Abstract
Termination of transcription by RNA polymerase II (Pol II) is a poorly understood yet essential step in eukaryotic gene expression. Termination of pre-mRNA synthesis is coupled to recognition of RNA signals that direct cleavage and polyadenylation of the nascent transcript. Termination of nonpolyadenylated transcripts made by Pol II in the yeast Saccharomyces cerevisiae, including the small nuclear and small nucleolar RNAs, requires distinct RNA elements recognized by the Nrd1 protein and other factors. We have used genetic selection to characterize the terminator of the SNR13 snoRNA gene, revealing a bipartite structure consisting of an upstream element closely matching a Nrd1-binding sequence and a downstream element similar to a cleavage/polyadenylation signal. Genome-wide selection for factors influencing recogniton of the SNR13 terminator yielded mutations in the gene coding for the essential Pol II-binding protein Ssu72. Ssu72 has recently been found to associate with the pre-mRNA cleavage/polyadenylation machinery, and we find that an ssu72 mutation that disrupts Nrd1-dependent termination also results in deficient poly(A)-dependent termination. These findings extend the parallels between the two termination pathways and suggest that they share a common mechanism to signal Pol II termination.
A productive cycle of transcription requires the recruitment of RNA polymerase to a promoter, initiation of RNA synthesis, elongation of the nascent RNA chain, and termination of transcription to release the completed RNA and the polymerase from the DNA template (28). Accurate and efficient termination is critical to prevent interference between adjacent transcription units and to allow recycling of RNA polymerase. Distinct mechanisms of termination characterize the three eukaryotic nuclear RNA polymerases (Pols). Pol III, which synthesizes small noncoding RNAs including tRNAs, 5S rRNA, and U6 snRNA, has an intrinsic ability to terminate transcription upon incorporation of 4 to 6 contiguous U residues (1, 10). Pol I transcribes rRNA genes and, like Pol III, terminates within a U-rich region. However, Pol I termination is also dependent on an elongational pause caused by Reb1 protein bound to DNA downstream of the termination site (40).
Pol II synthesizes transcripts with a great diversity of sizes and functions, and the control of termination by Pol II is more complex and less well understood than termination by Pol I or Pol III. The mature 3′ ends of most protein-coding Pol II transcripts (mRNAs) are generated by endonucleolytic cleavage followed by addition of a poly(A) tail to the newly generated 3′ end (58). This 3′ processing is carried out by a complex machinery that associates with Pol II during elongation (37) and engages processing signals in the nascent transcript even as the polymerase continues to transcribe beyond the processing site. Thus, the production of a mature mRNA 3′ end does not require termination of transcription. Nevertheless, pre-mRNA 3′ processing and termination of Pol II transcription are coupled in vivo: the same RNA signals that direct 3′ processing also promote termination of transcription downstream of the processing site in mammalian cells (13, 30, 53) and in yeast (7, 44). Recent studies of the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe have begun to identify components of the cleavage and polyadenylation machinery that participate in coupling termination to 3′ processing (4, 7, 17), and their findings support a model in which a subset of the 3′-processing machinery is involved in the relay of the termination signal to the polymerase. The association of the 3′-processing machinery with the carboxyl-terminal domain (CTD) of Rpb1, the largest subunit of Pol II, may facilitate efficient coupling of termination and processing (6, 38). However, the nature of the termination signal and the mechanism of its transmission to Pol II remain unknown.
In addition to mRNAs, Pol II is responsible for synthesis of many noncoding RNAs, including small nuclear and nucleolar RNAs (snRNAs and snoRNAs). In the yeast S. cerevisiae, many of these RNAs are synthesized from discrete transcription units, but their 3′ ends are not processed by cleavage and polyadenylation. Instead, their mature 3′ ends are produced by the exosome, a complex of 3′-to-5′ exonucleases that acts upon 3′-extended snRNA and snoRNA precursor transcripts that have unpolyadenylated 3′ ends (2, 18, 52). We recently described a pathway for 3′-end formation of snRNA and snoRNA precursor transcripts in yeast that requires the RNA-binding proteins Nrd1 and Nab3 and the presumptive RNA helicase Sen1 (49). Mutations in the Nrd1-dependent pathway block synthesis of mature forms of several snoRNAs. Such mutants instead accumulate elongated snoRNA transcripts that include coding regions and 3′-processing signals of neighboring protein-coding genes and thus become polyadenylated. This readthrough transcription can interfere with transcription from adjacent downstream promoters (49), probably through a “promoter occlusion” mechanism (23). Thus, the Nrd1-dependent 3′-end formation pathway serves the dual functions of enabling 3′-end maturation of nonpolyadenylated RNAs and preventing transcriptional interference by directing termination of transcription.
The Nrd1-dependent termination pathway exhibits several parallels to poly(A)-dependent termination. Like components of the poly(A)-dependent pathway, Nrd1 associates with a phosphorylated form of the Rpb1 CTD (14, 45, 57) and recognizes sequence elements in the nascent transcript (47). Bozzoni and colleagues have proposed a model in which a subset of the pre-mRNA cleavage and polyadenylation factors, including the Rna14 and Rna15 proteins, is utilized to direct cleavage of snoRNA and snRNA precursor transcripts without concomitant polyadenylation of the newly generated 3′ ends (18, 33). It is not clear whether Rna14 and Rna15 function as part of the Nrd1-dependent mechanism or in a distinct snoRNA 3′-end formation pathway.
We are using a genetic approach to further characterize the Nrd1-dependent 3′-end formation pathway. In this study we have used a selectable reporter gene system to define the important sequence features of the 3′-end formation region of the SNR13 snoRNA gene. We find that the SNR13 3′-flanking region contains at least two functionally separable but closely spaced Nrd1-dependent 3′-end formation elements, one of which closely resembles a previously characterized Nrd1-binding site.
We have also used this selection scheme to identify additional trans-acting factors required for the Nrd1-dependent 3′-end formation pathway and have characterized novel mutations in an essential gene, SSU72, that disrupt this pathway. The Ssu72 protein was shown previously to interact genetically and physically with both Pol II and the general transcription initiation factor TFIIB (36, 51) and was found recently to copurify with the cleavage/polyadenylation machinery (16, 20). We present evidence that Ssu72 functions in both Nrd1-dependent and poly(A)-dependent termination of transcription. These results extend parallels between the two termination pathways and suggest a possible mechanism for signaling termination of Pol II transcription.
MATERIALS AND METHODS
Plasmids.
The insertion of the SNR13 125-232 sequence into the XhoI site of the ACT-CUP plasmid pGAC24 has been described elsewhere (49). The SNR13 125-182, 150-232, and 150-182 fragments were likewise introduced into the XhoI site after PCR amplification with primers that introduced XhoI sites flanking the SNR13 sequences. An XhoI-SalI fragment containing the minimal 83-bp CYC1 3′-processing element (43) was similarly cloned into the XhoI site of pGAC24 in both orientations, creating pGAC24-CYC83F and pGAC24-CYC83R.
