Summary
Background
Prior to chromosome segregation, the mitotic spindle bi-orients and aligns sister chromatids along the metaphase plate. During metaphase, spindle length remains constant, suggesting that spindle forces (inward and outward) are balanced. The contribution of microtubule motors, regulators of microtubule dynamics, and cohesin to spindle stability has been previously studied. In this study, we examine the contribution of chromatin structure on kinetochore positioning and spindle length control. Following nucleosome depletion, by either histone H3 or H4 repression, spindle organization was examined using live cell fluorescence microscopy.
Results
Histone repression led to a 2-fold increase in sister centromere separation and an equal increase in metaphase spindle length. Histone H3 repression does not impair kinetochores, while H4 repression disrupts proper kinetochore function. Deletion of outward force generators, kinesins Cin8p and Kip1p, shortens the long spindles observed in histone-repressed cells. Oscillatory movements of individual sister chromatid pairs are not altered following histone repression.
Conclusions
The increase in spindle length upon histone repression and restoration of wild-type spindle length by the loss of plus-end directed motors suggests that during metaphase, centromere separation and spindle length are governed in part by the stretching of pericentric chromatin. Chromatin is an elastic molecule that is stretched in direct opposition to the outward force generators Cin8p and Kip1p. Thus we assign a new role to chromatin packaging as an integral biophysical component of the mitotic apparatus.
Introduction
The mitotic spindle apparatus functions to segregate the replicated genome during cell division [1]. Accurate chromosome segregation is ensured by the monitoring of sister chromatid bi-orientation prior to anaphase. Once bi-oriented, sister chromatids align along the metaphase plate and are held under tension at their kinetochores, a cue that satisfies the spindle checkpoint and allows mitosis to proceed [2]. In Saccharomyces cerevisiae, metaphase alignment of chromosomes results in the formation of two distinct kinetochore clusters [3, 4]. These clusters represent the average position of sister centromeres separated by kinetochore microtubule dependent forces. It has been proposed that tension-dependent rescue and a microtubule catastrophe gradient determine kinetochore microtubule length, and kinetochore clustering has been attributed to Cin8p function [5, 6]. These results highlight the importance of microtubule dynamics regulation and microtubule motors in defining kinetochore position within the spindle.
In order for the spindle to hold sister chromatids under tension, the spindle must form a stable structure. Indeed, during metaphase in most organisms, the spindle maintains a stable spindle length despite the dynamics of individual microtubules and chromosomes [7–9]. Interpolar microtubules from opposing spindle pole bodies form an organized array that may be cross-linked by microtubule motor proteins, and/or other microtubule associated proteins [10–12]. This arrangement contributes to the stability of the two halves of the mitotic spindle during metaphase and provides the means by which spindle pole bodies are rapidly separated from each other during anaphase B.
Metaphase spindle stability suggests that once formed, spindles are under roughly equal and opposing forces. Deletions of either CIN8 or KIP1 lead to abnormally short metaphase spindles, suggesting that these plus-end directed motors generate outward spindle force (via sliding interpolar microtubules against each other) [13]. Cells lacking both Cin8p and Kip1p are inviable, but deletion of the minus-end directed motor KAR3 suppresses this lethality, suggesting that Kar3p provides an inward force that opposes and balances the outward force generated by Cin8p and Kip1p [13]. Inward spindle force has also been attributed to the cohesin complexes that link sister chromatids prior to anaphase onset [14]. Neither of these hypotheses is consistent with recent data. Spindles in kar3 mutants alone are short, a result in contrast to the prevailing model [15–17]. Likewise, loss of cohesin does not result in complete separation of sister chromatids [18].
An alternative model, based upon the physical properties of chromatin, is that chromosomes themselves behave as mechanical springs that resist outward spindle forces (Figure 1A). As pericentric chromatin is stretched, its resistive force increases until it is balanced with the pulling forces of the spindle. This force balance defines the separation of bi-oriented sister chromatid centromeres along the spindle, and spindle length. This model predicts that changes in chromatin structure would result in changes in kinetochore separation and spindle length.
