Abstract
V(D)J recombinase mediates rearrangements at immune loci and cryptic recombination signal sequences (cRSS), resulting in a variety of genomic rearrangements in normal lymphocytes and leukemic cells from children and adults. The frequency at which these rearrangements occur and their potential pathologic consequences are developmentally dependent. To gain insight into V(D)J recombinase-mediated events during human development, we investigated 265 coding junctions associated with cRSS sites at the hypoxanthine-guanine phosphoribosyltransferase (HPRT) locus in peripheral T cells from 111 children during the late stages of fetal development through early adolescence. We observed a number of specific V(D)J recombinase processing features that were both age and gender dependent. In particular, TdT-mediated nucleotide insertions varied depending on age and gender, including percentage of coding junctions containing N-nucleotide inserts, predominance of GC nucleotides, and presence of inverted repeats (Pr-nucleotides) at processed coding ends. In addition, the extent of exonucleolytic processing of coding ends was inversely related to age. We also observed a coding-partner-dependent difference in exonucleolytic processing and an age-specific difference in the subtypes of V(D)J-mediated events. We investigated these age- and gender-specific differences with recombination signal information content analysis of the cRSS sites in the human HPRT locus to gain insight into the mechanisms mediating these developmentally specific V(D)J recombinase-mediated rearrangements in humans.
The development of a mature and diverse immune system is generated by V(D)J recombinase-mediated genomic recombination in lymphoid cells occurring primarily during the late stages of fetal and early childhood development. These somatic DNA rearrangements are mediated by the recombinase-activating gene 1 and 2 proteins (RAG1/2)3 and nonhomologous end joining (NHEJ) proteins (reviewed in Refs. 1–3). Normal recombination events rearrange variable (V), diversity (D), and joining (J) segments of the Ig and TCR genes allowing for diversity in Ag-specific recognition. The sites of genomic rearrangement are specified by recombination signal sequences (RSS) that immediately flank each immune gene segment. Immune-specific RSS are composed of heptamer (consensus CACAGTG) and nonamer (consensus ACAAAAACC) sequences separated by a 12- or 23-nonconserved base pair spacer. Efficient joining of sites takes place between a 12- and a 23-bp RSS. The RAG1/2 proteins and the high-mobility group DNA-binding proteins HMG1 and HMG2 bind and align the RSS within a synaptic complex. The DNA is subsequently cleaved generating double-strand breaks at the borders between the coding gene segments and the RSS. The blunt 5′ phosphorylated signal ends, containing the RSS, ligate forming a signal joint that is usually a precise joining of the heptamer sequences with no addition or loss of nucleotides. The covalently sealed hairpin coding ends must be opened and processed before they can be joined (Fig. 1). The end processing and joining are conducted by the NHEJ protein complex along with the RAG 1/2 proteins. The hairpin coding ends are opened by nicking (Fig. 1). Nicking can occur a few bases from the terminal position creating a short single-stranded extension, which, when incorporated into the junction, generates a short palindromic region (P-nucleotides). The coding ends can then be subject to exonucleolytic activity that results in a loss of bases, the amount and extent of which is associated with the sequence context at the coding end (4–6). TdT adds nontemplated nucleotides (N-nucleotides) to the 3′ termini of DNA strands. TdT preferentially adds G nucleotides resulting in primarily G-C rich N-regions (7). Inverted repeats, dependent on TdT activity, have been observed at exonucleolytically processed coding ends. The current model for this occurrence suggests that they are the consequence of stem loop structures formed by complementary N-nucleotide sequences. The proposed processing of these structures by Artemis:DNA-PKcs is thought to result in the presence of recessed inverted repeats termed Pr-nucleotides (Fig. 1) (8, 9). After processing, there is alignment-based gap fill-in, believed mediated by the family X polymerases pol μ and pol λ, followed by end ligation resulting in a coding joint that is retained in the chromosome (8). The exonucleolytic activity as well as the P-nucleotide and N-nucleotide insertions add variability at the coding joint and contribute to the diversity of Ag-specific recognition. These reactions result in the molecular “signature” of V(D)J recombinase-mediated genomic events.
FIGURE 1.
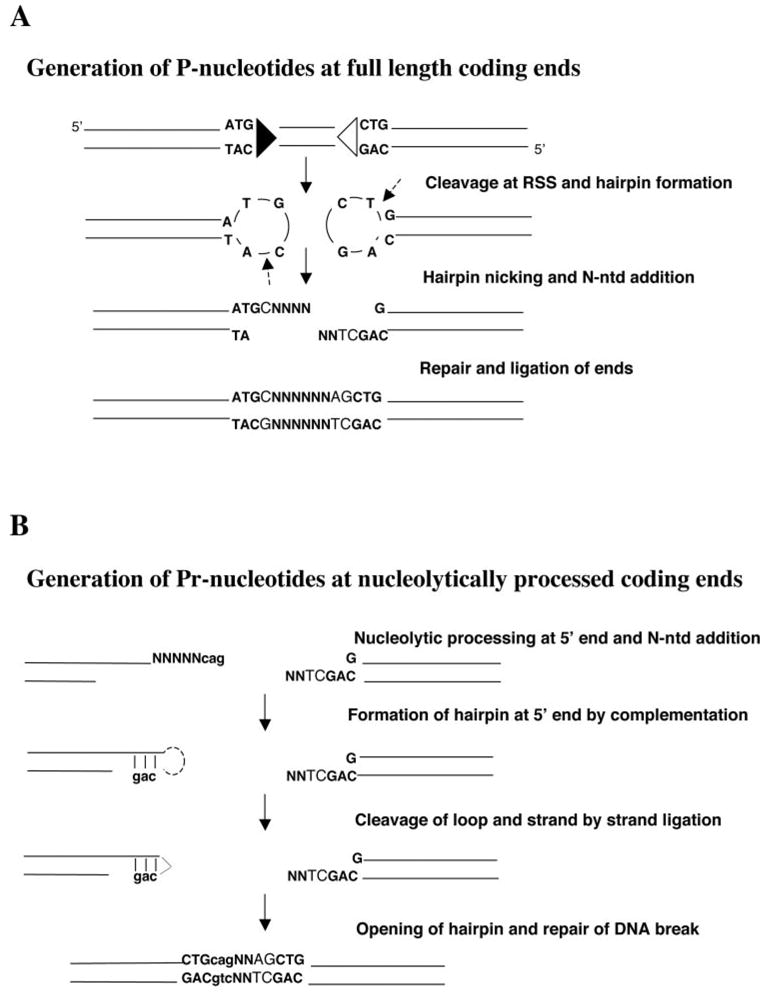
Schematic of coding end processing leading to P-nucleotide and Pr-nucleotide inserts at coding joints. A, The formation of a coding joint with P-nucleotides and N-nucleotide additions. B, The exonucleolytic processing at the 5′ coding end and the generation of Pr-nucleotides. Filled and open triangles represent the RSS sites. Bolded nucleotides are from the coding end. Uppercase unbolded nucleotides are P-nucleotides. Ns represent N-nucleotides whereas lowercase nucleotides are Pr-nucleotides (N-nucleotides that result in an inverted repeat).
RAG-mediated rearrangements have been observed at nonimmune cryptic RSS (cRSS) sites with both pathologic and non-pathologic consequences. Pathologic events have been associated with numerous cytogenetic alterations at clinically significant genes associated with T and B cell leukemia. These events include but are not limited to sil-tal1 deletions, as well as rearrangements at the MLL and MTS loci (10–18). Nonpathologic V(D)J recombinase rearrangements have also been observed and studied at the hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene. RAG-mediated rearrangements occur within the HPRT gene, resulting in the deletion of a DNA fragment of ~20 kb, which include exons 2 and 3 (Fig. 2) (19–22). Analysis of the breakpoints of these deletion events identified multiple cRSS sites that result in three different RAG-mediated deletions. The two most common HPRT RAG-mediated deletions involve a single 5′ cRSS adjacent to position 2197 in intron 1, in combination with a 3′ cRSS adjacent to either position 22251 or 22569 within intron 3 (termed class I and class III, respectively; Fig. 2). These specific RAG-mediated events have no clinical consequences and render the HPRT locus a useful, in vivo, unselected biomarker for studying cRSS-mediated V(D)J recombination events.
FIGURE 2.
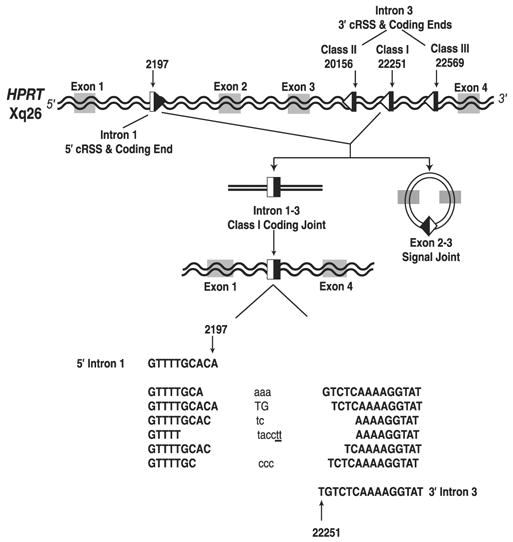
Diagram of the functional cRSS sites within the HPRT locus. RSS sites are represented with triangles and adjacent coding end sequences are represented with boxes. V(D)J recombination occurs between the 5′ cRSS at 2197 in intron 1 and one of the 3′ cRSS sites in intron 3, resulting in the ~20-kb deletion of exons 2 and 3. This schematic shows the formation of class I HPRT coding joints (coding ends 2197 and 22251). Representative class I coding joint sequences are shown. Inserted nucleotides include P-nucleotides (upper case un-bolded), N-nucleotides (lower-case), and Pr-nucleotides (lowercase and underlined).
