAD24 or RAD17 caused reduced ssDNA resection, whereas mutations in RAD9 had the opposite effect (Fig. 6B) (24).
In our system, RAD9, CHK1, and PDS1 operated separately from the RAD24 checkpoint pathway (Fig. 6). Previous reports concur with this model. In meiosis, the prophase arrest of dmc1 mutants depends on RAD24 but is independent of RAD9 and CHK1 (32). In addition, Maringele and Lydall (26) have shown that in response to telomere defects, Rad9p, Chk1p, and Pds1p act in a branch distinct from that of Rad24p, Rad17p, Rad53p, Ddc1p, Ddc2p, and Dun1p. Interestingly, these authors also found a role for the Mad2 spindle checkpoint protein in the response to DNA damage in telomeres (26).
Although the biochemical nature of the Rad9p branch of the checkpoint response is not fully understood, Rad9p is required for the activation of Chk1p, which acts to prevent cell cycle progression. Anaphase entry is prevented by the Chk1p-mediated phosphorylation of Pds1p, the yeast securin (8). In our experimental system, deletion of RAD9, CHK1, or PDS1 had a milder effect on DSB survival than mutations in the RAD24 epistasis group. These results agree with our conclusion that the ability to arrest in the cell cycle has little effect on survival.
Conclusions.
A broken chromosome elicits a complex response that includes cell cycle arrest, recruitment of repair proteins, homology search, and repair. Checkpoint proteins such as Rad24 play a central role in the coordination of this response. We have shown that in the absence of Rad24p, DNA end processing is defective. This affects the kinetics of repair and the choice of recombination partner as well as the ability to survive after repair. Although rad24 cells show no immediate cell cycle arrest, eventual G2/M arrest of rad24 cells depends on the spindle checkpoint protein Mad2p. rad24 mutants die irrespective of cell cycle arrest due to protracted kinetics of DSB repair.
Acknowledgments
We thank the anonymous reviewers whose suggestions helped to improve our manuscript. We thank Jasper Rine, Rodney Rothstein, and Anat Krauskopf for the generous gift of strains and plasmids. We are grateful to Nurit Paz and Lilach Chen for excellent ideas and technical help. We also thank all the members of the Kupiec laboratory for helpful comments and support.
This work was supported by a grant to M.K. from the Israel Science Foundation.
REFERENCES
- 1.Aboussekhra, A., J. E. Vialard, D. E. Morrison, M. A. de la Torre-Ruiz, L. Cernakova, F. Fabre, and N. F. Lowndes. 1996. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 15:3912-3922. [PMC free article] [PubMed] [Google Scholar]
- 2.Aylon, Y., B. Liefshietz, G. Bitan-Banin, and M. Kupiec. 2003. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 23:1403-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashkirov, V. I., J. S. King, E. V. Bashkirova, J. Schmuckli-Maurer, and W. D. Heyer. 2000. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 20:4393-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth, C., E. Griffith, G. Brady, and D. Lydall. 2001. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 29:4414-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr, A. M. 1997. Control of cell cycle arrest by the Mec1sc/Rad3sp DNA structure checkpoint pathway. Curr. Opin. Genet. Deversion 7:93-98. [DOI] [PubMed] [Google Scholar]
- 6.Frank-Vaillant, M., and S. Marcand. 2001. nonhomologous end joining regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Deversion 15:3005-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garber, P. M., and J. Rine. 2002. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics 161:521-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner, R., C. W. Putnam, and T. Weinert. 1999. RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 18:3173-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner, M., S. van Kreeveld, and T. T. Su. 2001. mei-31 and bub1 block mitosis at two distinct steps in response to incomplete DNA replication in Drosophila embryos. Curr. Biol. 11:1595-1599. [DOI] [PubMed] [Google Scholar]
- 10.Green, C. M., H. Erdjument-Bromage, P. Tempst, and N. F. Lowndes. 2000. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 10:39-42. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths, D. J., N. C. Barbet, S. McCready, A. R. Lehmann, and A. M. Carr. 1995. Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 14:5812-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grushcow, J. M., T. M. Holzen, K. J. Park, T. Weinert, M. Lichten, and D. K. Bishop. 1999. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haber, J. E., W. Y. Leung, R. H. Borts, and M. Lichten. 1991. The frequency of meiotic recombination in yeast is independent of the number and position of homologous donor sequences: implications for chromosome pairing. Proc. Natl. Acad. Sci. USA 88:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartwell, L., T. Weinert, L. Kadyk, and B. Garvik. 1994. Cell cycle checkpoints, genomic integrity, and cancer. Cold Spring Harb. Symp. Quant. Biol. 59:259-263. [DOI] [PubMed] [Google Scholar]
- 15.Heude, M., and F. Fabre. 1993. a/alpha-control of DNA repair in the yeast Saccharomyces cerevisiae: genetic and physiological aspects. Genetics 133:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inbar, O., and M. Kupiec. 1999. Homology search and choice of homologous partner during mitotic recombination. Mol. Cell. Biol. 19:4134-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inbar, O., B. Liefshitz, G. Bitan, and M. Kupiec. 2000. The relationship between homology length and crossing over during the repair of a broken chromosome. J. Biol. Chem. 275:30833-30838. [DOI] [PubMed] [Google Scholar]
- 18.Jinks-Robertson, S., and T. D. Petes. 1986. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics 114:731-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiser, G. L., and T. A. Weinert. 1996. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol. Biol. Cell 7:703-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein, H. L. 2001. Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics 159:1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294:867-870. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 23.Lowndes, N. F., and J. R. Murguia. 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Deversion 10:17-25. [DOI] [PubMed] [Google Scholar]
- 24.Lydall, D., and T. Weinert. 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270:1488-1491. [DOI] [PubMed] [Google Scholar]
- 25.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Deversion 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 28.Mercier, G., Y. Denis, P. Marc, L. Picard, and M. Dutreix. 2001. Transcriptional induction of repair genes during slowing of replication in irradiated Saccharomyces cerevisiae. Mutat. Res. 487:157-172. [DOI] [PubMed] [Google Scholar]
- 29.Myung, K., and R. D. Kolodner. 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paciotti, V., M. Clerici, M. Scotti, G. Lucchini, and M. P. Longhese. 2001. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol. 21:3913-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Reversion 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roeder, G. S., and J. M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395-403. [DOI] [PubMed] [Google Scholar]
- 33.Rouse, J., and S. P. Jackson. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9:857-869. [DOI] [PubMed] [Google Scholar]
- 34.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 35.Sung, P., K. M. Trujillo, and S. Van Komen. 2000. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 451:257-275. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, D. A., and F. W. Stahl. 1999. Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics 153:621-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valencia, M., M. Bentele, M. B. Vaze, G. Herrmann, E. Kraus, S. E. Lee, P. Schar, and J. E. Haber. 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414:666-669. [DOI] [PubMed] [Google Scholar]
- 38.Venclovas, C., M. E. Colvin, and M. P. Thelen. 2002. Molecular modeling-based analysis of interactions in the RFC-dependent clamp-loading process. Protein Sci. 11:2403-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinert, T. A., and L. H. Hartwell. 1990. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol. Cell. Biol. 10:6554-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 41.Yan, Y. X., R. H. Schiestl, and L. Prakash. 1995. Mating-type suppression of the DNA-repair defect of the yeast rad6 delta mutation requires the activity of genes in the RAD52 epistasis group. Curr. Genet. 28:12-18. [DOI] [PubMed] [Google Scholar]
- 42.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Deversion 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]