Figure 1. Increased centromere separation following histone repression.
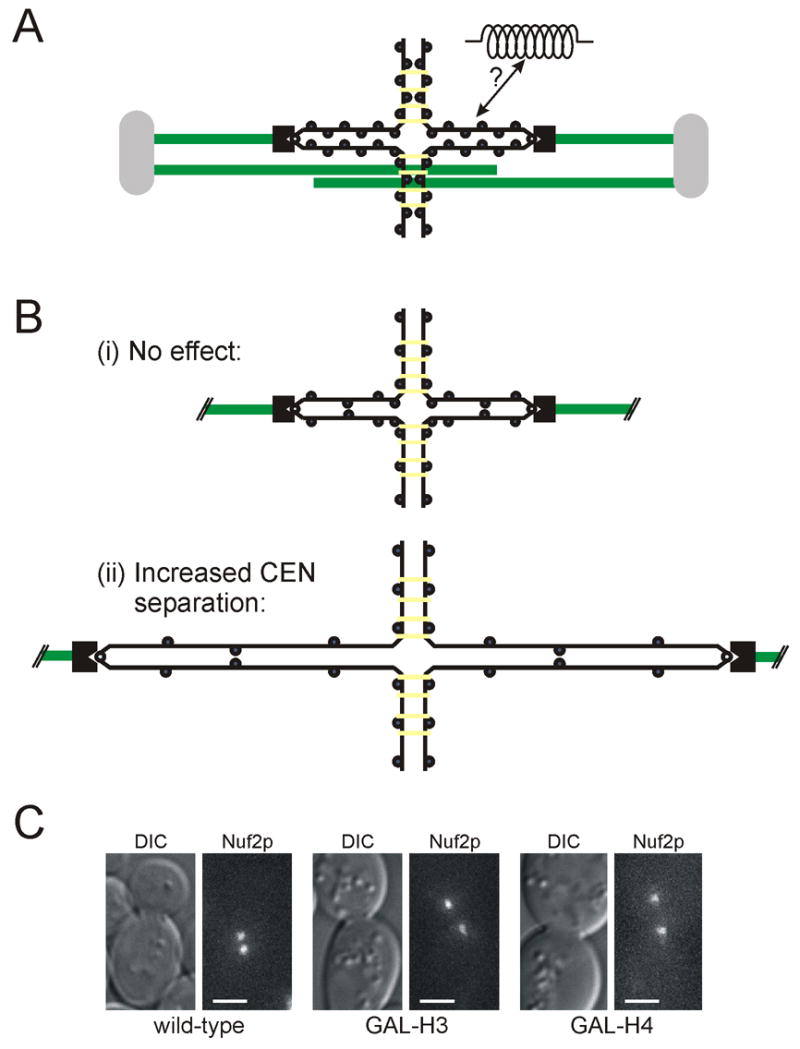
(A) Schematic model of the budding yeast spindle showing the separation of sister centromeres during metaphase. Sister chromatid arms are held together by cohesin complexes (yellow); centromeres, bound to kinetochores (black), are pulled apart by kinetochore microtubules. (B) Predicted outcomes for centromere separation following the lowering of nucleosome concentration: (i) no change indicates that chromatin structure does not affect centromere separation, or (ii) increased centromere separation indicates role of chromatin in determining centromere separation. (C) Nuf2p-GFP kinetochore clusters in wild-type, GAL-H3, and GAL-H4 cells following 3 h growth in repressive media (YPD). Histone repression results in ∼2-fold increase in sister centromere separation. (Scale bar, 2 μm)
To test this model, we have lowered chromatin packaging through the repression of histone proteins H3 and H4. Repression of histone H4 results in the decrease of nucleosome concentration within chromatin by approximately 2-fold [19]. The effect of histone H3 repression was also examined because centromeric nucleosomes contain a histone H3 variant, Cse4p (CENP-A) [20]. Repression of H3 would therefore affect nucleosomes throughout the genome except at the centromeric nucleosome where it is replaced by Cse4p, while H4 repression would affect nucleosomes at all loci, including the centromere.