We previously reported that the frequency of HPRT V(D)J recombinase-mediated deletions in T cells from preterm and full-term newborns is age and gender specific (23). Specifically, V(D)J recombinase-mediated HPRT deletions were shown to increase ~13% with each week of decreasing gestational age and were significantly higher in females (22, 23). We also observed that the percentage of RAG-mediated coding joints in the preterm newborns that did not contain N-nucleotide insertions was significantly higher compared with full-term newborns, suggesting that TdT activity increases with increasing gestational age (22).
Previous studies of V(D)J coding joints from unselected substrates have focused on processing of plasmid V(D)J substrates (4–6, 24). Our laboratory has generated a large database of 196 new and 69 previously published HPRT class I and class III RAG-mediated coding joints occurring in healthy children during human development (19, 20, 22). In this study, we examine gender- and age-dependent differences in V(D)J recombinase-mediated rearrangements occurring in vivo by analyzing coding joint processing at the HPRT locus, an unselected chromosomal V(D)J substrate during the late stages of fetal development through early adolescence.
Materials and Methods
Study populations and sample isolation
Heparinized umbilical cord blood samples were obtained from preterm infants (26–35 wk gestation), full-term infants (36–42 wk gestation), and heparinized peripheral blood samples from healthy children ≤ 12.5 years of age. These subjects were recruited from the labor and delivery unit at Fletcher Allen Health Care hospital at the University of Vermont College of Medicine and the general pediatric clinics of the Department of Pediatrics at the University of Vermont. Informed consent was obtained for all subjects, following the procedure approved by the Committee on Human Research at the University of Vermont. In addition, umbilical cord blood samples were also obtained from healthy newborn infants and healthy infants of HIV-infected mothers treated with Combivir or its components AZT (zidovudine) and 3TC (lamivudine) as part of a cohort study. The following clinical centers participated in this study under agreements with Westat (Albany Medical Center, San Juan City Hospital, Howard University Hospital, Harlem Hospital Center, Los Angeles County/University of Southern California Medical Center, and the University of Texas Southwestern Medical Center) and clinical centers in the Women and Infants Transmission Study. Institutional Review Board approval for this research protocol and the standardized informed consent forms for this study were obtained independently from each collaborating center. V(D)J coding junction information from samples obtained from healthy children treated in utero with anti-HIV therapy was only included in the deletion profiles, showing exonucleolytic processing of the coding end sequences.
Previously published V(D)J recombinase-mediated coding joint sequence of HPRT 2–3 deletion mutants were included from umbilical cord blood samples, healthy preterm and full-term infants (19, 22), and peripheral blood samples from healthy children (20) were included in this study.
Isolation of V(D)J recombination-mediated HPRT exon 2 and 3 deletion mutants
Mutants were isolated from peripheral or cord blood, using the HPRT T cell cloning assay that selects for individual T lymphocytes that have acquired an in vivo mutation resulting in a loss of HPRT activity as described previously (25, 26). Mutant clones are expanded in vitro, pelleted, and frozen at −80°C for future molecular analysis.
Cell pellets of HPRT mutant clones were used to generate cDNA and subjected to PCR analysis to amplify the coding region of HPRT as described previously (25). HPRT mutant clones showing loss of exons 2 and 3 in cDNA were then analyzed for evidence of V(D)J recombinase-mediated deletion. Genomic PCR analysis, across the breakpoints of the V(D)J recombinase-mediated exon 2–3 deletion mutants, was conducted as previously described using primers that anneal at nucleotides 1835–1851 (sense), 22742–22719 (antisense for class I and III), and 20327–20304 (antisense for class II) of the HPRT gene, which generates products of ~860 bp (class I), 600 bp (class II), or 570 bp (class III) (20, 21). Over 95% of the mutants that showed the loss of exons 2 and 3 in cDNA and their deletion in genomic DNA yielded a product consistent with a RAG-mediated event. Approximately 92% of the RAG-mediated events identified were class I and ~8% were class III. Although routinely screened for, the class II event has only been observed twice. All PCR products were analyzed on 1.4% agarose gels, the specific bands isolated and purified using QIAquick-spin columns (Qiagen). The PCR products were then sequenced by the Vermont Cancer Center Sequencing Facility at the University of Vermont, using Taq DyeDeoxy terminator cycle sequencing (ABI) on an ABI automated sequencer model 373A. HPRT nucleotide numberings are based on the HPRT genomic sequence submitted to DNA Data Base in Japan/European Molecular Biology Laboratory/GenBank database under accession number M26434.
Coding junction analysis
As described above, previously published HPRT V(D)J coding junctions as well as sequences from on going studies were gathered for this analysis. The number of coding joints examined varied from 1 to 9 coding junctions per patient. Our analysis was conducted on 40 V(D)J coding junctions from 19 preterm newborns, 170 coding junctions from 52 full-term newborns, and 55 coding junctions from 40 children ranging from 1 to 12.5 years of age. A complete summary of the HPRT V(D)J recombinase-mediated coding joint analysis of mutant isolates from these children, including templated 5′ and 3′ P-nucleotides, nucleotide nibbling, nontemplated N-nucleotide insertions, and nucleolytically processed termini Pr-nucleotides, is provided as an online Supplemental Table.4 Comparison of the HPRT V(D)J recombinase-mediated class I and class III coding junctions used the healthy preterm and full-term newborn samples composed of 192 class I and 18 class III coding junctions as well as 142 class I coding junctions and 10 class III coding junctions from 7 healthy preterm and 36 healthy full-term newborns treated in utero with anti-HIV drugs.
Similar to previous studies of coding end processing, systematic sets of rules were used in scoring deleted and inserted bases (4). First, nucleotide loss at each coding end was determined, as compared with the consensus (unprocessed) coding ends. Nucleotide loss at each coding partner was summed to give the overall number of bases excised within each coding joint. Second, the inserted bases were scored as either templated (P-nucleotides) or nontemplated (N-nucleotides). The N-nucleotides were subsequently analyzed for the presence of inverted repeats at nucleolytically processed termini (Pr-nucleotides). In two cases, one of three Pr-nucleotides in the coding joint could be assigned to either coding-end. These were scored as positive for the presence of Pr-nucleotides at both ends in this analysis. When analyzed for Pr-nucleotide inclusion within a coding joint these events were scored as one positive event. The coding junction information was then examined based on age and gender differences as well as the class of RAG-mediated HPRT deletion event. Statistical analyses to examine variations between different sample groups were performed via the unpaired Student’s t test.
Statistical analysis of TdT-mediated insertions
For the analysis of dinucleotide pairs, the total N-nucleotide sequences were analyzed for the overall frequency of each mononucleotide. We then calculated the expected frequency of each dinucleotide pair. For example, G nucleotides were observed at a frequency of 0.2861; therefore, the probability of a GG dinucleotide would be 0.2861 × 0.2861 or 0.0819. For the analysis of 1165 dinucleotide pairs, GG dinucleotides would be expected to be observed 95 times. In our analysis, we observed 151 GG dinucleotides. We then conducted a χ2 goodness-of-fit test with 1 degree of freedom and obtained the p value. For GG dinucleotides, p ≤ 0.001.
Statistical analysis of Pr-nucleotide insertions was conducted for the first base inserted via TdT activity in all samples. The frequency of processed coding termini ending in each mononucleotide was calculated for each age group analyzed. The frequency of each monomer in the N-nucleotides added for each age group was then used to determine the expected frequency of each palindromic dinucleotide pair. For example, in preterm samples, 13.47 complementary pairs would be expected of 52 processed coding ends. We observed only 5 complementary coding ends. A χ2 goodness-of-fit test with 1 degree of freedom was then conducted and for pre-term infants (p = 0.021). For samples from full-term newborns, a similar analysis was conducted for the first and second bases inserted via TdT activity.
Results
We investigated the relationship between V(D)J recombinase-mediated events at nonimmune cRSS sites and human development by examining coding junction processing at RAG-mediated HPRT class I and class III deletions in 265 peripheral T cell mutants from subjects 26 wk postgestation to 12.5 years of age.
Analyses of P-nucleotides at class I coding junctions
P-nucleotide calculations were conducted solely on the class I coding junctions as only 6% the class III coding junctions showed full-length coding ends (Supplemental Table). P-nucleotides were observed in 26% of all coding junctions and in 55% of all full-length coding ends (Table I). There is an increase in P-nucleotide insertions at the 5′ HPRT coding end, with 58% of the unprocessed termini having P-nucleotide inserts compared with 46% at the 3′ class I coding end. It has been previously shown that cleavage of the hairpin intermediate varies depending on the coding end sequence, which then results in variation of the size and amount of P-nucleotide insertions observed at an individual coding end (5, 6). These data suggest that the differences we see in P-nucleotide insertion are likely due to sequence variation at the coding ends. When we separated these samples by age and gender, we observed no apparent difference in the frequency of P-nucleotide insertions, suggesting that there is no developmental difference in the nicking mechanism responsible for the generation of these inverted repeats.
