Abstract
BAD, a proapoptotic molecule of the BCL2 family, is regulated by reversible phosphorylation. During survival, BAD is sequestered by 14-3-3 through serine 136 phosphorylation and is dissociated from BCL-XL through serine 155 phosphorylation. We report that phosphoserine 112 (pSer112) dephosphorylation functions as a gatekeeper for BAD-mediated apoptosis. During apoptosis, dephosphorylation of pSer112 preceded pSer136 dephosphorylation. Dephosphorylation of pSer112 accelerated dephosphorylation of pSer136, and inhibition of pSer112 dephosphorylation prevented pSer136 dephosphorylation, indicating that dephosphorylation of pSer112 is required for dephosphorylation of pSer136. Protein phosphatase 2A (PP2A) is the major pSer112 phosphatase. PP2A competed with 14-3-3 for BAD binding, and survival factor withdrawal enhanced PP2A association with BAD. Dephosphorylation of the critical residue, pSer136, could only be blocked by inhibition of all known subfamilies of serine/threonine phosphatases, suggesting that multiple phosphatases are involved in pSer136 dephosphorylation. Inhibition of PP2A rescued FL5.12 cells from apoptosis, demonstrating a physiologic role for PP2A-mediated pSer112 dephosphorylation. Thus, PP2A dephosphorylation of pSer112 is the key initiating event regulating the activation of BAD during interleukin-3 withdrawal-induced apoptosis.
BAD is a proapoptotic molecule of the BCL2 family of apoptosis regulators containing only the BH3 domain (1, 17, 21, 22). BAD promotes cell death by binding and inactivating the survival function of BCL-XL and BCL2 (43). Reversible phosphorylation regulates the activity of various members of the BCL2 family, including BCL2 itself (29), Bid (11), Bik (37), and BAD (44). Among these, the regulation of protein function by phosphorylation has been most clearly demonstrated for BAD. Phosphorylated BAD is sequestered in its inactive form in the cytosol by 14-3-3, while dephosphorylated BAD is targeted to the mitochondria, where it causes cell death by binding BCL-XL and BCL2 (43, 44).
Five phosphorylation sites have been reported for BAD. Phosphorylation at serine 112 (Ser112) and serine 136 (Ser136) is involved in 14-3-3 binding (44). Of these two sites, Ser136 appears to be dominant in determining 14-3-3 binding, whereas the role of Ser112 is less clear (42, 25). Phosphorylation of Ser136 is accomplished predominantly by Akt/protein kinase B or p70S6 kinase (8, 14), whereas mitochondrially localized protein kinase A, Rsk, and PAK1 have all been shown to phosphorylate Ser112 (15, 31, 32, 34). Dephosphorylation of residue Ser155 in the BH3 domain of BAD is key in mediating BCL-XL binding, and phosphorylation of this residue by protein kinase A or Rsk causes BAD dissociation from BCL-XL (9, 33). Ser170 is another site that is phosphorylated in cytokine-dependent cell survival (12). Recently, Ser128 was found to be phosphorylated by Cdc2 during induction of apoptosis in cerebellar granular neurons. Phosphorylation of Ser128 has also been implicated in dissociation of BAD from 14-3-3 (19).
The 14-3-3 proteins were identified as phosphoserine/threonine binding proteins (2, 24, 35). There are seven known mammalian 14-3-3 isoforms which bind and modify the functions of a wide variety of critical signaling molecules. 14-3-3 ligands include kinases such as Raf and protein kinase C, receptors such as the interleukin-3 (IL-3)/IL-5/granulocyte-macrophage colony-stimulating factor receptor βc chain, cytoskeletal proteins such as vimentin and keratins, cell cycle regulators such as Cdc25, transcription factors such as the forkhead family, and, importantly, apoptosis regulators, such as BAD. Structural studies of 14-3-3 indicate that the molecule is a dimer with two phosphopeptide-binding amphipathic grooves in antiparallel orientation (41), which could interact with the two 14-3-3 binding motifs on BAD.
It has been suggested that one function of 14-3-3 is to promote cell survival, as inhibition of apoptosis results from the binding of many 14-3-3 ligands, including BAD, ASK1, and forkhead factors (24). 14-3-3 forms a very stable complex with phosphorylated BAD and plays a significant role in the regulation of BAD function. Published data suggest that 14-3-3 binding leads to a conformation change in BAD which allows Ser155 to be phosphorylated (9). Moreover, we have shown that 14-3-3 prevents the ability of phosphatases to convert BAD into a death molecule (6).
Regulation by protein phosphatases has been shown for BCL2 and BAD. Phospho-Ser70 of BCL2 is dephosphorylated by mitochondrially localized protein phosphatase 2A (PP2A) in response to ceramide (28). The proapoptotic function of BAD is activated by serine/threonine phosphatases. Dephosphorylated BAD dissociates from 14-3-3 and inactivates BCL-XL or BCL2. Recent demonstration of increased lymphocyte developmental cell death in mice bearing knocked-in alleles of a phosphorylation-defective mutant underscores the physiologic importance of BAD dephosphorylation (10). Mammalian protein serine/threonine phosphatases consist of several families, including PP1, PP2A, the Ca2+-dependent PP2B or calcineurin, and the Mg2+-dependent PP2C. PP2B dephosphorylates BAD during Ca2+-induced apoptosis in prostate cell lines and in hippocampal neurons (40). PP1α has also been reported to be a BAD phosphatase in IL-2- and IL-4-dependent T-cell lines and IL-3-dependent 32D cells (3, 30).
PP2A comprises a family of highly regulated phosphatases (18). Each PP2A holoenzyme consists of a 36-kDa catalytic subunit (Cα or Cβ), a 65-kDa A structural subunit (Aα or Aβ), forming the AC core dimer, and a third highly diversified B regulatory subunit (B, B′, and B"). Several different isoforms make up each B subunit family, and the various combinations of the subunits can generate a large number of PP2A holoenzymes. Substrate specificity and subcellular localization are regulated by the B subunit in each holoenzyme complex. PP2A regulates a large number of signaling complexes, including extracellular signal-regulated kinase/mitogen-activated protein kinase, protein kinase A, protein kinase B, receptor complexes such as the ryanodine receptor calcium release channel, regulators of translation such as Tap42/α4, and cell cycle proteins such as cdc25.
We have found a PP2A activity that dephosphorylates BAD in the well-established IL-3-dependent FL5.12 cell line (6). Inhibition of BAD phosphatase activity in FL5.12 cells resulted in protection from IL-3 deprivation-induced cell death, indicating a physiologic role for this PP2A activity in cytokine-mediated apoptosis. Moreover, we found that successful dephosphorylation of BAD by PP2A required BAD to be dissociated from 14-3-3, suggesting that complex interactions between BAD, 14-3-3, and PP2A regulate the proapoptotic activity of BAD.
Previous studies have led to several questions regarding the molecular mechanisms regulating BAD dephosphorylation. Is dephosphorylation of pSer112, pSer136, and pSer155 regulated by the same or different phosphatase activities? Are the dephosphorylation events at these sites regulated independently or coordinately? What is the functional significance of dephosphorylation at each site? In this report, we present data pertaining to these questions. Our findings indicate a regulatory role for pSer112 dephosphorylation and suggest a model for the coordinately regulated dephosphorylation of BAD leading to apoptosis during survival factor deprivation.
MATERIALS AND METHODS
Plasmids and constructs.
With pBabeBADwt-112A, -136A, or -112A136A as the template (6), phosphorylation-defective alanine-substituted mutants of BAD at S155 were generated by standard PCR-based site mutagenesis. The mutant cDNAs were cloned in the retroviral expression vector pBabepuro, and viruses were prepared as described previously (6). The mammalian expression construct pEBG-BAD, expressing a glutathione S-transferase (GST) fusion of BAD, was from New England Biolabs. The bacterial expression construct GST-BAD was generated by cloning the mouse BAD cDNA into vector pGEX-4T-1 (Pharmacia). The bacterial expression construct GST-PP2A/c was prepared by cloning human PP2A/c into the pGEX-4T-1 vector by the PCR method with pHM6PP2A/c as the template (7).
Cell culture.
NIH 3T3 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% calf serum. FL5.12 cells were cultured as described previously (6). NIH 3T3 cells and FL5.12 BCL-XL cells expressing wild-type BAD or phosphorylation-defective mutants were constructed by infection with retroviruses carrying pBabeBAD wild-type or mutant constructs and selected with puromycin as described previously (6). FL5.12 cells expressing GST-BAD were prepared by electroporation of cells with pEBG-BAD plasmids as described previously (43). NIH 3T3 cells expressing BAD grown in 10% serum were treated with 5 μM fostriecin (gift from Shirish Shenolikar, Duke University) or other drugs for 3 h. For IL-3 deprivation and calyculin A treatment, FL5.12 cultures were treated as described previously (6).
Western blotting and immunoprecipitation.