Our results demonstrate that kinetochore cluster separation and spindle length are both dependent on proper chromatin packaging. Histone repression leads to both increased kinetochore separation and increased spindle length. By combining histone repression with CIN8 and KIP1 deletions, we find that chromatin stretching is proportional to applied force. Finally, we show that nucleosome depletion results primarily in a change in chromatin’s rest length. Together, these results are consistent with the model that chromatin plays a significant role as a structural element within the mitotic spindle by opposing outward, microtubule-based spindle forces.
Results
Pericentric chromatin constrains separated sister centromeres in metaphase
To test the hypothesis that pericentric chromatin restrains centromere separation, we lowered chromatin packaging through the repression of histone H3 or H4 and measured kinetochore cluster separation in the mitotic spindle. Two possible outcomes were predicted to follow histone repression (Figure 1B). If chromatin stretching does not resist the pulling forces from microtubules, then there would be no change in centromere separation following histone repression. Alternatively, if pericentric chromatin stretching is important in resisting pulling forces generated by depolymerizing kinetochore microtubules, then sister centromeres should be pulled further apart following histone repression.
To distinguish between these two outcomes, we examined kinetochore cluster separation in cells expressing Nuf2p-GFP (a kinetochore component) in which histone H3 or H4 levels could be controlled. One copy of the histone gene was deleted and the promoter of the second copy was replaced by the regulatable GAL1 promoter (see Material and Methods). Cells were grown in YPG (histone transcription on), arrested in G1 with the mating pheromone alpha factor, and then released from this arrest into YPD (histone transcription off) for 3 h. Repression of histones resulted in cell cycle arrest with large budded cells [19]. Following H3 repression, cells contained two clusters of Nuf2p-GFP, indicative of centromere separation observed in metaphase cells (Figure 1C). In wild-type cells, centromere clusters were separated by 0.84 μm (SD=0.23, n=71). The distance between centromeres increased to 1.60μm in H3-repressed cells (SD=0.34 μm, n=77). Fluorescence Recovery After Photobleaching (FRAP) analysis of bi-oriented Nuf2-GFP clusters in H3-repressed cells showed that Nuf2p-GFP remained stable, like wild-type cells (Supplementary Note 1) [21]. The stability of kinetochore attachments suggests that the increase in centromere separation is not a consequence of altered kinetochore function. Therefore, pericentric chromatin plays a physical role in determining the extent to which sister centromeres are stretched apart due to microtubule dependent pulling forces.
Unlike histone H3, histone H4 repression resulted in Nuf2p-GFP de-clustering in 55% of cells (Supplementary Figure 1). This defect is consistent with impaired kinetochore formation or function upon loss of the centromeric nucleosome (Supplementary Note 1). In H4-repressed cells with only two kinetochore clusters, Nuf2p-GFP foci were separated by 1.38 μm (SD=0.24 μm, n=60). Thus, H4 repression leads to greater separation of sister kinetochores in the fraction of cells with clustered kinetochores.
Chromatin structure regulates spindle length
The increased distance between sister centromeres could lead to changes in spindle structure, including shorter kinetochore microtubules, and/or increased spindle length (Figure 2A). To differentiate between these possibilities, we imaged Spc29p-CFP (a spindle pole body component) and Nuf2p-GFP to determine spindle length and the position of kinetochore clusters in the spindle upon histone repression. Consistent with previous studies, the average metaphase spindle length in wild-type cells was 1.47 μm (SD=0.28 μm, n=71) (Figure 2B). In contrast, metaphase spindles in cells with lowered H3 levels were 2.33μm (SD=0.40 μm, n=77), and cells with lowered H4 levels had even longer spindles (mean=2.69μm, SD=0.36 μm, n=60) (Figure 2B). The increase in spindle length in H3-repressed cells is equal to the increase in kinetochore separation. These results show that spindle length is directly affected by changes in chromatin structure.