Table I.
Age- and gender-specific processing of coding signatures at the HPRT locus
| Preterm Newborns
|
Full-Term Newborns
|
1–12.5 Year Olds
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Total | Male | Female | Total | All Newborns | Male | Female | Total | All Samples | |
| No. of patients | 11 | 8 | 19 | 29 | 23 | 52 | 71 | 22 | 18 | 40 | 111 |
| No. of coding joints | 21 | 19 | 40 | 104 | 66 | 170 | 210 | 34 | 21 | 55 | 265 |
| P-ntds at coding ends %a | 55.56 | 55.56 | 55.56 | 56.36 | 59.09 | 57.14 | 56.84 | 47.83 | 50.00 | 48.57 | 54.62 |
| 5′-P-ntds at coding endsa | 4/7 | 5/7 | 9/14 | 23/38 | 9/16 | 32/54 | 41/68 | 8/16 | 5/9 | 13/25 | 54/93 |
| 3′-P-ntds at coding endsa | 1/2 | 0/2 | 1/4 | 8/17 | 4/6 | 12/23 | 13/27 | 3/7 | 1/3 | 4/10 | 17/37 |
| 5′ and 3′ P-ntds | 1 | 0 | 1 | 3 | 0 | 3 | 4 | 1 | 0 | 1 | 5 |
| N-ntd free coding joints % | 23.80 | 10.53 | 17.50 | 13.46 | 7.60 | 11.17 | 12.38 | 8.82 | 0 | 5.45 | 10.94 |
| GC content of N-ntds % | 47 | 65 | 55 | 58 | 62 | 59 | 58 | 60 | 57 | 58 | 59 |
| Pr-ntds at coding ends %b | 3.70 | 14.29 | 9.09 | 30.00 | 35.78 | 32.43 | 28.34 | 25.00 | 30.30 | 27.16 | 28.10 |
| Pr-ntd in junctions %b | 6.25 | 23.53 | 15.15 | 43.33 | 55.74 | 48.34 | 42.39 | 32.26 | 47.62 | 38.62 | 41.53 |
| Ratio of Pr-ntds/junctionb | 1/16 | 4/17 | 5/33 | 39/90 | 34/61 | 73/151 | 10/31 | 10/21 | 20/52 | ||
| Average N-ntd inserts bpb | 6.2 (5.6) | 5.5 (3.1) | 5.9 (4.9) | 5.2 (3.5) | 5.1 (3.1) | 5.2 (3.4) | 5.3 (3.6) | 6.1 (3.1) | 6.1 (3.7) | 6.1 (3.3) | 5.5 (3.5) |
| Ntd excision at coding ends % | 83.33 | 76.32 | 80 | 73.33 | 83.33 | 77.22 | 77.75 | 66.18 | 71.43 | 68.18 | 75.76 |
| 5′ excisions % | 76.19 | 63.16 | 70 | 63.11 | 75.76 | 68.05 | 68.42 | 52.94 | 57.14 | 54.55 | 65.54 |
| 3′ excisions % | 90.49 | 89.47 | 90 | 83.50 | 90.91 | 86.39 | 87.08 | 79.42 | 85.71 | 81.18 | 85.86 |
| Average ntds lossc | 6.5 (6.7) | 6.8 (6.8) | 6.7 (6.7) | 7.7 (6.0) | 7.9 (7.1) | 7.8 (6.4) | 7.6 (6.5) | 5.9 (3.8) | 4.7 (2.6) | 5.4 (3.4) | 7.2 (6.0) |
| 5′ excisionsc | 2.3 (1.2) | 3.3 (2.5) | 2.7 (2.0) | 3.2 (3.2) | 4.2 (4.9) | 3.7 (4.0) | 3.5 (3.8) | 3.1 (2.3) | 3.3 (2.3) | 3.2 (2.3) | 3.4 (3.5) |
| 3′ excisionsc | 4.8 (6.1) | 5.4 (5.4) | 5.1 (5.7) | 6.2 (5.4) | 5.0 (4.4) | 5.7 (5.0) | 5.9 (5.1) | 4.5 (2.6) | 3.0 (2.1) | 3.9 (2.5) | 5.3 (4.8) |
Observed at unprocessed coding ends class I HPRT deletions, when separated into 5′ or 3′ P-ntds (number of ends with P-ntds/total unprocessed ends).
Calculated from coding junctions with N-nucleotide insertions.
Calculated from coding ends or junctions with nucleotide loss. SD are provided in parenthesis.
N-nucleotide additions at coding junctions
We evaluated TdT activity during pediatric development by examining the percentage of V(D)J coding junctions that were free of N-nucleotide additions. We observed an apparent significant age dependent increase in TdT activity as reflected by the decrease in the percentage of coding junctions without N-nucleotides with increasing age (Fig. 3 and Supplemental Table). There was also a gender-specific trend observed in all age groups examined suggesting that TdT activity is higher in females throughout pediatric development. However, due to the smaller sample size, this trend was not significant (p = 0.065). These results are consistent with and expand the previous observations seen in preterm and full-term newborns (20, 27).
FIGURE 3.
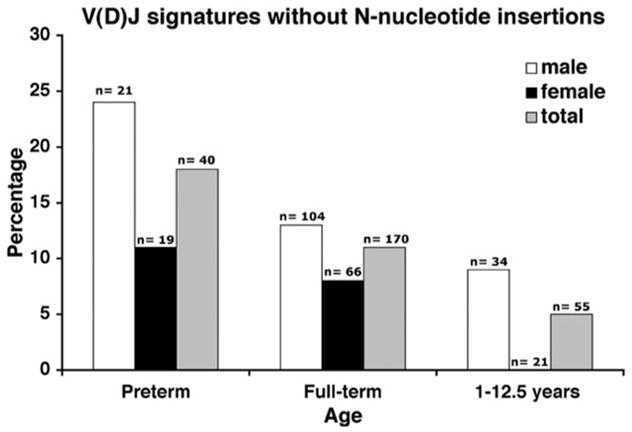
The percentage of coding junctions without N-nucleotide additions was determined in age- and gender-specific pediatric populations. The percentage of junctions without N-nucleotide inserts is likely inversely proportional to the amount of TdT activity. The apparent increase in TdT activity shows an age significant trend.
TdT has previously been shown to preferentially add G nucleotides, resulting in a higher GC content. Overall, the GC content of the N-region was 59%. However, when we separated these samples by age and gender, we observed that the GC content was significantly higher than 50% (55–65%, p ≤ 0.02) in all samples, except preterm males, which showed a GC content of 47% (Table II). In a concurrent study looking at coding joints at the TCR-β locus during the same age intervals, we observed a similar trend with N-regions from preterm males having an average GC content of 46% while all other samples remained significantly >50% (J. M. Murray, J. P. O’Neill, T. Messier, J. Rivers, V. E. Walker, P. M. Vacek, and B. A. Finette, unpublished observations). These data suggest that the TdT specificity for dGTP is lower in preterm males, compared with all other age- and gender-specific groups included in this study, further supporting an apparent age- and gender-specific difference in TdT activity and specificity.
Table II.
N-nucleotide dinucleotide frequencies
| Dinucleotide | No. of Observed | No. of Expecteda | p Value |
|---|---|---|---|
| AA | 54 | 48 | 0.394 |
| AC | 47 | 71 | 0.004*L |
| AG | 80 | 68 | 0.134 |
| AT | 52 | 50 | 0.729 |
| CA | 53 | 71 | 0.030*L |
| CC | 158 | 106 | <0.001*H |
| CG | 67 | 100 | <0.001*L |
| CT | 80 | 74 | 0.450 |
| GA | 79 | 68 | 0.169 |
| GC | 39 | 100 | <0.001*L |
| GG | 151 | 95 | <0.001*H |
| GT | 48 | 70 | 0.009*L |
| TA | 68 | 50 | 0.009*H |
| TC | 88 | 74 | 0.092 |
| TG | 50 | 70 | 0.018*L |
| TT | 51 | 51 | 1.000 |
Expected frequencies were calculated from the overall base composition of all N regions (A = 0.2031, C = 0.3015, G = 2861, T = 0.2094) and the total number of dinucleotides (n = 1165).
Values of p were calculated via χ2 goodness-of-fit test with 1 degree of freedom.
HSignificantly overrepresented with respect to the expected value.
LSignificantly underrepresented with respect to the expected value.
Dinucleotide insertions via TdT
Previous studies, examining plasmid V(D)J substrates, have shown that TdT insertions favor homopolymer tracts. In addition, homo-dinucleotide (purine-purine, RR and pyrimidine-pyrimidine, YY) inserts occurred at a higher than expected level and all hetero-dinucleotide inserts (RY and YR) occurred less often than expected (4). We conducted a similar analysis using the overall N-nucleotide base composition for our coding junctions (A = 0.2031, C = 0.3015, G = 0.2861, T = 0.2094) and the total number of dinucleotides in these junctions (n = 1165). An overall preference for RR and YY nucleotides was observed with all eight pairs being at or above the expected value (GG and CC pairs being significantly above expected values; Table II and Supplemental Table). For RY and YR pairs, six of eight were significantly below the expected levels. However, AT dinucleotides were observed at the expected levels, and TA dinucleotides were observed at significantly higher levels than expected, suggesting there is more tolerance for these hetero-dinucleotides than was observed using plasmid substrates. An analysis of dinucleotide insertions at different developmental stages did not demonstrate an observable difference in the preference for homo-dinucleotides (data not shown).