Cell lysates for Western blotting were prepared in radioimmunoprecipitation assay (RIPA) buffer as described previously (6). For immunoprecipitation, 107 cells were lysed in isotonic immunoprecipitation buffer as described previously (6). BAD was precipitated in the presence or absence of 1 μM microcystin-LR (a phosphatase inhibitor), R18 peptides (39), and 1% Empigen BB or phospho-Ser112, phospho-Ser136, or phospho-Ser155 peptide (gifts from Cell Signaling), as indicated, with rabbit polyclonal anti-BAD antibody C20 (Santa Cruz). BAD immunocomplexes were precipitated with protein A/G-Sepharose (Sigma), fractionated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to a polyvinylidene difluoride membrane. Western blots were detected by anti-14-3-3 K19 antibody (Santa Cruz), anti-BAD 31420 (Transduction Laboratories), anti-PP2A/c (Transduction Laboratories), or anti-PP2A/A (Santa Cruz), as indicated, and developed with enhanced chemiluminescence. Anti-phospho-Ser112 and phospho-Ser136 antibodies were purchased from Upstate Biotechnology, and anti-phospho-Ser155 antibody was purchased from Cell Signaling.
In vitro BAD dephosphorylation assay.
Cell lysates were prepared in phosphatase assay buffer (50 mM Tris-Cl, 150 mM NaCl, 0.25% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, and 10 μg of leupeptin per ml), and BAD dephosphorylation was analyzed in the presence and absence of 25 μM R18 as described previously (6). For profiling BAD phosphatases in vitro, cell lysates were incubated with 0.5 μM calyculin A, 1 μM microcystin, 0.5 μM okadaic acid, 1 μM fostriecin, 2.5 μM cyclosporine A, 500 ng of FK506 per ml, 5 mM EGTA, 10 mM NaF, or a cocktail (5 mM EGTA, 5 mM EDTA, 10 mM NaF, and 1 μM microcystin) on ice for 10 min before incubating at 30°C for 20 min.
GST pulldown assay.
GST, GST-BAD, GST-PP2A/A (gift from Kerry Campbell, Fox Chase Cancer Center, Philadelphia, Pa.), and GST-PP2A/c were expressed in Escherichia coli BL21 cells. Two to three hours after isopropyl-β-d-thiogalactopyranoside (IPTG) induction, bacteria were resuspended in lysis buffer (phosphate-buffered saline with 1% Triton X-100, 1% Tween 20, 1 mg of lysozyme per ml, and a bacterial protease inhibitor cocktail) and sonicated gently. GST fusion protein was purified with glutathione-Sepharose 4B (Pharmacia) under native conditions following the manufacture's protocols. From 1 to 2 μg of purified GST, GST-BAD, or GST-BAD phosphorylated by protein kinase A in vitro, GST-PP2A/A, or GST-PP2A/c was added to lysates of NIH 3T3 parental cells or of cells expressing wild-type BAD and incubated for 1 h at 4°C with or without R18. GST fusion protein complexes were pulled down by glutathione-Sepharose 4B, washed five times with isotonic immunoprecipitation buffer, fractionated by SDS-PAGE, and analyzed by Western blotting.
RESULTS
14-3-3 protects all three phosphorylated serine residues of BAD from the action of phosphatases, while only pSer136 is the major 14-3-3 recruitment site.
Survival and growth factors stimulate phosphorylation of BAD at Ser112, Ser136, and Ser155 and cause the sequestration of BAD in the cytosol by 14-3-3 (9, 44). In apoptosis induced by survival factor withdrawal, BAD is dephosphorylated and mitochondrial. We have previously shown that loss of the hyperphosphorylated form of BAD and dephosphorylation of pSer112 were catalyzed by a PP2A activity in IL-3-dependent FL5.12 cells (6). Dephosphorylation of pSer112 was prevented by 14-3-3 binding (6).
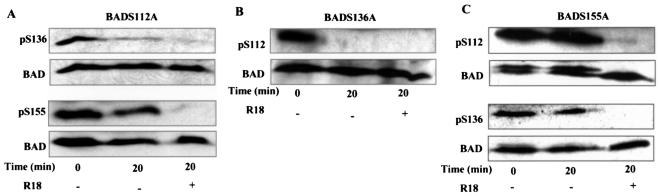
To determine whether 14-3-3 also protected other phosphoserine sites of BAD from phosphatases, we examined the dephosphorylation of key serine residues in BAD in the presence and absence of R18, a nonphosphorylated peptide which interacts with the amphipathic groove of 14-3-3 and disrupts substrate binding (39). Lysates from NIH 3T3 cells expressing wild-type mouse BAD were allowed to dephosphorylate in vitro by incubation at 30°C in isotonic buffer without exogenous ATP or phosphatase inhibitors so that phosphatases but not kinases were active. BAD dephosphorylation was assayed by SDS-PAGE and Western blotting with antibodies specifically recognizing each phosphoserine residue (Fig. 1A). The specificity of each antiphosphoserine antibody was confirmed by using phosphorylation-defective BAD mutants (Fig. 1B). Dephosphorylation of pSer136 and pSer155, as well as pSer112, occurred only when 14-3-3 was forced to dissociate from BAD by R18 (Fig. 1A), indicating that 14-3-3 protects all three phosphorylated serine residues.
FIG. 1.
14-3-3 prevents in vitro dephosphorylation of BAD pSer112, pSer136, and pSer155, but only pSer136 is the major 14-3-3 recruitment site. (A) Lysates were prepared from NIH 3T3 cells expressing wild-type (Wt) BAD in phosphatase buffer as described previously (6) and incubated at 30°C for 20 min in the absence or presence of 25 μM R18 peptide. Dephosphorylation reactions terminated by adding SDS sample buffer and boiling for 5 min were analyzed by SDS-PAGE and Western blotting with phospho-specific antibodies to pSer112, pSer136, and pSer155 and a mouse antibody against total BAD. (B) Specificity of antiphosphoserine residues was confirmed by Western blotting of lysates from parental NIH 3T3 cells (3T3), NIH 3T3 cells expressing wild-type BAD (3T3/WT), and the 112A, 136A, and 155A mutants (3T3/112A, 3T3/136A, and 3T3/155A, respectively) with antibodies to pSer112 (upper), pSer136 (middle), and pSer155 (lower) and an antibody against total BAD (Santa Cruz; 31420). Asterisks indicate nonspecific bands. (C) Alanine substitution of Ser136 results in significant loss of 14-3-3 association with BAD. BAD was immunoprecipitated with anti-BAD C-20 antibody from NIH 3T3 or FL5.12 cells expressing wild-type BAD or BAD with alanine substitutions at one, two, or three (3A) phosphorylation sites, as indicated. The BAD immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-14-3-3 antibody (upper) or anti-BAD 36420 antibody (lower). The asterisk indicates a species immunoreactive with BAD antibodies. (D) Top, phosphoserine 136 peptide displaces 14-3-3 from BAD immunocomplexes. BAD was immunoprecipitated from FL5.12 cells expressing wild-type BAD in the absence (−) or presence of 25 μM pSer112, pSer136, or pSer155 peptides (pS112p, pS136P, and pS155p, respectively) and immunoblotted for 14-3-3 and BAD as above. Bottom, phospho-Ser136 peptide blocks 14-3-3 association with phosphorylated GST-BAD. In vitro protein kinase A-phosphorylated GST-BAD was incubated with NIH 3T3 lysates in the presence of pSer112, pSer136, or pSer155 peptide, pulled down by glutathione agarose, and immunoblotted for 14-3-3 and BAD as above.
Studies with synthetic peptides and transient transfections of BAD mutants have shown that phosphorylation of Ser136 was essential, but phosphorylation of Ser112 was dispensable, for 14-3-3 binding to BAD (8, 23, 25). This is consistent with predictions of 14-3-3 binding from sequences flanking the two phosphorylation sites. The residues flanking Ser136, RSRpS136AP, match perfectly the 14-3-3 consensus binding motif, RSXpSXP, whereas the pSer112 domain, RHSpS112YP, is a suboptimal binding motif for 14-3-3 (26, 42). Nevertheless, Yaffe et al. found that peptides phosphorylated at two tandem motifs had 30-fold higher affinity for 14-3-3 than singly phosphorylated peptides and proposed that pSer112 and pSer136 of BAD cooperate in binding 14-3-3 (42). Although phosphorylation of Ser155 hindered the interaction of BAD with BCL-XL (9, 20, 33, 45), it is not clear whether pSer155 is involved in 14-3-3 binding.