Plasmids for transcription run-on (TRO) analysis were derived from plasmid pRL542, containing G-less cassettes of 131 and 262 nucleotides (nt) separated by a spacer region of 160 nt (31). The G-less cassette region was amplified by PCR and cloned by using an existing EcoRV site and an introduced SalI site into Ecl136II- and SalI-digested pGAC24. The tandem G-less cassettes thereby replace the ACT-CUP coding region and are transcribed from the TDH3 promoter. The SNR13 125-232 PCR product with 10 nt of primer sequence containing XhoI sites flanking the SNR13 sequence at each end was then cloned by blunt-end ligation into a StuI site located 105 nt downstream of the promoter-proximal 262-nt G-less cassette and 56 nt upstream of the distal 131-nt G-less cassette, generating pG-Leu-SNR13-125-232. The 83-bp CYC1 XhoI-SalI fragment was then cloned between the XhoI sites, replacing the SNR13 fragment and creating pG-Leu-CYCpAmin. A larger fragment of the CYC1 3′-flanking sequence, bounded by an introduced XhoI site flanking nt 425 and an existing SalI site at nt 829 (relative to the first nucleotide of the CYC1 coding region), was similarly introduced between the XhoI sites to create pG-Leu-CYCpAmax. This construct contains 80 nt of CYC1 sequence upstream and 324 nt downstream of the polyadenylation site at nt 505 (8). A smaller fragment containing only 243 nt of sequence downstream of the CYC1 poly(A) site (nt 587 to 829) was similarly introduced between the XhoI sites to create pG-Leu-CYCds. These fragments are diagrammed in Fig. 4A. The SNR13-plus-CYC1 spacer sequence plasmid, pG-Leu-SNR13-CYCds, was created by introducing the XhoI-flanked SNR13 125-232 fragment into the XhoI site at the upstream end of the CYCds fragment (see Fig. 5A).
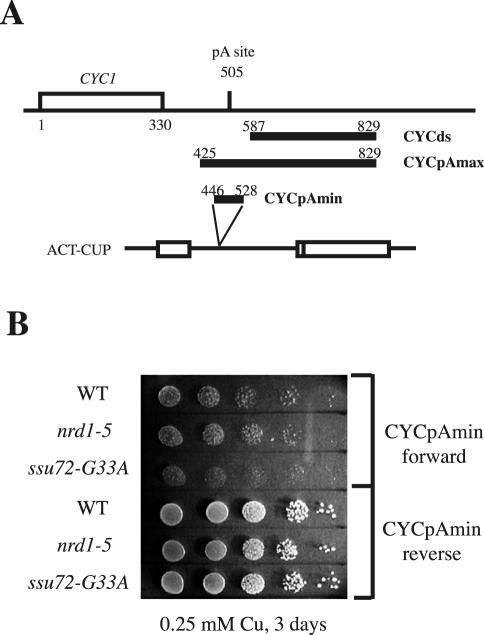
FIG. 4.
ssu72-G33A does not affect recognition of the CYC1 cleavage/polyadenylation site. (A) Schematic illustration of the CYC1 gene and 3′-flanking region showing the 83-bp CYCpAmin fragment inserted into the ACT-CUP reporter gene intron. Also shown are the CYCpAmax and CYCds fragments used in the TRO experiments for which results are shown in Fig. 5. (B) Copper sensitivity assay with wild type, nrd1-5, and ssu72-G33A strains harboring ACT-CUP reporter genes with the CYC1pAmin cleavage/polyadenylation element inserted into the intron in the forward or reverse orientation.
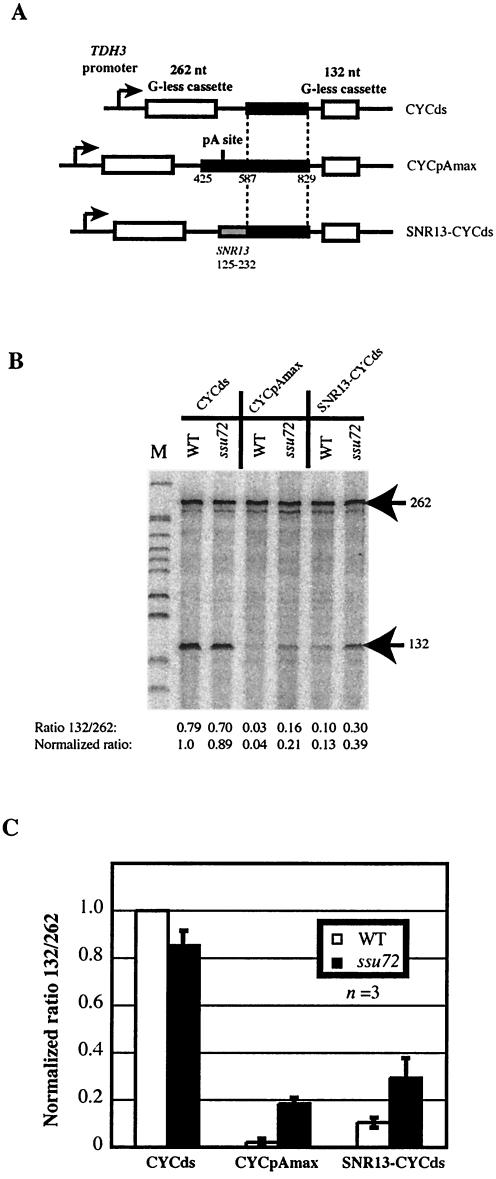
FIG. 5.
Ssu72 mediates both poly(A)-dependent and poly(A)-independent termination. (A) Schematic illustration showing the structure of the tandem G-less cassette constructs used for TRO analysis. Solid rectangles represent sequences derived from the CYC1 3′-flanking region, and the shaded rectangle represents the SNR13 125-232 sequence. (B) Results of G-less cassette TRO analysis of poly(A)-dependent and poly(A)-independent termination in wild-type (WT) and ssu72-G33A mutant strains. Strains were grown at 30°C and then shifted to 37°C for 3 h before the TRO procedure was conducted. Foreach sample, the ratio of total counts incorporated into the distal versus proximal G-less cassettes was determined and was normalized against the ratio for the inert spacer construct in the wild-type strain (CYCds, WT). (C) Bar graph showing averaged results of three experiments like that for which results are shown in panel B. Error bars, standard deviations.
Yeast strains and genetic methods.
The yeast cup1Δ strain 46α and the nrd1-5 mutant have been described previously (48). The cup1Δ strain 46a has the same genotype as 46α but the opposite mating type. Spontaneously arising copper-resistant mutants were selected in NRD1 strains 46a and 46α harboring pGAC24-SNR13 125-232 as well as a second, wild-type allele of NRD1 on the URA3-marked centromeric plasmid pRS316. Approximately 107 cells were spread onto each of 36 plates without uracil (−Ura plates) containing 0.15 mM CuSO4 (18 for each mating type), and plates were incubated at 30°C for ∼7 days. Several colonies from each plate were then screened for heat-sensitive (37°C) or cold-sensitive (16°C) growth defects on yeast extract-peptone-dextrose (YEPD) plates. Recessive temperature-sensitive (ts) and cold-sensitive (cs) mutants were tested for allelism to SEN1 by crossing against sen1Δ::TRP1 tester strains (15) harboring SEN1 on a URA3-marked plasmid, plating the resulting diploids on 5-fluoroorotic acid (5-FOA) plates to select for loss of the SEN1 plasmid, and scoring for ts or cs growth.