Inverted repeats at nucleolytically processed termini
It has been previously reported that TdT activity can add complementary nucleotides (termed Pr-nucleotides) to processed coding ends resulting in inverted repeats at the coding end borders and that the frequency of these additions is greater than predicted if these were the result of random nucleotide insertions (4). Gauss and Lieber (4) observed, using plasmid substrates, that coding junctions containing N-nucleotides had Pr-nucleotide inserts at 25% of the processed coding ends and in 40% of the coding joints. Our analysis showed similar results with 28% of the processed coding ends and 42% of the coding junctions in HPRT having these inverted repeats. However, when we separated these samples by age and gender, we observed age-specific differences in the amount of Pr-nucleotide insertions (Table I, Fig. 4 and Supplemental Table). Pr-nucleotides are observed most frequently in coding junctions from full-term newborns, with 32% of all processed coding ends and 48% of all N-nucleotide containing coding joints showing these inverted repeats. The lowest frequency was observed in preterm newborns with 9 and 15%, respectively, while 1–12.5 year olds showed an intermediate level with 27 and 39%, respectively. Statistical analysis demonstrated that preterm newborn samples had inverted repeats at processed coding ends at significantly lower levels than expected if the nucleotides were randomly inserted (first base inserted p = 0.021). Full-term newborn samples showed significantly higher levels of inverted repeats at processed coding ends than expected (first base inserted p = 0.005, first and second base inserted p = 0.003). In the samples from 1 to 12.5 year olds, the Pr-nucleotide inserts were not significantly different than expected due to random insertion via TdT activity. In all age groups, the coding junctions from females showed a higher level of Pr-nucleotides compared with males. The differences ranged from a 23% increase for females vs males in full-term newborns to a 75% increase in preterm newborns. However, due to small sample size the gender-specific variations were not significant (Table I). These data again suggest that the TdT activity may be regulated differently during human development.
FIGURE 4.
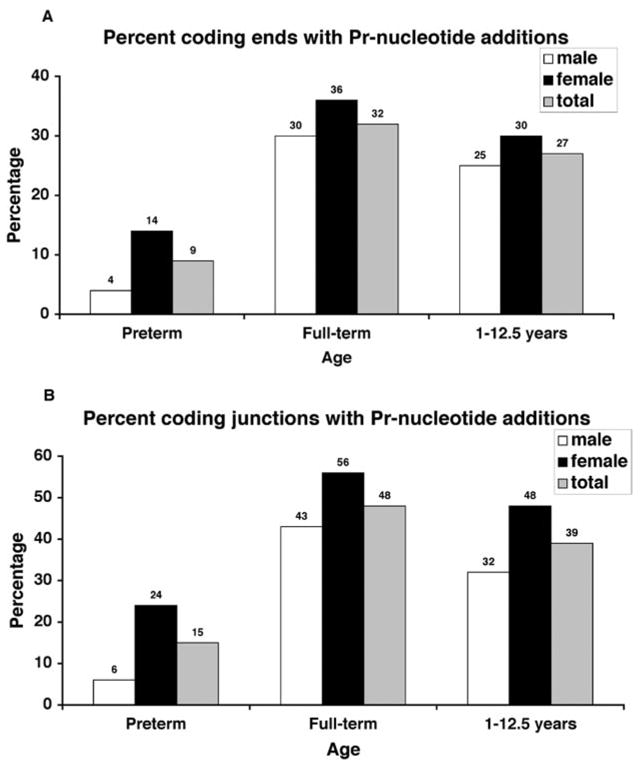
Pr-nucleotide additions at nucleolytically processed coding ends (A) or coding junctions (B) with N-nucleotide additions. Junctions were scored as positive if one or both ends showed evidence of these inverted repeats. The amount of Pr-nucleotides varies with age and appears to be higher in females compared with males in all age groups.-
Microhomology usage at coding junctions
Microhomology-directed end-joining as an alternate mechanism for double-strand break repair has been observed in both murine and human coding junctions in the absence of TdT activity. Twenty nine of 265 coding joints analyzed had no N-nucleotides. Of these 17% showed evidence of microhomology of either 1 or 2 bases demonstrating that in the absence of N-nucleotide additions microhomology can be used for the formation of coding junctions at the HPRT locus.
Nucleotide loss at the coding ends
We observed an increase in nucleotide loss at coding ends in newborns (78%) compared with 1–12.5 year olds (68%) (Table II and Supplemental Table). The extent of nucleotide loss per individual coding junction also varied significantly with age (p = 0.013); newborn coding joints average 7.6 bases deleted (preterm 6.7, full-term 7.8) and 1–12 year olds having an average of 5.4 bases deleted (Table II). There was less processing at 5′ coding ends (3.4 bases deleted) compared with 3′ coding ends (5.3 bases deleted). This was consistent for both genders and all ages studied. Coding end sequences have been shown to influence the extent of nucleolytic processing as well as the deletion profile (4 – 6). The 3′ coding end in both class I and class III coding junctions, positions 22251 and 22569, respectively, showed a larger percentage of base loss than the 5′ end at position 2197 of the HPRT gene (Table III).
Table III.
Processing in class I and class III coding junctions
| Class I | Class III | |
|---|---|---|
| No. of coding joints analyzed | 192 | 18 |
| Average N-nucleotide insertion bpa | 5.3 | 5.3 |
| Pr-nucleotides at processed ends (%)a | 29 | 39 |
| Pr nucleotide containing junctions (%)a | 40 | 71 |
| N-nucleotide free coding joints (%) | 13 | 6 |
| Nucleotide excision at coding ends (%) | 87 | 94 |
| Average bases excised/with processing | 6.7/8.5 | 12.9/13.3 |
| 5′ coding end average excision | 2.0/3.2 | 5.8/5.8 |
| 3′ coding end average excision | 4.7/5.4 | 7.1/7.5 |
| GC content of N-nucleotides | 59 | 56 |
With N-nucleotide insertions.
Comparison of V(D)J processing at HPRT class I and class III coding junctions
In this study, of 265 V(D)J-mediated deletion in HPRT, the class I event (2197–22251) is observed in ~92% of RAG-mediated deletions of exons 2 and 3 while the class III events (2197–22569) account for ~8% of these deletions (Fig. 2 and Supplemental Table). This is consistent with previous studies showing class III events accounting for 4 and 6% of the HPRT V(D)J deletions in newborns and 0–16 year olds, respectively. In our comparison of class I and class III coding junctions we limited our analysis to newborn samples as 86% of all class III events in healthy children have been identified in newborns. We compared 18 class III coding junctions to 192 class I coding junctions the percentage of these coding junctions from preterm individuals is 11 and 19%, respectively. We were unable to compare the extent of P-nucleotide insertion and microhomology usage between class I and class III coding junctions, as 94% of class III coding ends are nucleolytically processed. For both class I and class III coding junctions, the average number of N-nucleotides inserted was 5.3 bases (Table III). However, the percentage of Pr-nucleotides inserted is higher in class III coding joints, with 39% of coding ends with N-nucleotide addition and 71% of N-nucleotide containing coding junctions showing these inverted repeats compared with class I coding joints with 29 and 40%, respectively. The differences seen in Pr-nucleotide insertions were also observed at the shared 5′ coding end with 27% of the class I ends showing these inverted repeats compared with 41% at the class III ends. These data suggest there is a difference in the processing at the 5′ coding end depending on the 3′ coding partner.
Nucleolytic processing in the class III coding joints was higher with 94% of the coding ends demonstrating base loss compared with 87% of class I coding joints. Class III coding junctions had an average of 12.9 bases deleted compared with an average of 6.7 bases for class I coding joints. In addition, the increase in nucleolytic processing in the class III coding junctions is observed at both the 5′ coding end, which is conserved in class I and class III coding junctions, and the 3′ coding end (Table III). The variation observed at the different 3′ ends is expected, as the coding end sequence has been shown to effect end processing. The significant difference (p < 0.0001), observed at the conserved 5′ coding end, is unexpected and strongly suggests that the coding partner can affect nucleolytic processing.
To further evaluate the possible effect of different coding partners on coding end processing, we analyzed the deletion profile of the coding ends (Fig. 5). The 3′ coding ends have different coding profiles as would be expected since the coding end sequences vary. However, we also observed a clear difference in the deletion pattern at the common 5′ coding end depending on the 3′ joining partner used (Fig. 5). This analysis compared 18 class III coding junctions to 192 class I coding junctions all isolated from newborns. Due to the limited number of class III coding junctions, we included 142 class I and 10 class III junctions from an additional 43 healthy newborns of HIV-infected women that were treated with anti-HIV therapy in utero. Analysis of the coding junctions obtained from these newborns showed no significant difference in the overall processing of the ends as compared with the untreated newborns from this study (data not shown). When these coding ends were included in our analysis, the deletion profiles were conserved demonstrating that the nucleolytic processing of the 5′ coding end in HPRT is dependent on the 3′ coding partner.
FIGURE 5.