Having found that 14-3-3 protected all three phosphoserine sites in wild-type BAD protein (Fig. 1A), we wished to clarify the role of each phosphorylated residue, i.e., Ser112, Ser136, and Ser155, in mediating 14-3-3 association with full-length BAD protein. Phosphorylation-defective BAD mutants with alanine replacements of one, two, or three of the phosphoserine sites were expressed and immunoprecipitated from FL5.12 or NIH 3T3 cells and then immunoblotted for 14-3-3 with a pan-14-3-3 antibody (Fig. 1C). Alanine mutation at Ser136 (136A) resulted in a tremendous loss of 14-3-3 binding with BAD, but alanine substitutions at either Ser112 or Ser155 did not affect 14-3-3 binding. Double mutants containing 136A also did not bind 14-3-3 to any significant degree, while the 112A155A mutant bound 14-3-3 as well as wild-type BAD did. Thus, 14-3-3 binding was only significantly affected by mutation at Ser136.
Wild-type BAD from cells grown under steady-state conditions migrates as a doublet on SDS-PAGE. The upper band of the doublet was absent in lysates of cells expressing the 112A mutant, indicating that the “hyperphosphorylated” band seen in wild-type BAD consisted of the species containing phosphorylated Ser112 (Fig. 1C). Therefore, collapse of the doublet into the lower single band on SDS-PAGE reflects pSer112 dephosphorylation. A band higher than the doublet (indicated by an asterisk in Fig. 1C) was frequently seen on our BAD immunoblots. The exact identity of this band is unknown, but it is Ser136 dependent and possibly represents a posttranslationally modified form of BAD.
To confirm that pSer136 was the main 14-3-3 docking site, we tested the ability of synthetic phosphopeptides representing each of the phosphoserine sites to compete with BAD for 14-3-3 association. Immunoprecipitation of BAD from FL5.12 cells in the presence of phosphopeptides containing pSer112, pSer136, or pSer155 showed that only the pSer136 peptide abolished association of 14-3-3 with BAD (Fig. 1D, upper panels). Phosphorylated GST-BAD captured 14-3-3 from NIH 3T3 lysates in the presence of either pSer112 peptide or pSer155 peptide, but not in the presence of pSer136 peptide (Fig. 1D, lower panels). These experiments with stably expressed full-length BAD protein confirmed previous results from in vitro peptide studies that 14-3-3 is recruited to BAD mainly by the pSer136 domain of BAD and that the pSer112 or pSer155 domain does not play a significant role in 14-3-3 recruitment, even though pSer112 and pSer155 are protected from dephosphorylation by 14-3-3.
Dephosphorylation of pSer112 facilitates dephosphorylation of pSer136.
Binding of 14-3-3 to BAD is dependent on Ser136 phosphorylation, yet 14-3-3 also prevented dephosphorylation of pSer112 and pSer155 by phosphatases (Fig. 1). 14-3-3 binds ligands as a dimer, and Yaffe et al. proposed a bidentate association model of 14-3-3 binding to molecules containing multiple phosphorylated serine sites, such as Raf, BAD, and Cbl (41, 42). Given that peptides with multiple phosphorylated motifs result in higher 14-3-3 affinity than peptides with a single phosphorylated motif, we hypothesized that alanine substitution of either Ser112 or Ser136 should impair the stability of the 14-3-3/BAD complex and increase the accessibility of BAD to phosphatases.
Using BAD mutants containing alanine substitutions at each phosphorylation site, 112A, 136A, and 155A, we analyzed in vitro dephosphorylation of the other two sites. Whereas no dephosphorylation of wild-type BAD occurred unless 14-3-3 was forced to dissociate (Fig. 1A), significant dephosphorylation of pSer136 and some dephosphorylation of pSer155 occurred in the 112A mutant without forcing 14-3-3 dissociation with the R18 peptide (Fig. 2A). This suggests that dephosphorylation of pSer112 attenuated the ability of 14-3-3 to protect pSer136 and pSer155 against phosphatases. As expected, complete dephosphorylation of pSer112 in lysates of cells expressing the BAD136A mutant was observed by 20 min of incubation in the absence of R18 peptide, because 14-3-3 cannot bind and protect BAD when Ser136 is mutated (Fig. 2B). Very little to no Ser155 phosphorylation could be detected in the 136A lysates even at time zero of the dephosphorylation assay (data not shown), consistent with the previous report that Ser155 phosphorylation requires Ser136 phosphorylation (9). In lysates of cells expressing the 155A mutant, dephosphorylation of pSer112 and pSer136 still required displacement of 14-3-3 (Fig. 2C), suggesting that pSer155 does not play an active role in stabilizing 14-3-3 association with BAD. Taken together, these results suggest that the pSer112 domain cooperates with the pSer136 domain to maintain a stable complex of 14-3-3 and BAD and that these two sites synergistically block the access of phosphatases to BAD.
FIG. 2.
pSer112 and pSer136 cooperate in 14-3-3 binding to block dephosphorylation of BAD. (A) Lysates from NIH 3T3 cells expressing the BAD 112A mutant were incubated in phosphatase assay buffer in the absence or presence of R18 peptide and immunoblotted with anti-pSer136, anti-pSer155, and anti-BAD antibodies. (B) Lysates from NIH 3T3 cells expressing the BAD 136A mutant were subjected to in vitro dephosphorylation as in A and immunoblotted with anti-pSer112 and anti-BAD antibodies. Phosphorylation of Ser155 was barely detectable and is not shown. (C) Lysates from NIH 3T3 cells expressing the BAD 155A mutant were similarly subjected to in vitro dephosphorylation and assayed for pSer112 and pSer136 as above.
The finding that the phosphorylation status of Ser112 influenced dephosphorylation of pSer136 implied that dephosphorylation of BAD progressed in a tightly regulated manner during survival factor deprivation. To examine the timing of dephosphorylation of each phosphoserine site, the phosphorylation status of Ser112, Ser136, and Ser155 was assessed by Western blotting in FL5.12 lysates collected during the time course of IL-3 withdrawal. Significant dephosphorylation of Ser112 occurred between 1 and 2 h after IL-3 withdrawal and was nearly complete by 4 h (Fig. 3A, pS112 panel). Dephosphorylation of pSer112 was accompanied by a loss of the upper band of the BAD doublet at 2 to 4 h and a gain of the lower band at later times (Fig. 3A, BAD panels). This is consistent with our earlier finding that the upper band represents pSer112-phosphorylated BAD.
FIG. 3.
Dephosphorylation of pSer112 precedes and promotes pSer136 dephosphorylation during survival factor withdrawal. (A) Lysates of FL5.12 BCL-XL/BAD cells at the indicated times of IL-3 withdrawal were immunoblotted for pSer112, pSer136, pSer155, and total BAD (31420; Santa Cruz). Representative immunoblots are shown. Percentages of phosphorylation indicate the averages of pSer residue phosphorylation of two to three experiments as measured by densitometry. The ratio of each pSer residue phosphorylation to total BAD at time zero was set as 100%. (B) FL5.12 cells expressing the 112A BAD mutant were immunoblotted for pSer136 and compared to cells expressing wild-type (WT) BAD during IL-3 withdrawal. Percent phosphorylation was quantitated as in A. (C) Lysates of FL5.12 BCL-XL/BAD cells collected at the indicated times after IL-3 withdrawal were allowed to dephosphorylate in the absence or presence of R18 and immunoblotted for pSer136 and total BAD. (D) Graph of relative pSer136 dephosphorylation in the absence of R18, shown in C, as measured by densitometry. The ratio of pSer136 to total BAD at time zero was set as 100%. Percent pS136 phosphorylation is the ratio of pSer136 to total BAD after 20 min at 30°C in the absence of R18 compared to that at time zero in three experiments.
Nearly complete dephosphorylation of pSer136 occurred between 6 and 8 h after IL-3 withdrawal, significantly later than pSer112 dephosphorylation (Fig. 3A, pS136 panel). Significant dephosphorylation of pSer155 also occurred after pSer112 dephosphorylation, at 4 to 6 h of IL-3 withdrawal (Fig. 3A, pS155 panel). In repeated experiments, dephosphorylation of pSer136 and pSer155 always followed pSer112 dephosphorylation, indicating that phosphatase activity at pSer112 is the first dephosphorylation event in BAD during induction of cell death.
It is notable that when the doublet of BAD was present at 0, 0.5, and 1 h after IL-3 withdrawal, pSer136 and pSer155 immunoreactivity was found mainly in the upper band of the doublet. This indicates the existence of pSer112+ pSer136+ and pSer112+ pSer155+ BAD early during IL-3 withdrawal and conversion to pSer112− pSer136+or pSer112− pSer155+ BAD at 2 to 6 h. The timing of the appearance of these different phosphorylated BAD species is consistent with sequential dephosphorylation of BAD, dephosphorylation of pSer112 being first, followed by pSer136 and pSer155 dephosphorylation.
The order of dephosphorylation of the three phosphoserine sites on BAD suggested that pSer136 dephosphorylation might require prior pSer112 dephosphorylation. To test this hypothesis, we compared the timing of pSer136 dephosphorylation in wild-type and 112A mutant forms of BAD during IL-3 withdrawal (Fig. 3B). Whereas dephosphorylation of pSer136 in wild-type BAD occurred at 4 to 6 h, dephosphorylation of pSer136 in the 112A mutant was evident before 4 h. Acceleration of pSer136 dephosphorylation in the 112A mutant suggested that phosphorylation at Ser112 normally protected pSer136 dephosphorylation.