The SSU72 gene was cloned by complementation of the temperature-sensitive (37°C) growth defect of a 46α-derived copper-resistant mutant strain with a YCp50-based genomic library (42). The genomic SSU72 locus of this strain and that of a second mutant strain complemented by the same library plasmid were then amplified by PCR and sequenced, and point mutations altering the predicted Ssu72 amino acid sequence were identified in both strains. These strains were designated the ssu72-G33A and ssu72-G42V strains to reflect the amino acid substitutions.
Mutagenesis of the SNR13 125-232 element in the ACT-CUP intron was carried out by error-prone PCR and in vivo homologous recombination (34). Primers complementary to pGAC24 sequences were used to direct amplification by Tfl polymerase (Epicentre Technologies) of a 714-bp fragment containing the SNR13 125-232 sequence. Amplification conditions included 1 mM (each) dCTP, dGTP, and dTTP and 0.2 mM dATP to increase the error rate of synthesis. pGAC24 digested with BamHI and XhoI, creating a gap of 269 bp flanked by 152 and 293 nt of complementarity to the PCR product, was cotransformed with the mutagenized PCR product into yeast strain 46α. After selection for transformants on −Leu plates, colonies were replica plated onto −Leu plates containing 0.15 mM CuSO4. Plasmids were recovered from copper-resistant colonies for sequencing and retransformation into yeast to confirm the copper resistance phenotype.
RNA preparation and Northern blot analysis.
RNA samples were prepared by the glass bead-guanidinium isothiocyanate-hot phenol extraction method (54). Northern blot analysis was performed with 10 μg of total RNA per lane. SCR1 and snR13 RNAs were probed with 5′-32P-labeled oligonucleotides. Nrd1 mRNA was detected with a probe made by random priming of Klenow DNA polymerase on an 836-bp PCR product containing part of the NRD1 coding sequence in the presence of [α-32P]dCTP.
TRO analysis.
Our TRO method was adapted from procedures described elsewhere (7, 35, 56). Wild-type and ssu72-G33A mutant strains harboring G-less cassette TRO plasmids were inoculated to a starting A600 of ∼0.03 and grown in −Leu medium at 30°C. After they reached an A600 of ∼0.1 to 0.15, cultures were shifted to 37°C, and appropriate dilutions were made with 37°C −Leu medium to keep the A600 from exceeding 0.2. After 3 h at 37°C, 5 A600 units of culture (∼5 × 107 cells) were harvested by centrifugation at 3,000 × g for 3 min. Cells were resuspended in 5 ml of ice-cold H2O and then centrifuged at 4°C for 3 min at 3,000 × g. The cells were then resuspended in 950 μl of ice-cold H2O, and 50 μl of 10% Sarkosyl was added. The sample was gently mixed and incubated on ice for 20 min. The permeabilized cells were pelleted by microcentrifugation for 1 min, and the supernatant was carefully removed. Residual supernatant was removed after a second brief microcentrifugation.
Permeabilized cells were resuspended in 71 μl of ice-cold transcription mix composed of 60 μl of transcription buffer (50 mM Tris-HCl [pH 7.7], 500 mM KCl, 80 mM MgCl2), 4 μl of 20 mM ATP, 4 μl of 20 mM CTP, and 3 μl of 100 mM dithiothreitol. Labeling of nascent transcripts was initiated by the addition of 100 μCi of [α-32P]UTP (3,000 Ci/mmol; Amersham), and samples were incubated for 5 min at 30°C. GTP was excluded at this stage to restrict labeling to only one of the two G-less cassettes per transcript (35, 56). The labeling step was followed by a “chase” with excess unlabeled UTP and a small amount of GTP: 10 μl of 25 mM UTP-0.25 mM GTP was added, and incubation was continued for 10 min, allowing transcription of G-less segments to be completed. Reactions were stopped by addition of 900 μl of AE buffer (50 mM sodium acetate-10 mM EDTA [pH 5]) and microcentrifugation for 5 s. The supernatant was removed, and cells were resuspended in 400 μl of TES buffer (10 mM Tris-HCl [pH 7.5]-10 mM EDTA-0.5% sodium dodecyl sulfate) for extraction of RNA by the hot acidic phenol method (12). After precipitation with ethanol, samples were resuspended in 40 μl of H2O. RNA samples were digested for 2 h at 37°C with 5,000 U (5 μl) of RNase T1 (Ambion) in 100 μl of Tris-EDTA buffer, followed by treatment with 2.5 μl of 20% sodium dodecyl sulfate and 2.5 μl of 10-mg/ml proteinase K (Roche) at 37°C for 20 min to inactivate the RNase. Samples were precipitated with ethanol in the presence of 2 μg of Escherichia coli tRNA and 0.3 M sodium acetate and were resuspended in formamide gel-loading buffer before electrophoresis on denaturing (8.3 M urea) 6% polyacrylamide gels. Dried gels were analyzed with a phosphorimager using ImageQuant software (Molecular Dynamics). For each sample, the ratio of total counts in the 132-nt band divided by total counts in the 262-nt band was determined. The large and small cassettes contain 102 and 40 U residues, respectively.
RESULTS
Point mutations define tandem 3′-end formation elements for SNR13.
We showed previously that a Nrd1-dependent 3′-end formation element resides within 108 nt of genomic sequence immediately downstream of the mature 3′ end of the snR13 snoRNA, i.e., nt 125 to 232 with respect to the SNR13 transcription start site (49). In a yeast strain lacking the chromosomal CUP1 gene and relying on expression of a plasmid-borne ACT-CUP fusion gene for growth on copper-containing media, this 108-nt sequence causes RNA truncation and severe copper sensitivity when introduced into the intron of the ACT-CUP gene (Fig. 1A). Copper sensitivity due to the SNR13 Nrd1-responsive element (NRE) is partially relieved in a nrd1 mutant strain (49).
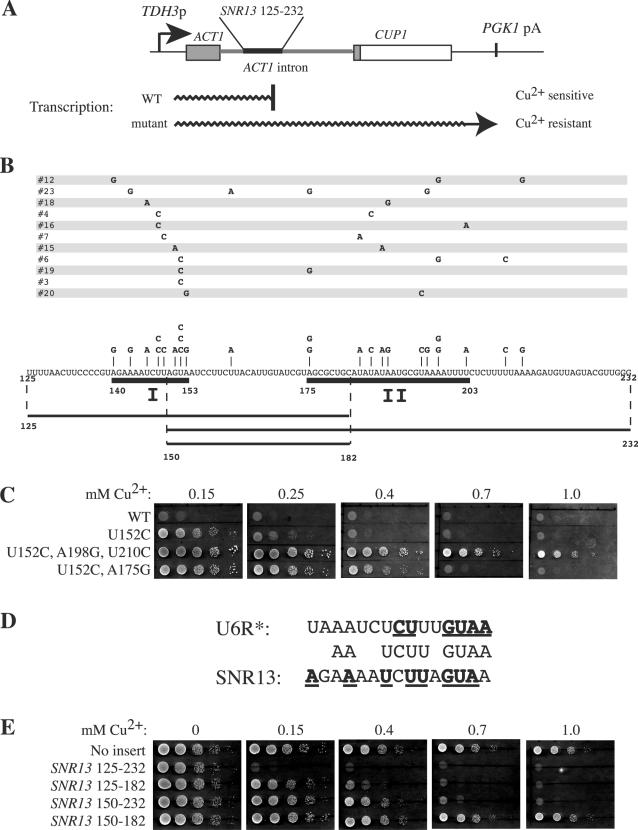
FIG. 1.