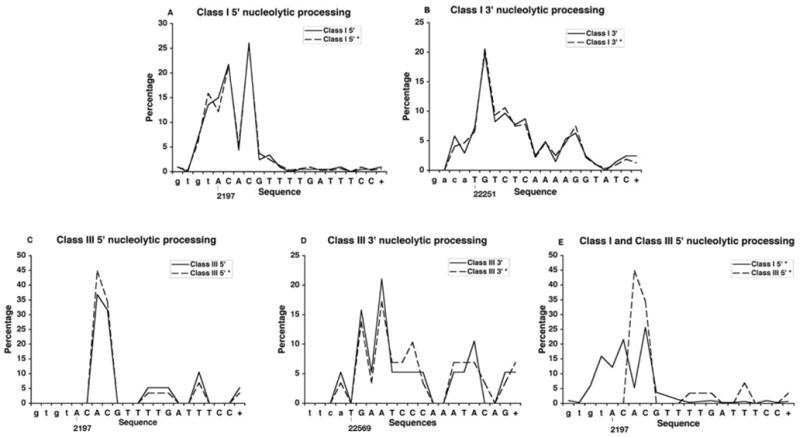
Distribution of the coding end processing in class I and class III coding junctions. Distributions that include junctions from healthy newborns treated in utero with anti-HIV drugs are denoted with a * sign. The shared 5′ coding end at position 2197 is shown for both class I and class III junctions (A and C, respectively). Class I 3′ coding end processing at position 22251 is shown in B, and the class III 3′ coding end processing at position 22569 is shown in D. E, The comparative distribution between the class I and class III coding ends at position 2197 demonstrating the difference in exonucleolytic processing depending on the coding partner. The coding sequences are orientated with 5′ to 3′ directionality labeled appropriately. The coding sequences are written in uppercase letters, and the first nucleotides are labeled with the HPRT genomic position (the + at the end of this sequence denotes nucleolytic processing that proceeds further than the sequence shown). Lowercase letters to the left correspond to the P-nucleotides.
Comparison of HPRT cRSS sequences to human RSS consensus sequences
As previously discussed, there is variable usage of the three 3′ coding ends at the HPRT locus. Of all the class III coding junctions identified in healthy patients, 86% (18 of 21) are observed in newborns while no class III coding junctions have been observed in children older than 1.4 years of age. These data suggest that some cRSS sites may only be available for RAG-mediated recombination during certain stages of childhood development.
To try to further understand the differential usage of these cRSS, we compared the one 5′ and three 3′ cRSS sites at the HPRT locus to consensus sequences constructed from 191 and 269 human, immunologic 12- and 23-RSS, respectively (available at www.duke.edu/~lgcowell). All RSS included in defining the consensus sequence were associated with V, D, or J gene segments and classified as functional (Immunogenetics Information System (http://imgt.cines.fr/)). In this set, the 12-RSS consensus sequence is CACAGTG ATACAGACCCTA ACAAAAACC; the 23-RSS consensus is CACAGTG AGGCAAAGTCATTGGGAACCTGG ACAAAAACC.
The four cRSS used in V(D)J recombinase-mediated HPRT deletions have varying degrees of similarity to the consensus human 12- and 23-RSS (consisting of 28 or 39 bp, respectively). All have the required CAC heptamer as well as an A nucleotide at position five of the spacer, which is well conserved in several species (28). Of the 28-bp cRSS, the 3′ class I cRSS at positions 22223–22250 in the HPRT locus is most similar to the human 12-RSS consensus, with 18 matches. The 3′ class II cRSS at 20126–20155 has 16 matches, the 5′ cRSS sequence at 2198–2226 has 11 matches, while the 3′ class III cRSS from 22541 to 22568 matches the consensus at five positions. Thus, nucleotide identity with consensus ranges from 19 to 67% in this cRSS set. Although this may appear low, many human immune-specific 12-RSS are comparably dissimilar: 73 human 12-RSS (38%) have ≤ 18 matches to consensus, 33 (17%) have ≤ 16 matches, and one immune-specific RSS matches the consensus at only 11 positions.
When compared with the 23-RSS consensus, the 3′ class II cRSS from 20117 to 20155 is most similar with 18 matches, the 5′ cRSS from 2198 to 2237 has 15 matches, the 3′ class III cRSS at 22430–22568 matches at 14 nt, and the 3′ class I cRSS at 22212–22250 matches at 12 nt. The percent similarities range from 32 to 47%. Eighteen immunologic 23-RSS (7%) match the human 23-RSS consensus at ≤ 18 positions, and two (~1%) match the consensus at only 12 positions.
The degree of similarity between the four cRSS at the HPRT locus and the 12- or 23-RSS human consensus are consistent with the cRSS serving as substrates for the V(D)J recombinase. However, consensus sequence representations of even moderately variable DNA motifs have limited utility for predictions of function (28, 29).
Recombination signal information content (RIC) analysis of HPRT cRSS sequences
Models of the correlation structure of mouse RSS, termed RIC, efficiently identify immunologic and cRSS and accurately predict recombination efficiencies (28, 29). Similar models for human RSS have not yet been fully developed, but the RIC model for mouse 12-RSS has been used to screen human VH gene segments for cRSS, and those cRSS tested supported detectable levels of recombination in extrachromosomal recombination assays (30). Thus, until verification of the human RIC12/23 models are completed, the murine RIC12/23 models may have utility in identifying and comparing human RSS and cRSS.
To evaluate the recombination potentials of the four cRSS in the human HPRT locus, we computed the RIC score for each 28- and 39-bp cRSS sequence (http://bioinformatics.duke.edu/dulci) or (http://bio.ieo-research.it/RSS/rss.html) (Fig. 6A). The lowest RICT12 score for any mouse immunologic 12-RSS is RIC12 = −48.16, thus we expect any 28-bp cRSS sequence scoring at or above this threshold to support RAG-mediated recombination. Similarly, the lowest RIC23 score for a immunologic mouse 23-RSS is RICT23 = −69.69, and 39-bp sequences with this score or better should also function. By these criteria, the putative 5′ cRSS at positions 2198 is predicted to act as either a 12- or 23-RSS substrate (Fig. 6A). The 3′ class I cRSS at nt 22250 is predicted to function as a 12-RSS but may also have marginal activity as a 23-RSS (Fig. 6A), as its RIC23 differs from the threshold by a value of only 4.79 while the range of scores for immunologic 23-RSS is 53.86 ((−69.69 – −74.48)/( −15.83 – −69.69) = 0.09). The 3′ class III cRSS at 22568 most likely functions only as a 23-RSS (Fig. 6A). Finally, the 3′ class II cRSS at 20155 is predicted to function as either a 12- or 23-RSS substrate.
FIGURE 6.
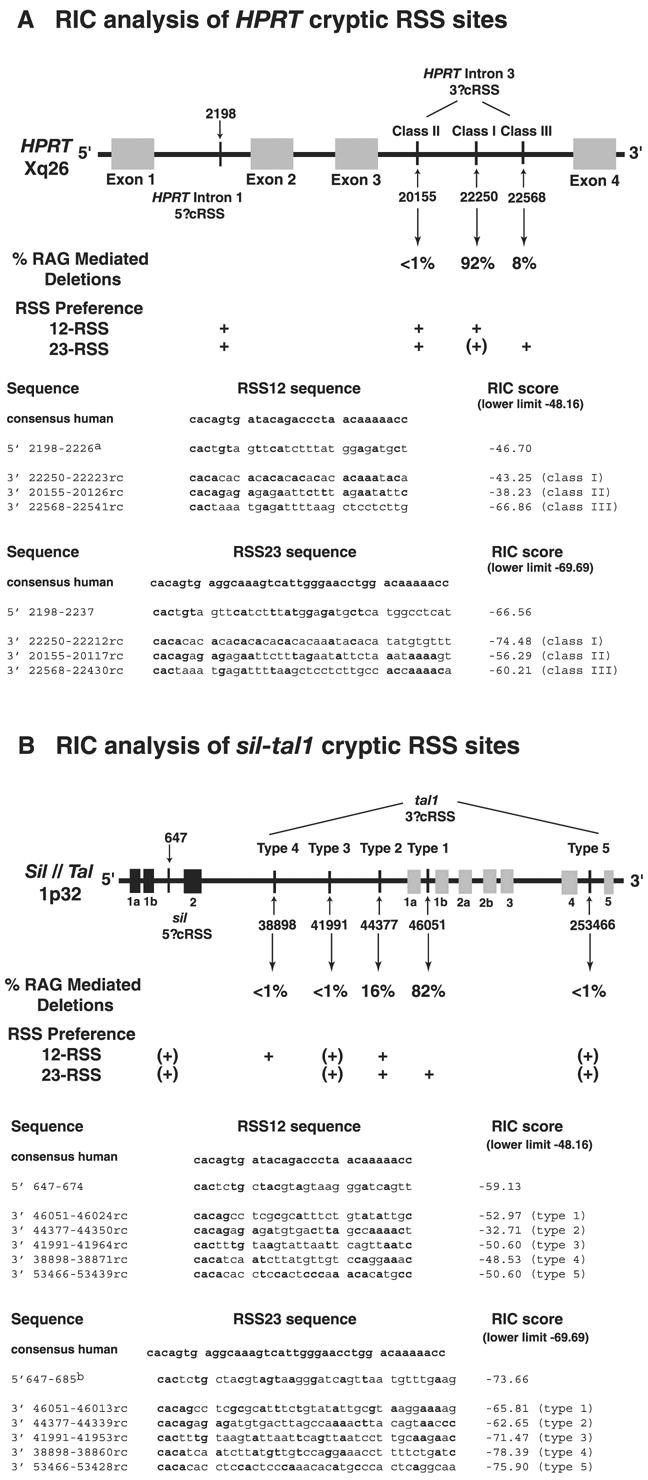
RIC analysis of functional cRSS sites in the HPRT and sal-tal1 loci. A, Schematic diagram of the intron/exon structure of the HPRT loci showing the genomic regions involved in V(D)J-mediated deletions. Percentage of V(D)J-mediated deletions for each class, and expected usage (12- or 23-RSS) for each of the cRSS as well as the sequence and RIC data for the HPRT cRSS sites are shown. B, Schematic diagram of the intron/exon structure of the sil-tal1 loci showing genomic regions involved in V(D)J-mediated deletions. Percentage of V(D)J-mediated deletions for each type and expected usage (12- or 23-RSS) for each of the cRSS as well as sequence and RIC data for the sil-tal1 cRSS sites are shown. Arrows and black lines indicate the various cRSS site. Genomic HPRT sequence DNA Data Base in Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank accession no. M26434. Genomic sil gene sequence DDBJ/EMBL/GenBank accession no. Y07540. Genomic SCL gene sequence DDBJ/EMBL/Gen-Bank accession nos. AJ131016 or AL135960. + sign indicates that the cRSS can be used with this specificity (12- or 23-RSS), and (+) indicate that there may be marginal use.