To determine whether pSer112 dephosphorylation promotes pSer136 dephosphorylation, lysates of FL5.12 cells expressing BAD collected after IL-3 withdrawal were assayed for pSer136 dephosphorylation in vitro without the addition of 14-3-3-displacing agents (Fig. 3C). In the presence of IL-3, pSer136 was dephosphorylated only when 14-3-3 was forcibly displaced from BAD by the R18 peptide (Fig. 3C, lanes 1, 2, and 3), as shown previously. At 1 h after IL-3 withdrawal, pSer112 was minimally dephosphorylated (Fig. 3A), and pSer136 dephosphorylation was also minimal in the absence of R18 (Fig. 3C, lanes 4 to 6; Fig. 3D). However, at 4 h after IL-3 withdrawal, when pSer112 has been significantly dephosphorylated (Fig. 3A), approximately 40% of pSer136 was dephosphorylated in the absence of R18 peptide (Fig. 3C, lanes 7 to 9; Fig. 3D). This result demonstrated that dephosphorylation of pSer112 rendered pSer136 accessible to phosphatases despite the presence of 14-3-3, suggesting that dephosphorylation at Ser112 destabilizes the interaction between 14-3-3 and BAD, facilitating phosphatase activity on pSer136.
PP2A competes with 14-3-3 for association with BAD.
We have previously shown that a PP2A activity catalyzed the dephosphorylation of BAD in FL5.12 cells and that purified PP2A catalytic subunit (PP2A/c) dephosphorylates pSer112 in vitro (6). To confirm that PP2A is a BAD phosphatase in cells, we coimmunoprecipitated BAD and PP2A from cell lysates. The PP2A core dimer, consisting of the A and C subunits, coimmunoprecipitated with BAD from FL5.12 and NIH 3T3 lysates, demonstrating physical association of PP2A and BAD (Fig. 4A). Since dissociation of 14-3-3 enhanced dephosphorylation of BAD, we predicted that displacement of 14-3-3 from BAD would increase PP2A association with BAD. Indeed, in the presence of 14-3-3 displacement reagents, including the detergent Empigen, R18 peptide, and pSer136 peptide, the amount of PP2A/c in the BAD immunocomplexes was substantially increased, while14-3-3 was undetectable (Fig. 4B, upper). Likewise, recombinant GST-BAD bound a significant amount of PP2A/c, but no 14-3-3, from NIH 3T3 lysates, whereas protein kinase A-phosphorylated GST-BAD interacted with abundant 14-3-3 but only a minimal amount of PP2A/c (Fig. 4C, left). When 14-3-3 was prevented from associating with GST-BAD by the R18 peptide, phosphorylated and nonphosphorylated GST-BAD bound PP2A/c equally well (Fig. 4C, right), indicating that 14-3-3 prevented PP2A/c from interacting with phosphorylated BAD. Reciprocally, in the presence of R18, recombinant GST-PP2A/c and GST-PP2A/A both captured BAD from lysates of cells expressing BAD (Fig. 4D). These results demonstrated that PP2A physically interacted with BAD and that 14-3-3 and PP2A compete for BAD binding.
FIG. 4.
PP2A competes with 14-3-3 to associate with BAD. (A) BAD was immunoprecipitated (IP) from FL5.12 cells or NIH 3T3 cells expressing vector or wild-type BAD in the presence of R18 and immunoblotted for the PP2A/cα and PP2A/Aα subunits of PP2A. (B) BAD was immunoprecipitated from NIH 3T3/BAD lysates in the absence (−) and presence of the 14-3-3 displacement reagents Empigen BB, R18 peptide, and pSer136 peptide and immunoblotted for PP2A/c, 14-3-3, and BAD. (C) Recombinant GST-BAD and in vitro protein kinase A-phosphorylated GST-BAD was incubated with NIH 3T3 lysates in the absence and presence of R18. Glutathione pulldowns were immunoblotted for 14-3-3 and PP2A/c as indicated. (D) Recombinant GST-PP2A/A and GST-PP2A/c were incubated with lysates from control NIH 3T3 cells or cells expressing wild-type BAD, and glutathione pulldowns were immunoblotted for BAD. Input recombinant GST and GST fusion proteins were detected with anti-GST, anti-PP2A/A, and anti-PP2A/c antibodies. (E) FL5.12 cells expressing GST-BAD were deprived of IL-3 for 30 min. GST-BAD was pulled down by glutathione-agarose in the absence (−R18) or presence (+R18) of R18 and immunoblotted for GST-BAD, PP2A/Cα, PP2A/Aα, and 14-3-3. One tenth of the total lysates from FL5.12 (FL) and FL5.12/GST-BAD (FL/GST-BAD) cells used for pulldown assays was loaded as a control.
Since we postulated that PP2A gains access to BAD following 14-3-3 dissociation, we expected that the interaction of PP2A and BAD should increase during apoptosis induction. To test this hypothesis, we used FL5.12 cells stably expressing GST-BAD, which is functional in these cells in that it can be phosphorylated at the expected serine sites and can bind BCL-XL (data not shown). IL-3 was withdrawn from FL5.12 cells expressing GST-BAD, and the relative amounts of PP2A in the GST-BAD complexes in the presence and absence of IL-3 were compared by Western blotting (Fig. 4E). In the presence of IL-3, PP2A/c and PP2A/A were minimally detectable, while 14-3-3 was abundant in the GST-BAD complex. As early as 30 min after IL-3 withdrawal, the amounts of PP2A/c and PP2A/A subunits in the GST-BAD complex were increased, consistent with PP2A's being a BAD phosphatase during induction of apoptosis. Since PP2A and 14-3-3 compete for BAD binding, this finding also suggests that the 14-3-3/BAD interaction is already altered at 30 min after IL-3 withdrawal.
PP2A dephosphorylation of pSer112 regulates dephosphorylation of pSer136 by multiple phosphatases.
PP1, PP2B, and PP2A have all been reported to dephosphorylate BAD in different cell types (3, 6, 30, 40). One possible explanation for the existence of multiple phosphatases for BAD is that different phosphoserine sites of BAD may be dephosphorylated by different phosphatases. To determine whether PP2A dephosphorylates one or all three phosphoserine sites in BAD, we tested the ability of the PP2A-selective inhibitor fostriecin to prevent dephosphorylation of each serine site. Fostriecin is highly selective for PP2A (50% inhibitory concentration = 3.2 nM) and PP2A-like enzymes such as PP4 (50% inhibitory concentration = 3 nM) (16). It inhibits PP2A and PP2A-like phosphatases with 40,000-fold higher potency than PP1 (50% inhibitory concentration = 131 μM) (38).
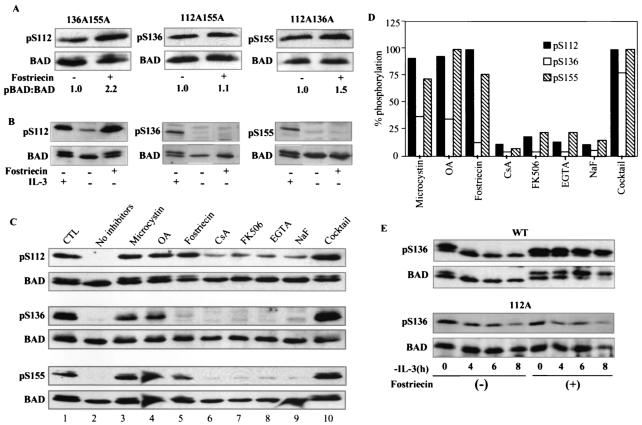
NIH 3T3 cells expressing double phosphorylation mutant 136A155A, 112A155A, or 112A136A were used to examine phosphorylation of the third site, Ser112, Ser136, and Ser155, respectively. Cells expressing these BAD constructs were treated with fostriecin, and the amounts of pSer112, pSer136, and pSer155 relative to total BAD detected by immunoblotting of lysates were compared to those of untreated lysates. Fostriecin treatment of cells caused a significant increase in the amount of pSer112 relative to total BAD (2.2-fold) but had a minimal effect on the phosphorylation of Ser136 (1.1-fold) and only a modest effect on the phosphorylation of Ser155 (1.5-fold) (Fig. 5A). The phosphorylation state of each site is the result of the action of kinases and phosphatases; therefore, the degree of phosphorylation enhancement by fostriecin can be influenced by the basal phosphorylation rate of each site. Nevertheless, the result that fostriecin affected the phosphorylation status of Ser112 the most suggests that PP2A acted differently on pSer112, pSer136, and pSer155 and that pSer112 was the preferred substrate for PP2A.
FIG. 5.