The SNR13 3′-flanking region contains tandem Nrd1-dependent 3′-end formation elements. (A) Schematic illustration of the ACT-CUP reporter gene containing the SNR13 3′-end formation element, and effects of the insert on transcription and copper sensitivity. Transcription of the fusion gene is driven by the TDH3 gene promoter (TDH3p), and the PGK1 gene polyadenylation signal (PGK1pA) is present downstreamof the CUP1 coding region. (B) cis-acting mutations in the SNR13 NRE that confer copper-resistant growth. Each line represents a unique allele that has one to four point mutations. All the alleles except no. 3 have mutations in both regions I and II, indicated by heavy underlining. The smaller fragments tested in the copper sensitivity assay in panel E are indicated below the sequence of the 125-232 fragment. (C) Copper sensitivity assay of three alleles bearing the U152C mutation. Tenfold serial dilutions (105 to 101 cells) of strains containing the indicated alleles were spotted on −Leu plates containing CuSO4 at the concentrations shown. (D) Comparison of the region I sequence to a portion of the U6R* NRE. Nucleotides at which point mutations conferring copper resistance have been isolated are boldfaced and underlined. (E) Copper sensitivity assay with SNR13 subfragments containing only region I (125-182) or region II (150-232).
In an attempt to identify important sequence features of the SNR13 NRE, we selected for spontaneously arising copper-resistant colonies. This selection yielded cis-acting mutants in which the SNR13 125-232 sequence was deleted from the reporter gene, but copper-resistant point mutant alleles were not readily recovered. The absence of spontaneous point mutants contrasts with results obtained with a 3′-end formation element from the U4 snRNA gene SNR14 (49) and with the artificial Nrd1-responsive element U6R* (48), suggesting that the SNR13 sequence may contain redundant 3′-end formation elements. Indeed, when the SNR13-derived sequence was subjected to mutagenesis by error-prone PCR, 10 of 11 copper-resistant mutant alleles characterized had at least two mutations, one in each of two discrete regions labeled I and II in Fig. 1B. Additional mutations occurred between regions I and II or downstream of region II, but only in alleles that also had mutations in both regions I and II. One allele contained a single point mutation in region I, U152 to C, that also occurred in two other alleles that had mutations in region II. Comparison of the copper resistance enabled by these three alleles (Fig. 1C) showed that those combining mutations in region II with the U152C mutation confer growth at higher copper concentrations than the U152C mutant alone, indicating that the mutations in regions I and II have additive effects.
The sequence of region I closely resembles that of an artificial Nrd1-binding site characterized previously (47, 48), exhibiting identity at 10 of 15 positions (Fig. 1D). All but one mutation selected in the SNR13 and U6R* NREs are in conserved residues. Therefore, mutations in region I likely disrupt Nrd1 interaction with the transcript. Region II contains several features similar to pre-mRNA 3′-processing elements (22, 58), including an AU rich “efficiency” element (nt 183 to 189), an A-rich “positioning” element (nt 195 to 199), and an extensive U-rich tract beginning at nt 200. To test if regions I and II can function independently, we subcloned fragments containing only one intact region into the ACT-CUP intron (Fig. 1B). The region I subfragment, nt 125 to 182, caused sensitivity to copper at concentrations above 0.4 mM, while a subfragment containing region II (nt 150 to 232) but lacking most of region I caused sensitivity to copper at concentrations above 0.7 mM (Fig. 1E). Copper sensitivity due to either subfragment was strongly relieved in the nrd1-5 mutant (see Fig. 2B). The 150-182 region, contained in both active subfragments, did not cause detectable copper sensitivity on its own (Fig. 1E). The latter result contrasts with the findings of Fatica et al. (18), who reported that this +150-182 region can function to enable 3′-end maturation when introduced into a chimeric snoRNA transcript. However, the finding that the 125-182 and 150-232 fragments can independently direct submaximal 3′-end formation is consistent with the results of Morlando et al. (33).
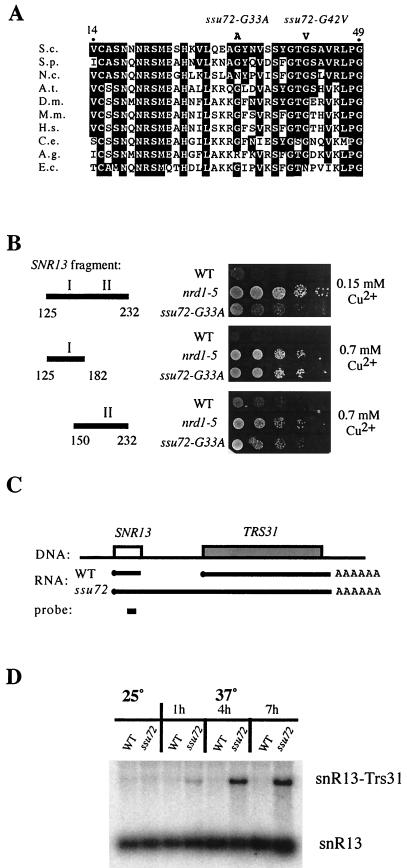
FIG. 2.
ssu72 mutants defective for SNR13 3′-end formation. (A) Sequences of an amino-terminal portion of the Ssu72 protein and homologues identified by BLAST searches, showing the identity of the ssu72-G33A and ssu72-G42V alleles. Residues identical to yeast Ssu72 are boxed. S.c., S. cerevisiae; S.p., S. pombe; N.c., Neurospora crassa; A.t., Arabidopsis thaliana; D.m., Drosophila melanogaster; M.m., Mus musculus; H.s., Homo sapiens; C.e., Caenorhabditis elegans; A.g., Anopheles gambiae; E.c., Encephalitozoon cuniculi. (B) Copper sensitivity assay of ssu72-G33A and nrd1-5 mutants harboring ACT-CUP reporter genes with the indicated SNR13 3′-end formation elements. (C) Schematic illustration of the SNR13 chromosomal locus, including the adjacent gene, TRS31. Lines labeled RNA indicate transcripts that accumulate in wild-type and ssu72 strains, and the position of the oligonucleotide probe used in panel D is shown. (D) Northern blot analysis of transcripts from the endogenous SNR13 locus in wild-type and ssu72-G33A strains, before and after a shift to the restrictive temperature (37°C) for 1, 4, or 7 h.
The SSU72 gene is required for normal SNR13 3′-end formation.