These RIC scores suggest that both class I and II deletions may represent RAG-driven excisions where the 5′ cRSS (at 2198) and the 3′ cRSS (at 22250 or 20155, respectively) can pair reciprocally as 12- and 23-RSS. This flexibility in synapsis may allow for multiple RAG-mediated events. In class III deletions, however, the 3′ cRSS (at 22568) can be used only as a 23-RSS restricting the molecular events that result in class III deletions. This could partially explain why the class I events are more common than the class III events, but does not explain the rarity of class II events.
Discussion
The goal of this study was to examine age and gender specific differences in the processing of nonselected V(D)J recombination events at cRSS at the HPRT locus during fetal and childhood development. This data can provide mechanistic insight into the developmentally specific V(D)J recombinase-mediated rearrangements observed in healthy children and the genetic changes in children with leukemia.
We observed several age and gender specific differences in the quantity and type of N-nucleotide additions. We observed that TdT-mediated insertions of N-nucleotides show a predominance of GC nucleotides and a preference for homopolymer insertions. In addition, the overall percent of coding junctions with N-nucleotide additions significantly increased with age, which may reflect an age dependent increase in TdT activity. An increase in coding junctions containing N-nucleotides in females compared with males also suggests that the activity of TdT is higher in females. The observed lack of preference for GC nucleotide insertions in preterm males suggests that TdT preference for dGTP addition is suppressed in male preterm newborns and is consistent with a concurrent study examining coding junctions at the TCR-β locus in these subjects (J. M. Murray, J. P. O’Neill, T. Messier, J. Rivers, V. E. Walker, P. M. Vacek, and B. A. Finette, unpublished observations). We also observed developmentally specific differences in the presence of inverted repeats (Pr-nucleotides) at nucleolytically processed coding ends. In contrast, analysis of Pr-nucleotide inserts in over 600 human VHDJH rearrangements by Soute-Carneiro et al. (32), showed no evidence of Pr-nucleotide insertions occurring at levels above that expected via random insertion by TdT activity (4, 31, 32). Pr-nucleotide insertions were only significantly different than expected when analyzed for age-specific differences. This suggests that the difference in Pr-nucleotide insertions is due to an age-specific change in TdT activity. Gauss and Lieber (4) reported that the presence of inverted repeats at processed ends was dependent on TdT activity, supporting the idea that the occurrence of Pr-nucleotides is either a result of TdT-mediated random insertion events that generate these inverted repeats or a modulated TdT activity that varies depending on developmental age (4). Our data suggest that Pr-nucleotide insertions are an age specific developmental event.
The TdT protein has been shown to complex with many proteins, including the DNA-PK hetero-trimer, proliferating cell nuclear Ag, TdT-interacting factor (TDIF)1, and TDIF2, that in turn, regulate the activity of TdT. DNA-PK has been shown to limit the activity of TdT resulting in addition of fewer N-nucleotides and a preference for dGTP insertions (33). Proliferating cell nuclear Ag and TDIF2 have been reported to decrease TdT activity using in vitro assays while TDIF1 can enhance TdT activity 4-fold (34–36). Our data suggest that the specificity of TdT activity is also developmentally regulated. We observed a variation in the preference for GC addition in preterm males at the HPRT locus, which suggests a change in the regulation via DNA-PK. However, whether the suspected age- and gender-specific regulation of TdT activity occurs via a novel mechanism or through differential regulation of one or more of these associated proteins remains to be elucidated.
Recently, two family X polymerases, pol μ and pol λ, have been shown to be recruited by the NHEJ core proteins and participate in gap filling of NHEJ-repaired double-strand breaks (8, 37, 38). It is thought that theses proteins are also responsible for rare N-nucleotides observed in V(D)J coding junctions in the absence of TdT activity (39, 40). The specific ability of these polymerases to use adjacent sequences through a looping mechanism to fill gaps suggests that with TdT these polymerases may have a role in the addition of Pr-nucleotide observed at coding junctions (41–43). Therefore age-specific expression of pol μ and pol λ could account for our developmental specific findings for N-nucleotide and Pr-nucleotide insertions.
Using extra-chromosomal substrates, Gauss and Lieber (4) demonstrated that microhomology was used in 57% of coding junctions from human cells in the absence of TdT activity. In the presence of TdT activity, microhomology use was reduced to 47% in junctions with no N-nucleotide additions (4). We observed that 17% of HPRT coding end junctions with no N-nucleotide additions appear to be resolved via microhomology. Repasky et al. (31) have reported that microhomology usage varied depending on the type of substrate used. Specifically, microhomology was observed in 38 and 63% of coding junctions, respectively, for plasmid substrates in murine cells expressing the full-length RAG1/2 proteins with and without expressing full-length TdT. Using integrated chromosomal substrates, microhomology levels drop to 8 and 16%, respectively (we calculated the percent microhomology with TdT expression using coding junctions with no N-nucleotides). This suggests that the ability of coding ends to use a microhomology mechanism of resolving the coding joint varies depending on the overall structure of the complex.
We also observed that newborns have increased nucleolytic processing compared with children 1–12.5 years of age. The Artemis protein, associated with NHEJ, has been shown to be capable of exonucleolytic activity at coding ends and our data could suggest a modulation of Artemis activity with age. However, Artemis-deficient cell lines have residual exonuclease activity, suggesting that there are alternate protein(s) involved in coding end processing (44). Recently, Thai and Kearney (45) identified three splice variants of human TdT termed TdTS, TdTL1, and TdTL2. TdTL1 appears to be expressed primarily in transformed cells while TdTS and TdTL2 are expressed in pro- and pre-B cells as well as in all thymocyte subpopulations (DN, DP, CD4+, and CD8+) during human fetal development. Their analysis demonstrated that TdTL1 and TdTL2 isoforms are capable of 3′–5′ exonuclease activity on ends with either 3′ or 5′ extensions. Expressing these variants with RAG1/2 core proteins resulted in an overall difference in the amount of nucleolytic processing. Expression of TdTS alone resulted in an average 2.3 nt being deleted at the coding junctions. This increased to 5.7 nt in the presence of TdTL2 and was at 3.1 nt when both isoforms were expressed. Age-dependent differences in the expression of these TdT isoforms could result in a modulation of TdT activity that may explain our observed age specific differences in the amount and extent of nucleolytic processing of coding ends in newborn infants compared with children 1–12.5 years of age.
The reasons for our differences in nucleolytic processing of the 5′ HPRT coding end depending on the 3′ HPRT coding partner are unclear and differ from that observed for plasmid substrates (6). Although a difference was observed in the amount of Pr-nucleotides inserted in class I vs class III deletion events, these values were not significantly above that predicted due to random insertion via TdT activity. A portion of the increase seen in the class III coding junctions appears to be due to the strict pattern of nucleolytic processing observed at the 5′ coding end, with 79% of the coding ends having 2 or 3 bases deleted limiting the coding end variation (Fig. 5).