PP2A dephosphorylation of pSer112 regulates dephosphorylation of pSer136 by multiple phosphatases. (A) NIH 3T3 cells expressing BAD 136A155A (left), BAD 112A155A (middle), or BAD 112A136A (right) were treated with 5 μM fostriecin for 3 h and immunoblotted with antibodies specific to pSer112 (left), pSer136 (middle), pSer155 (right), and total BAD. The ratios of each phosphorylated BAD band to total BAD quantitated by densitometry (pBAD:BAD) are shown below. The ratios for untreated samples were set at 1.0. (B) FL5.12 cells untreated or treated with 5 μM fostriecin and deprived of IL-3 for 4 h were immunoblotted for pSer112 and total BAD. For pSer136 and pSer155 immunoblots, IL-3 was withdrawn for 4 h before adding fostriecin for another 4 h. (C) Lysates of NIH 3T3 cells expressing wild-type BAD were incubated with the indicated phosphatase inhibitors for 10 min on ice, allowed to dephosphorylate at 30°C in the presence of R18 peptides, and immunoblotted with phospho-specific antibodies as indicated. Data shown represent one of two experiments. (D) Densitometry measurements of phosphorylated to total BAD in inhibitor assays, as shown in B, with the Bio-Rad Molecular Analyst, are graphed. Data are the averages of two experiments and were normalized by setting the ratio of phosphorylated BAD to total BAD at time zero to 100%. (E) Fostriecin sustains phosphorylation of Ser136 in wild-type BAD but not 112A mutant BAD during IL-3 withdrawal. FL5.12 cells expressing wild-type or 112A mutant BAD were treated with 5 μM fostriecin for 2.5 h or untreated, and IL-3 was withdrawn. Lysates collected at the indicated times after IL-3 withdrawal were immunoblotted for pSer136 and total BAD.
Treatment with okadaic acid, a phosphatase inhibitor which is at least 10 times more potent against PP2A than against PP1, resulted in changes in the phosphorylation of the three sites similar to those seen with fostriecin (data not shown). To confirm that PP2A also preferentially dephosphorylated pSer112 during apoptosis, FL5.12 cells deprived of IL-3 were treated with fostriecin, and each phosphorylation site on BAD was examined by immunoblotting (Fig. 5B). Only dephosphorylation at Ser112 was inhibited by fostriecin, further indicating that PP2A or a PP2A-like enzyme is the major pSer112 phosphatase in these cells.
To further characterize the phosphatases responsible for dephosphorylation of each phosphoserine site, we examined the sensitivity of each site to phosphatase inhibitors in vitro. Dephosphorylation of each site was assayed in lysates from NIH 3T3 cells expressing wild-type BAD in the presence of a variety of phosphatase inhibitors (Fig. 5C). In the absence of phosphatase inhibitors, all three phosphoserine sites were completely dephosphorylated after 20 min (Fig. 5C, compare lane 2 to lane 1). Addition of either 1 μM microcystin or 500 nM okadaic acid, both of which inhibit PP1 and PP2A at these doses, inhibited dephosphorylation of pSer112 to levels comparable to those achieved by 1 μM fostriecin (Fig. 5C, top panel, lanes 3, 4, and 5, and Fig. 5D). Inhibition of PP2B by cyclosporin A or FK506 or of PP2C by EGTA had minimal effect on pSer112 dephosphorylation (Fig. 5C, top panel, lanes 6, 7, and 8, and Fig. 5D), consistent with PP2A's being the main phosphatase for pSer112.
In contrast, less than 50% of pSer136 dephosphorylation was inhibited by 1 μM microcystin or 500 nM okadaic acid (Fig. 5C, middle panel, lanes 3 and 4, and Fig. 5D), and none of the other inhibitors individually affected dephosphorylation of pSer136 (Fig. 5C, lanes 5 to 9, and Fig. 5D). Remarkably, a cocktail of the inhibitors of the PP1 and PP2A subfamilies, as well as Ca2+- and Mg2+-dependent serine/threonine phosphatases, prevented any dephosphorylation of pSer136 (Fig. 5C, middle panel, lane 10, and Fig. 5D), suggesting that no individual phosphatase plays a major role in the dephosphorylation of pSer136 but that multiple phosphatases of different subfamilies participate in the dephosphorylation of this site. It is also possible that the pSer136 phosphatase is a novel enzyme which is only susceptible to a cocktail of inhibitors.
Dephosphorylation of pSer155 was substantially inhibited by microcystin and fostriecin and completely inhibited by 500 nM okadaic acid (Fig. 5C, bottom panel, lanes 3, 4, and 5, and Fig. 5D), suggesting that PP2A or PP2A-like enzymes also play a major role in dephosphorylating pSer155. Inhibition of phosphatases other than PP1 or PP2A had minimal effect in preventing dephosphorylation of pSer155 (Fig. 5C, lanes 6 to 9; Fig. 5D). Taken together, these inhibitor assays are consistent with the conclusions that (i) PP2A is the main phosphatase for pSer112; (ii) multiple phosphatases, but no specific phosphatase, dephosphorylate pSer136; and (iii) PP2A family members are involved in pSer155 dephosphorylation.
To test whether PP2A dephosphorylation of pSer112 was required for pSer136 dephosphorylation, we inhibited pSer112 dephosphorylation during IL-3 withdrawal with the PP2A inhibitor fostriecin and monitored pSer136 dephosphorylation. In the absence of fostriecin, significant dephosphorylation of pSer136 occurred at 6 to 8 h of IL-3 deprivation, as expected (Fig. 5E, top left). However, when phosphorylation of pSer112 was maintained by fostriecin during IL-3 deprivation, as indicated by the presence of the upper band, no significant pSer136 dephosphorylation was detected up to 8 h (Fig. 5E, top right). In contrast, in cells expressing BAD 112A, substantial dephosphorylation of pSer136 occurred at 4 h in the absence of fostriecin, and fostriecin did not sustain the phosphorylation of pSer136 during IL-3 withdrawal (Fig. 5E, bottom). Since fostriecin had no effect on pSer136 dephosphorylation (Fig. 5A), these results demonstrated that phosphorylation of Ser112 prevented dephosphorylation of pSer136, confirming that pSer136 dephosphorylation required prior pSer112 dephosphorylation. Our data strongly suggest that after pSer112 is selectively dephosphorylated by PP2A, pSer136 becomes susceptible to multiple phosphatases.
Inhibition of phosphatase activity on specific serine sites disrupts the proapoptotic function of BAD in FL5.12 cells.
Based on our findings that dephosphorylation of pSer136 followed by dephosphorylation of pSer112, we postulated that apoptosis is initiated by pSer112 dephosphorylation, which facilitates pSer136 dephosphorylation and 14-3-3 dissociation as well as pSer155 dephosphorylation and binding to BCL-XL (9) (see Fig. 7). Immunoprecipitates of BCL-XL from FL5.12 cells contained only BAD, no 14-3-3, confirming that each BAD molecule binds either BCL-XL or 14-3-3 but not both at the same time (Fig. 6E). Phosphatases would promote apoptosis in FL5.12 cells by causing BAD to dissociate from 14-3-3 and bind to BCL-XL. Conversely, both increased association with 14-3-3 and inhibition of BCL-XL binding by BAD would result in protection from apoptosis.
FIG. 7.
Schematic model of regulation of the proapoptotic activity of BAD by 14-3-3, PP2A, and multiple phosphatases. Phosphorylation of BAD at Ser136 recruits the 14-3-3 dimer to BAD. Phosphorylation of Ser112 stabilizes 14-3-3 binding, which protects BAD dephosphorylation by PP2A and other serine/threonine phosphatases. Although pSer155 in the BH3 domain does not bind 14-3-3 directly, a presumed conformation change or spatial constraint induced by 14-3-3 binding blocks pSer155 from phosphatases. Survival factor withdrawal promotes destabilization of the BAD/14-3-3 complex. PP2A initiates dephosphorylation of BAD at the low-affinity 14-3-3-binding site, pSer112. This leads to further destabilization of 14-3-3 binding, allowing multiple phosphatases to act on the high-affinity 14-3-3 binding site, pSer136, and exposure of pSer155 to dephosphorylation. Dephosphorylation of pSer136, resulting in 14-3-3 dissociation, and dephosphorylation of pSer155, enabling binding of BAD to BCL-XL via BH3, together lead to cell death.
FIG. 6.