We also used the ACT-CUP reporter gene system to select for trans-acting mutations that allow readthrough of the SNR13 terminator. In preliminary experiments, complementation analysis of spontaneously arising copper-resistant mutant strains with recessive temperature-sensitive growth defects revealed that many of these strains contain mutations allelic to NRD1. Therefore, to facilitate the identification of mutations in other factors involved in this pathway, further selections were carried out in the presence of a second, plasmid-borne NRD1 allele. Approximately half of the recessive copper-resistant, temperature-sensitive mutants selected under these conditions were allelic to SEN1, as revealed by crosses to a sen1Δ strain carrying the SEN1 gene on a URA3-marked plasmid followed by selection against the plasmid on plates containing 5-FOA. From the remaining copper-resistant mutant strains, we chose for further analysis two that had a tight temperature-sensitive growth defect at 37°C and one with a cold-sensitive growth defect at 16°C. A clone obtained by complementation of the cold-sensitive mutant with a genomic library contained the NAB3 gene, whose product is an RNA-binding protein that associates with Nrd1 (14) and is important for Nrd1-dependent 3′-end formation (49).
Analysis of clones obtained by genomic library complementation of the 37°C growth defect of one the two remaining temperature-sensitive mutants revealed that the SSU72 gene was responsible for complementation, and we found subsequently that the SSU72 gene also complemented the second temperature-sensitive mutant. The SSU72 gene product is a small (206-residue) acidic protein that is well conserved among eukaryotes over most of its length and is essential for viability in yeast (36, 51). It was first identified through a mutation, ssu72-1, that exacerbates the growth defect and transcription initiation site selection phenotypes caused by a mutation in the SUA7 gene, which encodes the general transcription initiation factor TFIIB. Subsequent studies identified complex genetic interactions between SSU72 and RPB2, which encodes the second largest subunit of Pol II: an rpb2 mutation in combination with another (unidentified) mutation can suppress the growth defect of the ssu72-2 mutation. Ssu72 protein produced by in vitro translation can bind to recombinant TFIIB as well as to purified Pol II (36, 55). These studies suggested a role for Ssu72 during initiation of transcription. However, recent studies have established that Ssu72 copurifies with the pre-mRNA 3′-processing machinery and may function to couple termination to pre-mRNA processing (16, 20).
Sequencing of the SSU72 gene in the two temperature-sensitive, copper-resistant strains identified distinct mutations, each altering a glycine residue in a highly conserved region near the amino terminus of the protein (Fig. 2A). We have named these alleles ssu72-G33A and ssu72-G42V. Since our selection scheme employed the 108-nt SNR13-derived fragment containing tandem 3′-end formation elements, we tested the ssu72 mutants with the region I and region II elements individually. The copper sensitivity caused by either element was strongly relieved by the ssu72 mutations, but they suppressed the full-length NRE less well than the nrd1-5 mutation (Fig. 2B and data not shown). The ssu72-G33A allele appears to be a stronger suppressor than ssu72-G42V, so in subsequent experiments only the ssu72-G33A allele was used.
To confirm the involvement of Ssu72 in snoRNA 3′-end formation, we analyzed transcripts from the endogenous SNR13 locus. Previous studies showed that disruption of the Nrd1-dependent 3′-end formation pathway leads to the accumulation of extended SNR13 transcripts that read through the neighboring gene downstream, TRS31, before being cleaved and polyadenylated at the TRS31 pre-mRNA processing site (39, 49) (Fig. 2C). The ssu72-G33A mutant strain similarly shows strong accumulation of chimeric SNR13-TRS31 transcripts after a shift to the restrictive temperature (Fig. 2D). Thus, Ssu72 is required for normal termination of transcripts from the endogenous SNR13 locus.
General requirement for Ssu72 in the Nrd1-dependent 3′-end formation pathway.
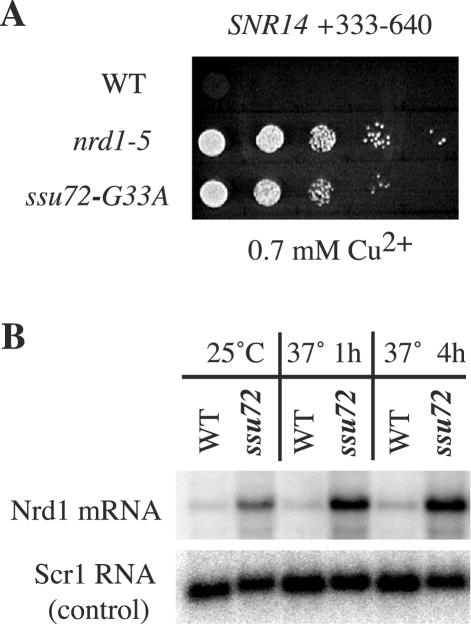
In addition to snoRNA transcripts, the Nrd1 pathway is used for 3′-end formation of primary transcripts of the spliceosomal snRNA U4, encoded by the SNR14 gene in yeast (49). To test if Ssu72 is required for the function of the SNR14 3′-end formation element, we used an ACT-CUP reporter gene containing the NRE from the SNR14 locus. The ssu72-G33A mutation relieved the copper sensitivity caused by the SNR14 NRE, albeit to a slightly lesser degree than the nrd1-5 mutation (Fig. 3A).
FIG. 3.
General requirement for SSU72 in the Nrd1-dependent 3′-end formation pathway. (A) Copper sensitivity assay of nrd1-5 and ssu72-G33A mutants harboring an ACT-CUP reporter gene with an SNR14 3′-end formation element in the intron. (B) Northern blot analysis of Nrd1 mRNA in wild-type and ssu72-G33A strains at permissive (25°C) and restrictive (37°C for 1 or 4 h) temperatures. Scr1 is the signal recognition particle RNA and is synthesized by Pol III.
The Nrd1-dependent 3′-end formation pathway also functions to autoregulate the synthesis of the Nrd1 mRNA, through an element in the 5′ untranslated region of the transcript. Thus, mutations that disrupt the Nrd1-dependent 3′-end formation pathway result in the increased accumulation of full-length Nrd1 mRNA (49). As shown in Fig. 3B, the ssu72-G33A mutant accumulates increased amounts of Nrd1 mRNA, even at the permissive temperature of 25°C, and this accumulation increases further to ∼6-fold after a shift to 37°C. These results strongly support the conclusion that Ssu72 is a general component of the Nrd1-dependent 3′-end formation pathway.
The ssu72-G33A mutant does not disrupt pre-mRNA processing at the CYC1 poly(A) site.
Our copper resistance selection scheme requires partial loss of function on the SNR13 terminator but sufficient function at the PGK1 poly(A) site to allow proper 3′-end formation of the ACT-CUP mRNA. This suggests that the ssu72-G33A mutant is not significantly impaired in cleavage and polyadenylation, despite the association of the Ssu72 protein with the cleavage/polyadenylation machinery (16). Northern blot analysis of Nrd1 mRNA shows that the NRD1 poly(A) site remains functional in the ssu72-G33A mutant, even after a shift to the restrictive temperature of 37°C (Fig. 3B). To further investigate whether the ssu72-G33A mutation confers a defect in pre-mRNA 3′-end formation, we introduced a well-characterized minimal 83-bp pre-mRNA cleavage/polyadenylation element from the CYC1 gene into the ACT-CUP intron (Fig. 4). Consistent with previous studies demonstrating that this minimal CYC1 element can direct cleavage and polyadenylation when located in the intron of a similar reporter gene construct (43), the CYCpAmin insertion resulted in sensitivity to a copper concentration of 0.25 mM. No increase in copper-resistant growth was observed with this construct in ssu72 or nrd1 mutant strains incubated for 3 days at 30°C. Note that robust copper-resistant growth was observed with the SNR13 element under these conditions in the ssu72 and nrd1 mutants (Fig. 2B). We conclude that the ssu72-G33A mutation does not significantly disrupt pre-mRNA cleavage and polyadenylation for at least two different processing sites. We cannot rule out the possibility that cleavage at other poly(A) sites is impaired in the ssu72-G33A mutant, as recently demonstrated for the ssu72-2, ssu72-3, and ssu72-7 alleles at the GAL7 processing site in vitro (25) (see Discussion).