To gain insight into the differential usage of the 3′ cRSS in RAG-mediated deletions of exons 2 and 3 in HPRT we computed RIC scores (http://bioinformatics.duke.edu/dulci) for the 28- and 39-nt sequences adjacent to each breakpoint. The RIC scores indicate that the conserved 5′ cRSS site can be used as both a 12-RSS and 23-RSS. Of clinical importance is that RAG-mediated deletions at the HPRT locus are analogous to sil-tal1 deletions observed in 15–26% of T cell acute lymphoblastic leukemias (27, 46, 47). RAG-mediated rearrangements at the sil-tal1 loci result in a ~90-kb deletion altering the expression of the TAL1 transcription factor. Analogous to RAG-mediated recombination at the HPRT locus a single 5′ cRSS site is used in combination with multiple 3′ cRSS sites (Fig. 6B). Type 1 and 2 sil-tal1 deletions are most frequently identified accounting for ~82 and 16% of these deletion events, respectively. The three other 3′ cRSS sites account for ~3% of the events, each having been observed only one time. We computed RIC scores for the 28- and 39-bp cRSS sites used in RAG-mediated sil-tal1 deletions as either 12- or 23-RSS using the RSS Project Database Search (http://bioinformatics.duke.edu/dulci) or (http://bio.ieo-research.it/RSS/rss.html) (Fig. 6B). RIC scores for the 5′ cRSS predict that its recombination efficiency would be very low whether used as a 12- or 23-RSS. The RIC scores for the type 1 3′ cRSS predicted it to function as a 23-RSS, while the type 2 3′ cRSS indicate that this cRSS could function as both a 12- and 23-RSS. The type 4 3′ cRSS is predicted to function as a 12-RSS, and the remaining two 3′ cRSS are predicted to have low recombination efficiencies whether used as 12- or 23-RSS. In vitro recombination frequency (Rf) experiments using plasmid substrates suggest that the 5′ cRSS functions as a 12-RSS (48). If this is the case and the 3′ cRSS are used as 23-RSS, then the RIC scores accurately predict that the type 3, 4, and 5 deletions would be rare events while the type 1 and 2 deletions would be limited by the poor recombination efficiency of the 5′ cRSS. The RIC scores do not reflect, however, the relative frequencies of the type 1 and 2 RAG-mediated events at the sil-tal1 loci. To investigate the difference between RIC scores and observed cRSS usage, we conducted systematic RIC analyzes of introns 1 and 3 at the HPRT locus. We identified putative cRSS predicted to function in V(D)J-mediated deletions of exons 2 and 3. We used more stringent RIC cutoffs to identify cRSS predicted to be the most functional (12-RSS < −39.0 and 23-RSS < −59.0). We identified six putative cRSS (two 12-RSS and four 23-RSS) within intron 1 and three putative cRSS (all 23-RSS) within intron 3 (data not shown). In-tron 3 analysis also identified the 3′ cRSS sites previously described for class II deletion events (Fig. 6). The newly identified putative cRSS sites are predicted to generate exon 2–3 deletions in conjunction with each other (a 5′ cRSS in intron 1 with a 3′ cRSS in intron 3) or with the cRSS sites described in this study. However, of the exon 2 and 3 exclusions observed in cDNA analysis of the HPRT locus in children, ~93% are class I or class III V(D)J mediated deletions, ~5% are splice site or other mutations affecting exon 2 and 3 splicing and ~2% have not been completely characterized. These data suggest that the putative cRSS identified are either not used in vivo or that they are used at such a low level they have not been observed to date. The fact that the putative cRSS sites have better RIC scores than the used HPRT cRSS class I and III sites suggests that the RIC analysis using the mouse model is not applicable for human RSS and/or cRSS sites or that accessibility plays a crucial role in usage of in vivo sites. The 93 and 77% identity between the mouse and human 12- and 23-RSS consensus sequences suggests the latter.
The RIC score thresholds for immunologic mouse RSS predict that there are some 0.5–1.0 × 106 cRSS sites in mammalian genomes that, if accessible to the recombinase, would be capable of rearranging as efficiently as RSS at immune loci (30). Several of the cRSS at the HPRT and sil-tal1 loci have lower RIC scores indicating that the number of sites potentially involved in RAG-mediated rearrangement at biologically relevant levels is probably much higher.
We present the first large-scale analysis of in vivo V(D)J recombinase coding-end processing in peripheral T cells at functional cRSS during various stages of human development. We demonstrate that there are significant age- and gender-specific differences in V(D)J recombinase coding-end processing. These changes suggest there are alterations in the processing machinery that could also affect the immune repertoire of children. These data are consistent with other gender- and development-specific differences that occur during fetal ontogeny for neural differentiation (49), growth (50), lung maturation (51), cardiac development (52), and glutathione-reductase expression (53). In addition, an increasing number of genomic rearrangements have been identified in leukemic cells from children, many of which appear to be mediated by V(D)J recombinase during fetal development (54–57). Of importance is the correlation between the significant increase in V(D)J recombinase mediated HPRT deletions at functional cRSS during the late stages of fetal development, and the epidemiologic data that show a gender-specific female predominance for infant leukemia (22, 23). These observations suggest that functional cRSS sites at pathologic and nonpathologic loci could be more accessible during specific phases of fetal and childhood development.
Acknowledgments
We thank Pamela M. Vacek for her assistance with the statistical analysis.
Disclosures
The authors have no financial conflict of interest.
Abbreviations used in this paper
- RAG
recombinase-activating gene
- cRSS
cryptic RSS
- HPRT
hypoxanthine-guanine phosphoribosyltransferase
- NHEJ
nonhomologous end joining
- RIC
recombination signal information content
- RSS
recombination signal sequence
- TDIF
TdT-interacting factor
Footnotes
This research was supported by Grant 6042 from the Leukemia and Lymphoma Society of America, Grant HD35309 from the Institute for Child Health and Human Development, National Institutes of Health Grant HD33648-01, Grant 1003312 from the Burroughs Wellcome Fund, and National Cancer Institute Grants CA77737, CA09094013, and CA22435 to the University of Vermont Cancer Center DNA Analysis Facility. L.G.C. is supported by a Career Award at the Scientific Interface from the Burroughs-Wellcome Fund.
4 The online version of this article contains supplemental material.
References
- 1.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Ann Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 3.Roth DB, Craig NL. VDJ recombination: a transposase goes to work. Cell. 1998;94:411–414. doi: 10.1016/s0092-8674(00)81580-9. [DOI] [PubMed] [Google Scholar]
- 4.Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadel B, Feeney AJ. Influence of coding-end sequence on coding-end processing in V(D)J recombination. J Immunol. 1995;155:4322–4329. [PubMed] [Google Scholar]
- 6.Nadel B, Feeney AJ. Nucleotide deletion and P addition in V(D)J recombination: a determinant role of the coding-end sequence. Mol Cell Biol. 1997;17:3768–3778. doi: 10.1128/mcb.17.7.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basu M, Hegde MV, Modak MJ. Synthesis of compositionally unique DNA by terminal deoxynucleotidyl transferase. Biochem Biophys Res Commun. 1983;111:1105–1112. doi: 10.1016/0006-291x(83)91413-4. [DOI] [PubMed] [Google Scholar]
- 8.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 9.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair. 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, Kirsch IR. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250:1426–1429. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 11.Bash RO, Crist WM, Shuster JJ, Link MP, Amylon M, Pullen J, Carroll AJ, Buchanan GR, Smith RG, Baer R. Clinical features and outcome of T cell acute lymphoblastic leukemia in childhood with respect to alterations at the TAL1 locus: a Pediatric Oncology Group study. Blood. 1993;81:2110–2117. [PubMed] [Google Scholar]
- 12.Breit TM, Mol EJ, Wolvers-Tettero IL, Ludwig WD, van Wering ER, van Dongen JJ. Site-specific deletions involving the tal-1 and sil genes are restricted to cells of the T cell receptor α/β lineage: T cell receptor δ gene deletion mechanism affects multiple genes. J Exp Med. 1993;177:965–977. doi: 10.1084/jem.177.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cline MJ. The molecular basis of leukemia. N Engl J Med. 1994;330:328–336. doi: 10.1056/NEJM199402033300507. [DOI] [PubMed] [Google Scholar]
- 14.Duro D, Bernard O, Valle VD, Leblanc T, Berger R, Larsen CJ. Inactivation of the P16INK4/MTS1 gene by a chromosome translocation t(9; 14)(p21–22;q11) in an acute lymphoblastic leukemia of B-cell type. Cancer Res. 1996;56:848–854. [PubMed] [Google Scholar]
- 15.Gu Y, Cimino G, Alder H, Nakamura T, Prasad R, Canaani O, Moir DT, Jones c, Nowell PC, Croce CM, Canaani E. The (4;11)(q21;q23) chromosome translocations in acute leukemias involve the VDJ recombinase. Proc Natl Acad Sci USA. 1992;89:10464–10468. doi: 10.1073/pnas.89.21.10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haluska FG, Finver S, Tsujimoto Y, Croce CM. The t(8;14) chromosomal translocation occurring in B cell malignancies results from mistakes in V-D-J joining. Nature. 1986;324:158–161. doi: 10.1038/324158a0. [DOI] [PubMed] [Google Scholar]