Calyculin A increases 14-3-3 association and enhances survival of cells expressing wild-type BAD and 112A BAD with intact Ser155. (A to D) Cells expressing wild-type BAD (A), phosphorylation-defective BAD mutant 112A (B), 112A155A BAD (C), or 112A136A155A BAD (D) were treated with 5 nM calyculin A or dimethyl sulfoxide (DMSO) for 3 h and deprived of IL-3. BAD was immunoprecipitated at the indicated times after IL-3 withdrawal and immunoblotted for 14-3-3. Graphs show percent viability as measured by trypan blue exclusion in the absence (−) or presence (+) of calyculin A at 18 h after IL-3 withdrawal. Standard deviations are derived from three experiments. (E) 14-3-3 is absent from BAD/BCL-XL complexes. BCL-XL was immunoprecipitated from FL5.12 cells expressing BCL-XL or BCL-XL and BAD with monoclonal antibody 7B2.5 and immunoblotted for BCL-XL (H-5), BAD (C-20), and 14-3-3 (K19). One-tenth of the supernatant after the immunoprecipitations was loaded as a control. (F) Calyculin A inhibits dephosphorylation of pSer112, pSer136, and pSer155. Lysates of NIH 3T3 cells expressing wild-type BAD were subjected to dephosphorylation in vitro in the absence and presence of calyculin A and immunoblotted for pSer112, pSer136, pSer155, and BAD, as described earlier. (G) FL5.12 cells expressing wild-type BAD were untreated or treated with 5 μM fostriecin for 3 h and deprived of IL-3. Graphs show percent viability as measured by trypan blue exclusion at 12 h after IL-3 withdrawal, normalized to viability in the presence of IL-3. Data are means ± standard deviations of triplicate assays and are representative of two experiments. The viability difference between assays with IL-3 and without IL-3 in the presence and absence of fostriecin has a P value of <0.001 by analysis of variance.
We previously reported that calyculin A, which inhibits both PP1 and PP2A, rescued FL5.12 cells expressing wild-type BAD during survival factor deprivation (6). Calyculin A inhibited dephosphorylation of pSer112, pSer136, and pSer155 (Fig. 6F). To address the role of phosphatase activities at individual phosphoserine residues of BAD in regulating cell survival, we measured the viability of calyculin A-treated FL5.12 cells expressing wild-type BAD or phosphorylation-defective BAD mutants during IL-3 withdrawal. The effect of calyculin A on the interaction of 14-3-3 with BAD and the BAD mutants was assayed by coimmunoprecipitation of 14-3-3 with BAD.
In wild-type and 112A cells, survival enhancement correlated with significant increases in 14-3-3 association (Fig. 6A and B), most likely due to moderately enhanced phosphorylation of Ser136 and Ser155. Calyculin A treatment did not enhance survival of cells expressing the 112A155A mutant (Fig. 6C), because mutation at Ser155 abolished the ability of BAD to dissociate from BCL-XL by Ser155 phosphorylation. Phosphatase inhibition had no survival effect on cells expressing the triple mutant because neither 14-3-3 association nor BCL-XL dissociation could occur. The results of these experiments with calyculin A are consistent with the predictions that (i) 14-3-3 dissociation, resulting from sequential dephosphorylation of pSer112 and pSer136, and (ii) binding to BCL-XL, resulting from dephosphorylation of pSer155, are both necessary for BAD-mediated apoptosis (Fig. 7).
To assess the physiologic relevance of PP2A dephosphorylation of pSer112, we treated FL5.12 cells with the PP2A-selective inhibitor fostriecin. Viability of cells during IL-3 withdrawal was increased from 60% in untreated cells to 80% in cells treated with fostriecin (Fig. 6G). Inhibition of apoptosis by fostriecin is consistent with PP2A's playing a proapoptotic role in cytokine-dependent FL5.12 cells.
DISCUSSION
We have identified a mechanism by which death stimulation triggers sequential dephosphorylation of BAD phosphoserine sites, which is regulated by competitive binding of PP2A and 14-3-3 to BAD. We have previously shown that 14-3-3 protected BAD from dephosphorylation in FL5.12 cells, particularly at pSer112 (6). Now we report that pSer136 is the major recruitment site for 14-3-3, which protects not only pSer112 but also pSer136 and pSer155 from phosphatases. Lack of 14-3-3 association in the 136A mutant allowed complete dephosphorylation of pSer112 without addition of R18 peptide. The low level of Ser155 phosphorylation in the 136A mutant was also consistent with the previous report by Datta et al. that phosphorylation of Ser155 is dependent on phosphorylation of Ser136 (9).
How does 14-3-3 protect these three sites with only one major binding site? We demonstrated that the phosphorylated Ser112 domain itself cannot recruit 14-3-3 but plays a cooperative role in stabilizing the 14-3-3/BAD complex. Although the pSer112 domain of BAD (RHSpSYP) is an imperfect 14-3-3 binding motif, we found that the phosphorylation-defective 112A mutation rendered 14-3-3 less effective at protecting pSer136 and pSer155 from phosphatases (Fig. 2). Moreover, phosphorylation mutation at Ser112 enhances the proapoptotic activity of BAD, and phosphorylation of Ser112 promotes cell survival (4, 5, 34, 44).
Our experiments with full-length protein expressed in cells substantiate the proposition by Yaffe et al., based on in vitro peptide studies, that one consensus 14-3-3 binding site in some ligands may serve as a gatekeeper whose phosphorylation is necessary for 14-3-3 binding but may not be sufficient for full biological activity (41). Our results also explain findings by Haian Fu's group that the pSer112 peptide was unable to bind 14-3-3 in vitro (25). Dephosphorylation of p155 is prevented by 14-3-3, but the pSer155 domain does not contain a consensus 14-3-3 binding motif (26), nor does phosphorylation mutation at this site affect 14-3-3 binding at pSer112 and pSer136. We believe that 14-3-3 binding induces a conformational change in BAD or causes steric hindrance so that pSer155 becomes inaccessible to phosphatases. Since pSer155 must be dephosphorylated for BAD to bind BCL-XL, BAD which is bound to 14-3-3 cannot be dephosphorylated at pSer155 and therefore cannot bind BCL-XL. This is consistent with our assessment that BAD binds to either 14-3-3 or BCL-XL but not both simultaneously (Fig. 6).
Previously, we reported that an activity of the PP2A subfamily was involved in BAD dephosphorylation. Here we show that the PP2A core dimer and BAD interact in a complex both in cells and in vitro, unequivocally identifying PP2A as an enzyme dephosphorylating BAD during IL-3 withdrawal. The presence of PP2A in the BAD complex was significantly increased when 14-3-3 was displaced, illustrating the competition between PP2A and 14-3-3 for BAD binding. This suggests that in survival, the affinity of 14-3-3 for BAD phosphorylated at Ser112 and Ser136 is high and PP2A cannot successfully compete for BAD binding. During cell death induction, we presume that a change in affinity between 14-3-3 and BAD allows PP2A to bind BAD to initiate dephosphorylation. The substrate specificity of PP2A holoenzymes is usually determined by a regulatory subunit of the B, B′, or B" subfamily. We detected both the C catalytic and the A structural subunit of the PP2A core dimer in BAD complexes, but the regulatory subunit of the PP2A for BAD is unknown.
We found that in both FL5.12 lymphoid cells and NIH 3T3 fibroblasts, PP2A preferentially dephosphorylated pSer112. In addition to PP2A, BAD phosphatase has been identified by others as PP1 and PP2B in other cell types (3, 30, 40). We considered that different phosphatases may target different phosphoserine sites in BAD. Our inhibitor studies in cells and in vitro demonstrate that PP2A or a PP2A-like phosphatase plays a role in pSer155 dephosphorylation but that PP1 may also be involved (Fig. 5). Interestingly, the inhibitor data strongly suggest that while Ser136 is key in mediating apoptosis, pSer136 is not dephosphorylated by a specific known phosphatase; rather, it is vulnerable to many serine/threonine phosphatases or a novel phosphatase. Our data that multiple phosphatases dephosphorylate BAD and that different phosphatases target different phosphoserine residues provide a possible explanation for why multiple phosphatases have been reported to be BAD phosphatases.
The observation that, in the 112A mutant, displacement of 14-3-3 was no longer necessary for pSer136 dephosphorylation led us to postulate that pSer112 dephosphorylation regulated subsequent pSer136 dephosphorylation. In our sequential dephosphorylation model (Fig. 7), pSer112 dephosphorylation by PP2A is an early regulatory step in activation of the proapoptotic function of BAD. Loss of the phosphate moiety on Ser112 results in decreased stability between 14-3-3 and BAD, because 14-3-3 now binds BAD via one phosphorylation site only. This allows access to pSer136 by multiple phosphatases.
This model is supported by four different experimental results. First, if dephosphorylation of pSer112 weakens the ability of 14-3-3 to protect pSer136, then pSer136 dephosphorylation should occur earlier in the 112A mutant. We found that pSer136 dephosphorylation in the 112A mutant occurred 2 h earlier than in wild-type cells (Fig. 3B). Second, if dephosphorylation of pSer112 reduces the stability of the 14-3-3/BAD complex, then the BAD complex after pSer112 but before pSer136 dephosphorylation should be susceptible to phosphatases in the absence of 14-3-3 displacement reagents. Indeed, at 4 h after IL-3 withdrawal, endogenous phosphatases dephosphorylated pSer136 (Fig. 3C and D). Third, if pSer136 dephosphorylation depends on pSer112 dephosphorylation, then pSer136 should not be dephosphorylated if pSer112 dephosphorylation is prevented. As expected, inhibition of PP2A dephosphorylation of pSer112 by fostriecin resulted in a marked delay of pSer136 dephosphorylation during IL-3 withdrawal from FL5.12 cells (Fig. 5E). This correlated with a survival advantage in cells treated with fostriecin (Fig. 6G). Fourth, PP2A interaction with BAD should increase upon induction of cell death. Both PP2A catalytic and structural subunits were found in higher amounts in the BAD complex at 30 min after IL-3 withdrawal, which was the earliest time we assayed (Fig. 4E). Thus, the model that PP2A activity at pSer112 is required for dephosphorylation of pSer136 was experimentally validated by these criteria.