Ssu72 mediates both poly(A)-dependent and poly(A)-independent termination.
To investigate more directly the role of Ssu72 in the termination of transcription, we adopted a TRO approach. This technique has been used extensively to study termination in response to cleavage/polyadenylation signals in yeast (5, 7, 16, 17, 24) and to show that the Nrd1 pathway directs termination of a snoRNA transcript (49). In a TRO experiment, cells are permeabilized to allow the limited incorporation of labeled nucleotides into nascent transcripts. The labeled transcripts are typically hybridized to a series of immobilized probes to reveal the distribution of polymerases along a transcription unit in a population of cells.
In order to simplify and standardize the analysis of different 3′-end formation elements, we developed a variation of the TRO method that exploits the use of G-less cassettes (35, 56), commonly employed for in vitro studies of transcription (see Materials and Methods). This adaptation allows TRO products to be analyzed directly after treatment with T1 RNase and denaturing gel electrophoresis, thus avoiding the requirement for hybridization to immobilized single-stranded DNA probes. Our G-less TRO constructs use the strong, constitutive TDH3 promoter (from which the ACT-CUP reporter gene is also expressed) to drive transcription of two G-less cassettes of 262 and 132 nt, separated by a spacer region of 161 nt (Fig. 5A). Poly(A)-dependent and Nrd1-dependent 3′-end formation elements were introduced into the space between the two G-less cassettes, and the ratios of radioactivity incorporated into the 262- and 132-nt products in wild-type and mutant strains were compared.
Preliminary results indicated that minimal termination elements elicited only modest termination within the short distance (60 nt) between the site of terminator insertion and the second G-less cassette, necessitating the insertion of additional “spacer” sequence to allow efficient termination before the second cassette. We used a 243-nt CYC1 fragment beginning 83 nt downstream of the cleavage/polyadenylation site (CYCds) as an inert spacer sequence, since previous studies have shown that termination occurs in this region only when a functional 3′-processing signal is present upstream (5, 7). As expected, the CYCds insertion resulted in undiminished labeling of the promoter-distal cassette relative to the construct with no insert (Fig. 5B and data not shown). Inclusion of the 5′-adjacent 162 bp of CYC1, which encompasses the cleavage and polyadenylation site (Fig. 5A, CYCpAmax), resulted in greatly diminished labeling of the distal G-less cassette (Fig. 5B). By use of the CYCds construct as a control to normalize the ratio of 32P incorporated into distal versus proximal G-less cassettes, the efficiency of termination directed by the CYC1 cleavage/polyadenylation element was found to be greater than 95% (Fig. 5C).
Transcriptional readthrough of the CYC1 poly(A)-dependent terminator was strongly increased in the ssu72-G33A mutant strain shifted to the restrictive temperature of 37°C (Fig. 5C). The ratio of label incorporated into distal versus proximal cassettes was increased approximately four- to fivefold compared to that for the wild type, supporting the conclusion that Ssu72 is required for efficient poly(A)-dependent termination. No increased labeling of the distal cassette was seen in the ssu72-G33A mutant when only the CYCds fragment was present. This termination defect distinguishes the ssu72-G33A from the ssu72-2 mutant, which showed little or no increase in transcriptional readthrough of the CYC1 processing site (16, 25) (see Discussion).
We next analyzed termination directed by the SNR13 125-232 element by inserting this sequence in place of the CYC1 processing signal upstream of the inert CYCds spacer sequence (Fig. 5A). The SNR13 sequence directed efficient termination upstream of the 132-nt G-less cassette, with the ratio of distal to proximal G-less cassette transcription reduced to only ∼10 to 15% of that for the spacer-only (CYCds) control (Fig. 5B and C). The ssu72-G33A mutant strain showed a clear defect in termination directed by the SNR13 125-232 element. Increased transcription of the distal cassette was observed in the mutant strain after a shift to 37°C either with the SNR13 element insert alone (data not shown) or with the SNR13-CYCds construct (Fig. 5C). We conclude that Ssu72 functions in both Nrd1-dependent and poly(A)-dependent termination.
DISCUSSION
Multiple cis-acting elements direct SNR13 3′-end formation.
Our genetic approach provides a powerful tool for the identification and analysis of cis-acting elements and trans-acting factors involved in 3′-end formation of Pol II transcripts. Mutational analysis reveals a surprising complexity of the 3′-flanking region of the SNR13 gene, which contains at least two closely spaced 3′-end formation elements with disparate sequences. One of these elements strongly resembles a previously characterized Nrd1-binding site, and the response to this element is dependent on the Nrd1 protein. A second element contains several features characteristic of cleavage/polyadenylation sites. Polyadenylated precursors would not be expected to yield functional snR13 snoRNA; however, the presence of a polyadenylation site may serve to help prevent readthrough transcription from interfering with expression of the adjacent gene, TRS31. Surprisingly, the response to the second SNR13 element is also dependent on Nrd1, unlike the CYC1 cleavage/polyadenylation element, suggesting that the second SNR13 element does not function simply to direct cleavage and polyadenylation. Region II does contain a weak match to the Nrd1-binding consensus, including a UxxGUAA sequence found both in region I and in U6R* (Fig. 1B and D). Alternatively, protein-protein interactions with other RNA-binding proteins, such as Nab3, may recruit Nrd1 to region II in the absence of a high-affinity Nrd1-binding site. Interestingly, the artificial NRE with which Nrd1 was discovered, U6R*, contains tandem Nrd1-binding sites, both of which are required for full function (47, 48). It remains to be determined how common this tandem arrangement is in natural NREs. The similarities between region II and yeast cleavage/polyadenylation signals are consistent with the hypothesis that Nrd1-dependent termination may use a subset of cleavage/polyadenylation factors.
Functions of Ssu72 in 3′-end formation of Pol II transcripts.
It is important to draw clear distinctions between different molecular events in 3′-end formation of Pol II transcripts, which may include endonucleolytic cleavage of a nascent transcript as well as termination of transcription. Cleavage of nascent pre-mRNA transcripts by the cleavage and polyadenylation machinery frees the functional portion of the pre-mRNA from the transcription complex, but termination is still required to achieve release of the polymerase and the remaining transcript fragment from the template. Although transcript cleavage and termination are thus distinct events, termination is at least in some cases coupled to recognition of pre-mRNA 3′-processing sequences (7, 13, 30, 53).