- 17.Thandla S, Alashari M, Green DM, Aplan PD. Therapy-related T cell lymphoblastic lymphoma with t(11;19)(q23;p13) and MLL gene rearrangement. Leukemia. 1999;13:2116–2118. doi: 10.1038/sj.leu.2401558. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Jani-Sait SN, Escalon EA, Carroll AJ, de Jong PJ, Kirsch IR, Aplan PD. The t(14;21)(q11.2;q22) chromosomal translocation associated with T cell acute lymphoblastic leukemia activates the BHLHB1 gene. Proc Natl Acad Sci USA. 2000;97:3497–3502. doi: 10.1073/pnas.97.7.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finette BA, O’Neill JP, Vacek PM, Albertini RJ. Gene mutations with characteristic deletions in cord blood T lymphocytes associated with passive maternal exposure to tobacco smoke. Nat Med. 1998;4:1144–1151. doi: 10.1038/2640. [DOI] [PubMed] [Google Scholar]
- 20.Finette BA, Poseno T, Albertini RJ. V(D)J recombinase-mediated HPRT mutations in peripheral blood lymphocytes of normal children. Cancer Res. 1996;56:1405–1412. [PubMed] [Google Scholar]
- 21.Fuscoe JC, Zimmerman LJ, Lippert MJ, Nicklas JA, O’Neill JP, Albertini RJ. V(D)J recombinase-like activity mediates hprt gene deletion in human fetal T lymphocytes. Cancer Res. 1991;51:6001–6005. [PubMed] [Google Scholar]
- 22.Yoshioka M, O’Neill JP, Vacek PM, Finette BA. Gestational age and gender specific in utero V(D)J recombinase mediated deletions. Cancer Res. 2001;61:3432–3438. [PubMed] [Google Scholar]
- 23.Finette BA, Kendall H, Vacek PM. Mutational spectral analysis at the HPRT locus in healthy children. Mutat Res. 2002;505:27–41. doi: 10.1016/s0027-5107(02)00119-7. [DOI] [PubMed] [Google Scholar]
- 24.Gerstein RM, Lieber MR. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev. 1993;7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 25.Messier T, O’Neill JP, Hou SM, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 2003;22:1381–1388. doi: 10.1093/emboj/cdg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Neill JP, McGinniss MJ, Berman JK, Sullivan LM, Nicklas JA, Albertini RJ. Refinement of a T lymphocyte cloning assay to quantify the in vivo thioquanine-resistant mutant frequency in humans. Mutagenesis. 1987;2:87–94. doi: 10.1093/mutage/2.2.87. [DOI] [PubMed] [Google Scholar]
- 27.van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, Gottardi E, Rambaldi A, Dotti G, Griesinger F, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease: report of the BIOMED-1 concerted action—investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 28.Cowell LG, Davila M, Kepler TB, Kelsoe G. Identification and utilization of arbitrary correlations in models of recombination signal sequences. Genome Biol. 2002;3:RESEARCH0072.1–0072.20. doi: 10.1186/gb-2002-3-12-research0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee AI, Fugmann SD, Cowell LG, Ptaszek LM, Kelsoe G, Schatz DG. A functional analysis of the spacer of V(D)J recombination signal sequences. PLoS Biol. 2003;1:56–69. doi: 10.1371/journal.pbio.0000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowell LG, Davila M, Yang K, Kepler TB, Kelsoe G. Prospective estimation of recombination signal efficiency and identification of functional cryptic signals in the genome by statistical modeling. J Exp Med. 2003;197:207–220. doi: 10.1084/jem.20020250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Repasky JA, Corbett E, Boboila C, Schatz DG. Mutational analysis of terminal deoxynucleotidyltransferase-mediated N-nucleotide addition in V(D)J recombination. J Immunol. 2004;172:5478–5488. doi: 10.4049/jimmunol.172.9.5478. [DOI] [PubMed] [Google Scholar]
- 32.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOIN-SOLVER. J Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 33.Mickelsen S, Snyder C, Trujillo K, Bogue M, Roth DB, Meek K. Modulation of terminal deoxynucleotidyltransferase activity by the DNA-dependent protein kinase. J Immunol. 1999;163:834–843. [PubMed] [Google Scholar]
- 34.Fujita K, Shimazaki N, Ohta Y, Kubota T, Ibe S, Toji S, Tamai K, Fujisaki S, Hayano T, Koiwai O. Terminal deoxynucleotidyltransferase forms a ternary complex with a novel chromatin remodeling protein with 82 kDa and core histone. Genes Cells. 2003;8:559–571. doi: 10.1046/j.1365-2443.2003.00656.x. [DOI] [PubMed] [Google Scholar]
- 35.Ibe S, Fujita K, Toyomoto T, Shimazaki N, Kaneko R, Tanabe A, Takebe I, Kuroda S, Kobayashi T, Toji S, et al. Terminal deoxynucleotidyltransferase is negatively regulated by direct interaction with proliferating cell nuclear antigen. Genes Cells. 2001;6:815–824. doi: 10.1046/j.1365-2443.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita N, Shimazaki N, Ibe S, Kaneko R, Tanabe A, Toyomoto T, Fujita K, Hasegawa T, Toji S, Tamai K, et al. Terminal deoxynucleotidyltransferase directly interacts with a novel nuclear protein that is homologous to p65. Genes Cells. 2001;6:641–652. doi: 10.1046/j.1365-2443.2001.00449.x. [DOI] [PubMed] [Google Scholar]
- 37.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 38.Ramadan K, I, Shevelev V, Maga G, Hubscher U. De novo DNA synthesis by human DNA polymerase λ, DNA polymerase μ and terminal deoxyribonucleotidyl transferase. J Mol Biol. 2004;339:395–404. doi: 10.1016/j.jmb.2004.03.056. [DOI] [PubMed] [Google Scholar]
- 39.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase λ: implications for function. J Biol Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 40.Ramadan K, Maga G, Shevelev IV, Villani G, Blanco L, Hubscher U. Human DNA polymerase lambda possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J Mol Biol. 2003;328:63–72. doi: 10.1016/s0022-2836(03)00265-1. [DOI] [PubMed] [Google Scholar]
- 41.Jager U, Bocskor S, Le T, Mitterbauer G, Bolz I, Chott A, Kneba M, Mannhalter C, Nadel B. Follicular lymphomas’ BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- 42.Maga G, Villani G, Ramadan K, Shevelev I, Tanguy Le Gac N, Blanco L, Blanca G, Spadari S, Hubscher U. Human DNA polymerase λ functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J Biol Chem. 2002;277:48434–48440. doi: 10.1074/jbc.M206889200. [DOI] [PubMed] [Google Scholar]
- 43.Welzel N, Le T, Marculescu R, Mitterbauer G, Chott A, Pott C, Kneba M, Du MQ, Kusec R, Drach J, et al. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11semi]14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001;61:1629–1636. [PubMed] [Google Scholar]
- 44.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 45.Thai TH, Kearney JF. Distinct and opposite activities of human terminal deoxynucleotidyltransferase splice variants. J Immunol. 2004;173:4009–4019. doi: 10.4049/jimmunol.173.6.4009. [DOI] [PubMed] [Google Scholar]
- 46.Carlotti E, Pettenella F, Amaru R, Slater S, Lister TA, Barbui T, Basso G, Cazzaniga G, Rambaldi A, Biondi A. Molecular characterization of a new recombination of the SIL/TAL-1 locus in a child with T cell acute lymphoblastic leukaemia. Br J Haematol. 2002;118:1011–1018. doi: 10.1046/j.1365-2141.2002.03747.x. [DOI] [PubMed] [Google Scholar]
- 47.Delabesse E, Bernard M, Landman-Parker J, Davi F, Leboeuf D, Varet B, Valensi F, Macintyre EA. Simultaneous SIL-TAL1 RT-PCR detection of all tal(d) deletions and identification of novel tal(d) variants. Br J Haematol. 1997;99:901–907. doi: 10.1046/j.1365-2141.1997.4833286.x. [DOI] [PubMed] [Google Scholar]
- 48.Raghavan SC, I, Kirsch R, Lieber MR. Analysis of the V(D)J recombination efficiency at lymphoid chromosomal translocation breakpoints. J Biol Chem. 2001;276:29126–29133. doi: 10.1074/jbc.M103797200. [DOI] [PubMed] [Google Scholar]
- 49.Hutchison JB. Gender-specific steroid metabolism in neural differentiation. Cell Mol Neurobiol. 1997;17:603–626. doi: 10.1023/A:1022581902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smulian JC, Campbell WA, Rodis JF, Feeney LD, Fabbri EL, Vintzileos AM. Gender-specific second trimester biometry. Am J Obstet Gynecol. 1995;173:1195–1201. doi: 10.1016/0002-9378(95)91352-1. [DOI] [PubMed] [Google Scholar]
- 51.Hanley K, Rassner U, Jiang Y, Vansomphone D, Crumrine D, Komuves L, Elias PM, Feingold KR, Williams ML. Hormonal basis for the gender difference in epidermal barrier formation in the fetal rat: acceleration by estrogen and delay by testosterone. J Clin Invest. 1996;97:2576–2584. doi: 10.1172/JCI118706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fleisher LA, Dipietro JA, Johnson TR, Pincus S. Complementary and noncoincident increases in heart rate variability and irregularity during fetal development. Clin Sci. 1997;92:345–349. doi: 10.1042/cs0920345. [DOI] [PubMed] [Google Scholar]
- 53.Lovoie JC, Chessex P. Gender and maturation affect glutathione status in human neonatal tissues. Free Radic Biol Med. 1997;23:648–657. doi: 10.1016/s0891-5849(97)00011-7. [DOI] [PubMed] [Google Scholar]
- 54.Gale KB, Ford AM, Repp R, Borkhardt A, Keller C, Eden OB, Greaves MF. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci USA. 1997;94:13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gill-Super HJ, Rothberg PG, Kobayashi H, Freeman AI, Diaz MO, Rowley JD. Clonal, nonconstitutional rearrangements of the MLL gene in infant twins with acute lymphoblastic leukemia: in utero chromosome rearrangement of 11q23. Blood. 1994;83:641–644. [PubMed] [Google Scholar]
- 56.Sandler DP, Ross JA. Epidemiology of acute leukemia in children and adults. Semin Oncol. 1997;24:3–16. [PubMed] [Google Scholar]
- 57.Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, Saha V, Biondi A, Greaves MF. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]