Our model, based on expression of native and phosphorylation mutants of BAD, is well supported and explains most published data on BAD, although the possibility exists that endogenous BAD may be regulated differently. The sequential dephosphorylation model presented above predicts that constitutive phosphorylation of Ser112 would prevent dephosphorylation of pSer136. However, this could not be tested directly because a mutant carrying a glutamic acid or aspartate substitution at Ser112 either was unstable or behaved like the alanine mutant (data not shown), and these did not function as phospho-mimetic mutations in this case.
Based on cooperativity between pSer112 and pSer136 in 14-3-3 binding, sequential dephosphorylation, and the finding that PP2A preferentially dephosphorylate pSer112 while multiple phosphatases act on pSer136, we propose the following scenario for activation of BAD's proapoptotic activity (Fig. 7). In the presence of IL-3, phosphorylation at Ser112 cooperates with that at Ser136 to capture 14-3-3 in a stable complex with BAD, preventing PP2A as well as other phosphatases from dephosphorylating BAD. Upon IL-3 deprivation, a change in the 14-3-3/BAD interaction allows PP2A to successfully compete for binding to BAD and dephosphorylate pSer112. Once pSer112 is dephosphorylated, 14-3-3 is further destabilized, and pSer136 becomes susceptible to many phosphatases, or a novel phosphatase and is rapidly dephosphorylated. Destabilization of 14-3-3/BAD also exposes pSer155 to PP2A or PP2A-like phosphatases. BAD which is dephosphorylated at pSer155 binds BCL-XL, causing cell death. This is supported by viability data and 14-3-3 binding data from cells expressing wild-type and phosphorylation-defective BAD in FL5.12 cells, in which PP2A and PP1 have been inhibited (Fig. 6).
Rescue from apoptosis could only occur if enhancement of 14-3-3 association and inhibition of BCL-XL binding were both possible. Since the binding affinity between BAD and BCL-XL is much higher than that between BAD and 14-3-3 (9), any mutant containing 155A would remain tightly bound to BCL-XL, and survival would not be enhanced by calyculin A. We found that phosphorylation of Ser155 was variably increased by phosphatase inhibitors. This implies that, in vivo, Ser155 can be phosphorylated in the absence of Ser136 phosphorylation, but phosphorylation of Ser155 would be unprotected by 14-3-3 and it would be constantly dephosphorylated in the absence of phosphatase inhibitors. This is consistent with the observation of Michael Greenberg's group that pSer155 phosphorylation is dependent on Ser136 phosphorylation (9). Whereas their model is that 14-3-3 facilitates Ser155 phosphorylation, our data indicate that 14-3-3 prevents dephosphorylation of pSer155. Greenberg's group proposed a model for BAD inactivation by sequential phosphorylation starting with BAD bound to BCL-XL. Our model describes activation of BAD by sequential dephosphorylation starting with phosphorylated BAD already bound to 14-3-3. Thus, the models are complementary.
Our data indicate that 14-3-3 and PP2A compete for BAD binding. In survival conditions, 14-3-3 excludes PP2A from binding to BAD, but in apoptosis, PP2A successfully competes with 14-3-3 for BAD binding. It is not clear what changes after IL-3 withdrawal to allow binding of PP2A to BAD. We found no increase in BAD phosphatase activity in lysates prepared from surviving or apoptotic FL5.12 cells (data not shown), suggesting that the PP2A activity against BAD is always present but that BAD is not always susceptible. Therefore, we hypothesize that upon apoptosis induction, a conformation change occurs in the 14-3-3/BAD complex which favors PP2A binding to BAD.
14-3-3 is known to be cleaved by caspases, but this occurs late in FL5.12 cell apoptosis, and is not likely involved in the early destabilization of the 14-3-3/BAD complex. In addition, we observed a smaller 14-3-3-immunoreactive band, which could be a cleavage product, coprecipitating with BAD at 4 and 8 h in some of the IL-3-deprived cells (Fig. 6C). Phosphorylation of 14-3-3 is a possibility, as phosphorylation of 14-3-3ζ negatively regulates c-Raf (13, 36). Modification of BAD can also enable the pSer112 site to become an accessible substrate for PP2A. For example, BAD phosphorylation at Ser128 by Cdc2 disrupts interaction with 14-3-3 in cerebellar granular neurons induced to die (19). Recently, Bax was shown to bind 14-3-3, and a change in cytosolic pH induced by apoptosis may contribute to the conformation change in Bax that results in dissociation from 14-3-3 and relocation to the mitochondria (27). It is possible that a similar mechanism might apply to BAD.
In summary, we have shown that PP2A dephosphorylation of pSer112 functions as the gatekeeper in the activation of BAD's proapoptotic function. Our data confirm that phosphorylation of Ser136 is essential in inactivating BAD and that dephosphorylation of Ser136 is key in mediating apoptosis. Paradoxically, we discovered that specific regulation of BAD dephosphorylation occurs at pSer112, not pSer136, and once the “gate” at Ser112 is open, pSer136 is vulnerable to multiple phosphatases. The model of BAD dephosphorylation presented here encompasses many existing observations of BAD phosphorylation and expands the mechanism of regulation of BAD by reversible phosphorylation.
Acknowledgments
We are grateful to Brian Wadzinski for constant advice and support in this project. We thank Roger Colbran, David Hockenbery, and Brian Wadzinski for critical reading of the manuscript.
This work was supported by NIH grant RO1 CA92498 (E.Y.) and a Grant-in-Aid (E.Y.) and a postdoctoral fellowship (C.W.C.) from the American Heart Association.
REFERENCES
- 1.Adams, J. M., and S. Cory. 2001. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem. Sci. 26:61-66. [DOI] [PubMed] [Google Scholar]
- 2.Aitken, A., H. Baxter, T. Dubois, S. Clokie, S. Mackie, K. Mitchell, A. Peden, and E. Zemlickova. 2002. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem. Soc. Trans. 30:351-360. [DOI] [PubMed] [Google Scholar]
- 3.Ayllon, V., A. C. Martinez, A. Garcia, X. Cayla, and A. Rebollo. 2000. Protein phosphatase 1alpha is a Ras-activated Bad phosphatase that regulates interleukin-2 deprivation-induced apoptosis. EMBO J. 19:2237-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertolotto, C., L. Maulon, N. Filippa, G. Baier, and P. Auberger. 2000. Protein kinase C theta and epsilon promote T-cell survival by a Rsk-dependent phosphorylation and inactivation of BAD. J. Biol. Chem. 275:37246-37250. [DOI] [PubMed] [Google Scholar]
- 5.Bonni, A., A. Brunet, A. E. West, S. R. Datta, M. A. Takasu, and M. E. Greenberg. 1999. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286:1358-1362. [DOI] [PubMed] [Google Scholar]
- 6.Chiang, C. W., G. Harris, C. Ellig, S. C. Masters, R. Subramanian, S. Shenolikar, B. E. Wadzinski, and E. Yang. 2001. Protein phosphatase 2A activates the proapoptotic function of BAD in interleukin-3-dependent lymphoid cells by a mechanism requiring 14-3-3 dissociation. Blood 97:1289-1297. [DOI] [PubMed] [Google Scholar]
- 7.Chuang, E., T. S. Fisher, R. W. Morgan, M. D. Robbins, J. M. Duerr, M. G. Vander Heiden, J. P. Gardner, J. E. Hambor, M. J. Neveu, and C. B. Thompson. 2000. The CD28 and CTLA-4 receptors associate with the serine/threonine phosphatase PP2A. Immunity 13:313-322. [DOI] [PubMed] [Google Scholar]
- 8.Datta, S. R., H. Dudek, X. Tao, S. Masters, H. Fu, Y. Gotoh, and M. E. Greenberg. 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91:231-241. [DOI] [PubMed] [Google Scholar]
- 9.Datta, S. R., A. Katsov, L. Hu, A. Petros, S. W. Fesik, M. B. Yaffe, and M. E. Greenberg. 2000. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell 6:41-51. [PubMed] [Google Scholar]