Recent studies have demonstrated that mutations in the cleavage/polyadenylation machinery can result in the uncoupling of RNA processing and termination. For example, mutations affecting Yhh1, a component of the cleavage/polyadenylation factor CPF, result in defects in poly(A)-dependent termination but do not disrupt cleavage and polyadenylation (17). On the other hand, mutations affecting the Rna14, Rna15, or Pcf11 protein (proteins which, together with the Clp1 protein, constitute cleavage/polyadenylation factor CFIA) disrupt both RNA processing and termination, while mutations affecting other CPF subunits such as Fip1 or Yth1 disrupt processing but not termination (7). The ssu72-G33A mutant is deficient for poly(A)-dependent termination but competent for cleavage and polyadenylation. Thus, Ssu72 may function like Yhh1 to couple termination to recognition of cleavage/polyadenylation signals.
A cleavage mechanism related to that which occurs at pre-mRNA polyadenylation sites, but which does not result in polyadenylation of the 3′ ends produced by cleavage, has been proposed to function in snoRNA 3′-end formation (18, 33). Interestingly, the putative cleavage site identified in the SNR13 transcript by these studies maps precisely to the 3′ end of region I. Thus, termination of pre-snoRNA transcripts, like that of pre-mRNA transcripts, may be coupled to recognition of signals that also direct RNA cleavage. An unpolyadenylated primary 3′ end is thought to be a prerequisite for appropriate snoRNA 3′-end maturation by the exosome. However, unpolyadenylated primary 3′ ends produced by termination rather than cleavage may serve equally well as exosome substrates. Indeed, if the exosome associates with elongating Pol II in yeast, as was recently reported for Drosophila (3), this may allow for efficient coupling of snoRNA 3′-end maturation to termination of transcription.
The results of TRO experiments presented here clearly demonstrate that the SNR13 3′-end formation element directs efficient termination of transcription and that Ssu72 contributes to this termination event. Our results do not directly address the issue of whether or not endonucleolytic cleavage is an essential step in snoRNA 3′-end formation. The simplest model would posit a role for Ssu72 in the snoRNA 3′-end formation pathway similar to that observed for the pre-mRNA pathway; i.e., the ssu72-G33A mutation would disrupt termination but not pre-snoRNA cleavage. However, the ssu72-G33A mutant allows copper-resistant growth in our selection scheme and accumulates extended chimeric SNR13-TRS31 transcripts, suggesting that a significant proportion of snoRNA transcripts normally terminate without cleavage of the transcript. Alternatively, the ssu72-G33A mutation may inhibit pre-snoRNA cleavage but not pre-mRNA cleavage.
While this report was in preparation and under review, two reports were published demonstrating roles for Ssu72 in 3′-end formation of snoRNA and mRNA transcripts (19, 25). In the first of these, Ganem et al. (19) identified within Ssu72 a signature motif found in a family of low-molecular-weight tyrosine phosphatases and demonstrated that a recombinant Ssu72 protein can dephosphorylate the small-molecule substrate para-nitrophenyl phosphate (PNPP). A similar conclusion was also reported by Meinhart et al. (32). Ganem et al. also observed the accumulation of chimeric snoRNA/mRNA and mRNA/mRNA transcripts in an ssu72-ts mutant strain, indicative of a termination defect, and consistent with our results.
The second report, by He at al. (25), demonstrates a requirement for Ssu72 in pre-mRNA cleavage at a polyadenylation site in vitro: depletion of Ssu72 protein or heat inactivation of extracts prepared from temperature-sensitive ssu72-2, ssu72-3, and ssu72-7 mutant strains results in failure to cleave a GAL7-derived 3′-processing substrate. However, the ssu72-2 mutant strain did not show a defect in CYC1 poly(A) site-dependent termination, as was also reported by Dichtl et al. (16). The discrepancy between these reports and our demonstration of termination defects in the ssu72-G33A mutant is likely a reflection of allele specificity. Both the G33A and G42V mutations in the strains we isolated occur near the amino terminus of the protein and the phosphatase signature motif and were recovered from a selection for defects in SNR13 termination. In contrast, the ssu72-2 mutation (R129A) was selected only for temperature sensitivity (36). The identities of the ssu72-3 and ssu72-7 mutations (25) have not been reported.
He et al. also used a ts-degron in vivo depletion strategy to examine the role of Ssu72 in termination (25). As noted by the authors, the SSU72-td allele shows significant transcriptional readthrough of the CYC1 poly(A)-dependent terminator relative to wild-type SSU72. While the authors conclude that Ssu72 is dispensable for termination, based on the observation that readthrough of the CYC1 terminator does not increase further upon a temperature shift and depletion of Ssu72, the observation of significant readthrough due to the N-terminal degron fusion is consistent with a role for Ssu72 in termination.
Taken together, our results and those presented in other recent reports indicate that the Ssu72 protein functions both in poly(A) site cleavage and in termination of Pol II transcription. Ssu72 mutations differentially affecting one or the other of these functions can uncouple processing and termination. A precedent for distinct mutations within a single factor having differential effects on termination and processing is provided by a recent study of the CFIA subunit Pcf11 (45).
Termination signal transduction from the transcript to Pol II.
It is interesting to consider previously described genetic interactions involving SSU72 in light of its role in termination. Although identified initially in a selection for suppressors of a mutation in the SUA7 gene, which encodes the general transcription initiation factor TFIIB, the ssu72-1 mutation exacerbates the growth defect and transcription start site selection phenotypes of the sua7-2 allele and other sua7 alleles (50, 55). These results were interpreted in terms of a direct role for Ssu72 in initiation, and dual functions in initiation and termination cannot be ruled out, particularly since Ssu72 binds directly to TFIIB (55). However, defective initiation might also be an indirect consequence of faulty termination, since readthrough polymerases from adjacent transcription units may displace preinitiation complexes already weakened by TFIIB mutations. Intriguingly, the SUB1 gene, whose product associates with the cleavage/polyadenylation machinery (9, 25) and has been proposed to function as an antiterminator in the poly(A)-dependent termination pathway (9), interacts genetically with a similar subset of sua7 alleles (55).
Recent studies have focused much attention on the association of RNA processing factors and elongation factors with the Pol II CTD (5, 17, 26, 29, 41). The state of phosphorylation of the CTD correlates with the progression of Pol II through the transcription cycle. A hypophosphorylated CTD is thought to be required for initiation of transcription, while phosphorylation of serine-5 of the CTD heptad repeat correlates with entry into elongation and binding of capping enzymes, and subsequent phosphorylation of serine-2 correlates with binding of 3′-processing factors and termination (11, 29). The discovery of an intrinsic phosphatase activity of Ssu72 is intriguing given the importance of CTD phosphorylation in the transcription cycle, but the natural substrates of this proposed phosphotyrosine phosphatase activity remain to be identified (19, 32).
Interactions with the CTD may facilitate the transmission of termination signals from 3′-processing factors to the polymerase, but additional contacts with the polymerase core may be required to effect disruption of the transcript elongation complex. The interaction between Ssu72 and Rpb2, the second largest subunit of Pol II (16, 36), may provide one such contact, since elements of the Rpb1 and Rpb2 subunits have been proposed to help stabilize the elongation complex through contacts with the RNA-DNA duplex and the unwound portion of the DNA (21). Termination-altering mutations have been identified in Rpb2-homologous subunits of bacterial RNA polymerase (27) and yeast Pol III (46), consistent with the possibility that Rpb2 is an important target for termination signals that may be transmitted through Ssu72. Further study is needed to elucidate the role of Ssu72 and its potential phosphatase activity in the transmission of the termination signal and to identify the site of action of this signal within Pol II.
Acknowledgments
We thank T. Platt and R. Landick for plasmids, M. Culbertson for strains, J. Cloute and D. Ebacher for technical assistance, and members of the Brow lab for discussions.
This work was supported by Public Health Service grant GM44665.
REFERENCES
- 1.Allison, D. S., and B. D. Hall. 1985. Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 4:2657-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrulis, E. D., J. Werner, A. Nazarian, H. Erdjument-Bromage, P. Tempst, and J. T. Lis. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420:837-841. [DOI] [PubMed] [Google Scholar]
- 4.Aranda, A., and N. Proudfoot. 2001. Transcriptional termination factors for RNA polymerase II in yeast. Mol. Cell 7:1003-1011. [DOI] [PubMed] [Google Scholar]
- 5.Barilla, D., B. A. Lee, and N. J. Proudfoot. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. [DOI] [PubMed] [Google Scholar]
- 7.Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280:298-301. [DOI] [PubMed] [Google Scholar]
- 8.Butler, J. S., and T. Platt. 1988. RNA processing generates the mature 3′ end of yeast CYC1 messenger RNA in vitro. Science 242:1270-1274. [DOI] [PubMed] [Google Scholar]
- 9.Calvo, O., and J. L. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7:1013-1023. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, F. E., Jr., and D. R. Setzer. 1992. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 12:2260-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collart, M. A., and S. Oliviero. 1993. Saccharomyces cerevisiae, p. 13.12.1-13.12.2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, suppl. 23. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 13.Connelly, S., and J. L. Manley. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 2:440-452. [DOI] [PubMed] [Google Scholar]
- 14.Conrad, N. K., S. M. Wilson, E. J. Steinmetz, M. Patturajan, D. A. Brow, M. S. Swanson, and J. L. Corden. 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154:557-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeMarini, D. J., M. Winey, D. Ursic, F. Webb, and M. R. Culbertson. 1992. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2154-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dichtl, B., D. Blank, M. Ohnacker, A. Friedlein, D. Roeder, H. Langen, and W. Keller. 2002. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10:1139-1150. [DOI] [PubMed] [Google Scholar]
- 17.Dichtl, B., D. Blank, M. Sadowski, W. Hubner, S. Weiser, and W. Keller. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 21:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fatica, A., M. Morlando, and I. Bozzoni. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19:6218-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganem, C., F. Devaux, C. Torchet, C. Jacq, S. Quevillon-Cheruel, G. Labesse, C. Facca, and G. Faye. 2003. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 22:1588-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 21.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292:1876-1882. [DOI] [PubMed] [Google Scholar]
- 22.Graber, J. H., G. D. McAllister, and T. F. Smith. 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greger, I. H., A. Aranda, and N. Proudfoot. 2000. Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:8415-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greger, I. H., and N. J. Proudfoot. 1998. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, X., A. U. Khan, H. Cheng, D. L. Pappas, Jr., M. Hampsey, and C. L. Moore. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 17:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landick, R., J. Stewart, and D. N. Lee. 1990. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 4:1623-1636. [DOI] [PubMed] [Google Scholar]
- 28.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 29.Licatalosi, D. D., G. Geiger, M. Minet, S. Schroeder, K. Cilli, J. B. McNeil, and D. L. Bentley. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 30.Logan, J., E. Falck-Pedersen, J. E. Darnell, Jr., and T. Shenk. 1987. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc. Natl. Acad. Sci. USA 84:8306-8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.London, L., R. G. Keene, and R. Landick. 1991. Analysis of premature termination in c-myc during transcription by RNA polymerase II in a HeLa nuclear extract. Mol. Cell. Biol. 11:4599-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meinhart, A., T. Silberzahn, and P. Cramer. 2003. The mRNA transcription/processing factor ssu72 is a potential tyrosine phosphatase. J. Biol. Chem. 278:15917-15921. [DOI] [PubMed] [Google Scholar]
- 33.Morlando, M., P. Greco, B. Dichtl, A. Fatica, W. Keller, and I. Bozzoni. 2002. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 22:1379-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8:79-82. [DOI] [PubMed] [Google Scholar]
- 35.Orozco, I. J., S. J. Kim, and H. G. Martinson. 2002. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J. Biol. Chem. 277:42899-42911. [DOI] [PubMed] [Google Scholar]
- 36.Pappas, D. L., Jr., and M. Hampsey. 2000. Functional interaction between Ssu72 and the Rpb2 subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:8343-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proudfoot, N., and J. O'Sullivan. 2002. Polyadenylation: a tail of two complexes. Curr. Biol. 12:R855-R857. [DOI] [PubMed] [Google Scholar]
- 38.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen, T. P., and M. R. Culbertson. 1998. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6885-6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeder, R. H., P. Guevara, and J. G. Roan. 1999. Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol. Cell. Biol. 19:7369-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez, C. R., E. J. Cho, M. C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose, M. D., P. Novick, J. H. Thomas, D. Botstein, and G. R. Fink. 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60:237-243. [DOI] [PubMed] [Google Scholar]
- 43.Ruohola, H., S. M. Baker, R. Parker, and T. Platt. 1988. Orientation-dependent function of a short CYC1 DNA fragment in directing mRNA 3′ end formation in yeast. Proc. Natl. Acad. Sci. USA 85:5041-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russo, P., and F. Sherman. 1989. Transcription terminates near the poly(A) site in the CYC1 gene of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86:8348-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadowski, M., B. Dichtl, W. Hubner, and W. Keller. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 22:2167-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaaban, S. A., B. M. Krupp, and B. D. Hall. 1995. Termination-altering mutations in the second-largest subunit of yeast RNA polymerase III. Mol. Cell. Biol. 15:1467-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinmetz, E. J., and D. A. Brow. 1998. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. USA 95:6699-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinmetz, E. J., and D. A. Brow. 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16:6993-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinmetz, E. J., N. K. Conrad, D. A. Brow, and J. L. Corden. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413:327-331. [DOI] [PubMed] [Google Scholar]
- 50.Sun, Z.-W., and M. Hampsey. 1996. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell. Biol. 16:1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, Z. W., A. Tessmer, and M. Hampsey. 1996. Functional interaction between TFIIB and the Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 24:2560-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitelaw, E., and N. Proudfoot. 1986. Alpha-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α2 globin gene. EMBO J. 5:2915-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wise, J. A. 1991. Preparation and analysis of low molecular weight RNAs and small ribonucleoproteins. Methods Enzymol. 194:405-415. [DOI] [PubMed] [Google Scholar]
- 55.Wu, W. H., I. Pinto, B. S. Chen, and M. Hampsey. 1999. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153:643-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeung, G., L. M. Choi, L. C. Chao, N. J. Park, D. Liu, A. Jamil, and H. G. Martinson. 1998. Poly(A)-driven and poly(A)-assisted termination: two different modes of poly(A)-dependent transcription termination. Mol. Cell. Biol. 18:276-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuryev, A., M. Patturajan, Y. Litingtung, R. V. Joshi, C. Gentile, M. Gebara, and J. L. Corden. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. USA 93:6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]