- 10.Datta, S. R., A. M. Ranger, M. Z. Lin, J. F. Sturgill, Y. C. Ma, C. W. Cowan, P. Dikkes, S. J. Korsmeyer, and M. E. Greenberg. 2002. Survival factor-mediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Dev. Cell 3:631-643. [DOI] [PubMed] [Google Scholar]
- 11.Desagher, S., A. Osen-Sand, S. Montessuit, E. Magnenat, F. Vilbois, A. Hochmann, L. Journot, B. Antonsson, and J. C. Martinou. 2001. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell 8:601-611. [DOI] [PubMed] [Google Scholar]
- 12.Dramsi, S., M. P. Scheid, A. Maiti, P. Hojabrpour, X. Chen, K. Schubert, D. R. Goodlett, R. Aebersold, and V. Duronio. 2002. Identification of a novel phosphorylation site, Ser-170, as a regulator of bad proapoptotic activity. J. Biol. Chem. 277:6399-6405. [DOI] [PubMed] [Google Scholar]
- 13.Dubois, T., C. Rommel, S. Howell, U. Steinhussen, Y. Soneji, N. Morrice, K. Moelling, and A. Aitken. 1997. 14-3-3 is phosphorylated by casein kinase I on residue 233. Phosphorylation at this site in vivo regulates Raf/14-3-3 interaction. J. Biol. Chem. 272:28882-28888. [DOI] [PubMed] [Google Scholar]
- 14.Harada, H., J. S. Andersen, M. Mann, N. Terada, and S. J. Korsmeyer. 2001. p70S6 kinase signals cell survival as well as growth, inactivating the proapoptotic molecule BAD. Proc. Natl. Acad. Sci. USA 98:9666-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada, H., B. Becknell, M. Wilm, M. Mann, L. J. Huang, S. S. Taylor, J. D. Scott, and S. J. Korsmeyer. 1999. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell 3:413-422. [DOI] [PubMed] [Google Scholar]
- 16.Hastie, C. J., and P. T. Cohen. 1998. Purification of protein phosphatase 4 catalytic subunit: inhibition by the antitumour drug fostriecin and other tumour suppressors and promoters. FEBS Lett. 431:357-361. [DOI] [PubMed] [Google Scholar]
- 17.Huang, D. C., and A. Strasser. 2000. BH3-Only proteins-essential initiators of apoptotic cell death. Cell 103:839-842. [DOI] [PubMed] [Google Scholar]
- 18.Janssens, V., and J. Goris. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353:417-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konishi, Y., M. Lehtinen, N. Donovan, and A. Bonni. 2002. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol. Cell 9:1005-1016. [DOI] [PubMed] [Google Scholar]
- 20.Lizcano, J. M., N. Morrice, and P. Cohen. 2000. Regulation of BAD by cAMP-dependent protein kinase is mediated via phosphorylation of a novel site, Ser155. Biochem. J. 349:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Lutz, R. J. 2000. Role of the BH3 (Bcl-2 homology 3) domain in the regulation of apoptosis and Bcl-2-related proteins. Biochem. Soc. Trans. 28:51-56. [DOI] [PubMed] [Google Scholar]
- 22.Marsden, V. S., and A. Strasser. 2003. Control of apoptosis in the immune system: Bcl-2, BH3-only proteins and more. Annu. Rev. Immunol. 21:71-105. [DOI] [PubMed] [Google Scholar]
- 23.Maslyar, D. J., M. Aoki, and P. K. Vogt. 2001. The growth-promoting activity of the Bad protein in chicken embryo fibroblasts requires binding to protein 14-3-3. Oncogene 20:5087-5092. [DOI] [PubMed] [Google Scholar]
- 24.Masters, S. C., R. R. Subramanian, A. Truong, H. Yang, K. Fujii, H. Zhang, and H. Fu. 2002. Survival-promoting functions of 14-3-3 proteins. Biochem. Soc. Trans. 30:360-365. [DOI] [PubMed] [Google Scholar]
- 25.Masters, S. C., H. Yang, S. R. Datta, M. E. Greenberg, and H. Fu. 2001. 14-3-3 inhibits Bad-induced cell death through interaction with serine-136. Mol. Pharmacol. 60:1325-1331. [DOI] [PubMed] [Google Scholar]
- 26.Muslin, A. J., J. W. Tanner, P. M. Allen, and A. S. Shaw. 1996. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84:889-897. [DOI] [PubMed] [Google Scholar]
- 27.Nomura, M., S. Shimizu, T. Sugiyama, M. Narita, T. Ito, H. Matsuda, and Y. Tsujimoto. 2003. 14-3-3 interacts directly with and negatively regulates proapoptotic Bax. J. Biol. Chem. 278:2058-2065. [DOI] [PubMed] [Google Scholar]
- 28.Ruvolo, P. P., X. Deng, T. Ito, B. K. Carr, and W. S. May. 1999. Ceramide induces Bcl2 dephosphorylation via a mechanism involving mitochondrial PP2A. J. Biol. Chem. 274:20296-20300. [DOI] [PubMed] [Google Scholar]
- 29.Ruvolo, P. P., X. Deng, and W. S. May. 2001. Phosphorylation of Bcl2 and regulation of apoptosis. Leukemia 15:515-522. [DOI] [PubMed] [Google Scholar]
- 30.Salomoni, P., F. Condorelli, S. M. Sweeney, and B. Calabretta. 2000. Versatility of BCR/ABL-expressing leukemic cells in circumventing proapoptotic BAD effects. Blood 96:676-684. [PubMed] [Google Scholar]
- 31.Schurmann, A., A. F. Mooney, L. C. Sanders, M. A. Sells, H. G. Wang, J. C. Reed, and G. M. Bokoch. 2000. p21-activated kinase 1 phosphorylates the death agonist Bad and protects cells from apoptosis. Mol. Cell. Biol. 20:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimamura, A., B. A. Ballif, S. A. Richards, and J. Blenis. 2000. Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr. Biol. 10:127-135. [DOI] [PubMed] [Google Scholar]
- 33.Tan, Y., M. R. Demeter, H. Ruan, and M. J. Comb. 2000. BAD Ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell survival. J. Biol. Chem. 275:25865-25869. [DOI] [PubMed] [Google Scholar]
- 34.Tan, Y., H. Ruan, M. R. Demeter, and M. J. Comb. 1999. p90(RSK) blocks Bad-mediated cell death via a protein kinase C-dependent pathway. J. Biol. Chem. 274:34859-34867. [DOI] [PubMed] [Google Scholar]
- 35.Tzivion, G., and J. Avruch. 2002. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277:3061-3064. [DOI] [PubMed] [Google Scholar]
- 36.Van Der Hoeven, P. C., J. C. Van Der Wal, P. Ruurs, M. C. Van Dijk, and J. Van Blitterswijk. 2000. 14-3-3 isotypes facilitate coupling of protein kinase C-zeta to Raf-1: negative regulation by 14-3-3 phosphorylation. Biochem. J. 345:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verma, S., L. J. Zhao, and G. Chinnadurai. 2001. Phosphorylation of the proapoptotic protein BIK: mapping of phosphorylation sites and effect on apoptosis. J. Biol. Chem. 276:4671-4676. [DOI] [PubMed] [Google Scholar]
- 38.Walsh, A. H., A. Cheng, and R. E. Honkanen. 1997. Fostriecin, an antitumor antibiotic with inhibitory activity against serine/threonine protein phosphatases types 1 (PP1) and 2A (PP2A), is highly selective for PP2A. FEBS Lett. 416:230-234. [DOI] [PubMed] [Google Scholar]
- 39.Wang, B., H. Yang, Y. C. Liu, T. Jelinek, L. Zhang, E. Ruoslahti, and H. Fu. 1999. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38:12499-12504. [DOI] [PubMed] [Google Scholar]
- 40.Wang, H. G., N. Pathan, I. M. Ethell, S. Krajewski, Y. Yamaguchi, F. Shibasaki, F. McKeon, T. Bobo, T. F. Franke, and J. C. Reed. 1999. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science 284:339-343. [DOI] [PubMed] [Google Scholar]
- 41.Yaffe, M. B. 2002. How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513:53-57. [DOI] [PubMed] [Google Scholar]
- 42.Yaffe, M. B., K. Rittinger, S. Volinia, P. R. Caron, A. Aitken, H. Leffers, S. J. Gamblin, S. J. Smerdon, and L. C. Cantley. 1997. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91:961-971. [DOI] [PubMed] [Google Scholar]
- 43.Yang, E., J. Zha, J. Jockel, L. H. Boise, C. B. Thompson, and S. J. Korsmeyer. 1995. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell 80:285-291. [DOI] [PubMed] [Google Scholar]
- 44.Zha, J., H. Harada, E. Yang, J. Jockel, and S. J. Korsmeyer. 1996. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell 87:619-628. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, X. M., Y. Liu, G. Payne, R. J. Lutz, and T. Chittenden. 2000. Growth factors inactivate the cell death promoter BAD by phosphorylation of its BH3 domain on Ser155. J. Biol. Chem. 275:25046-25051. [DOI] [PubMed] [Google Scholar]