Figure 2. Spindle length increases following histone repression.
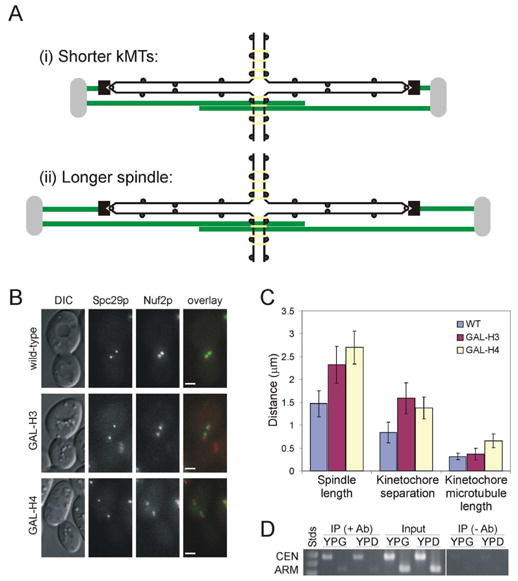
(A) Predicted effects of increased centromere separation on spindle structure: (i) kinetochore microtubules shorten (no change in spindle length), or (ii) the entire spindle length increases with no change in kinetochore microtubule length. (B) Spc29p-CFP (spindle pole bodies) and Nuf2p-GFP in wild-type, H3-repressed, and H4-repressed cells. (Scale bar, 2 μm) (C) Histone repression results in increased separation of both kinetochore clusters and spindle pole bodies. Error bars represent standard deviation. (D) ChIP of Mcd1/Scc1p-6HA in GAL-H3 cells grown in permissive (YPG) or repressive (YPD) media. Centromere and arm loci were assayed for Mcd1/Scc1p association.
Average kinetochore microtubule length (distance from spindle pole to corresponding kinetochore cluster) was nearly identical in wild-type and H3-repressed cells (0.31 vs. 0.36 μm), while the average kinetochore microtubule length in H4-repressed cells was 0.66μm. The small difference in kinetochore microtubule length observed between wild-type and H3-repressed cells indicates that the primary effect of increased centromere separation is increased spindle length rather than shorter kinetochore microtubules (Figure 2C). H4-repressed cells showed increased kinetochore cluster separation as well as longer kinetochore microtubules. In both cases, histone repression resulted in greater separation of sister kinetochore clusters, demonstrating that histone repression lowers chromatin packaging and allows for greater separation of sister centromeres in metaphase. Together, these results show that changes in chromatin structure affect spindle length and not spindle stability (Supplementary Note 1), suggesting that pericentric chromatin exerts an inward resistive force governing metaphase spindle length.
Cohesin mutants have been associated with altered spindle morphology [14]. To address the possibility that cohesin loading or function was altered by the change in chromatin structure caused by histone repression, we assayed the association and function of cohesin in H3-repressed cells. Chromatin immunoprecipation of cohesin subunits have previously shown an enrichment of cohesin near the centromere [22, 23]. We found that histone H3 repression did not affect Mcd1/Scc1p-6HA association with the centromere (Figure 2D). To determine whether cohesin function (cohesion) was changed by histone repression, we treated H3-repressed cells with nocodazole and measured the frequency of sister re-association. In cells with collapsed spindles (indicative of microtubule depolymerization), 100% of sister chromatids reassociated (n=83; data not shown). Together, these results demonstrate that cohesin is both present and functional at sister chromatids. Thus the long spindles observed after histone repression are not likely due to perturbed cohesin association or function.
Pericentric chromatin behaves as an elastic spindle component
Pericentric chromatin stretching could limit sister centromere separation as either an inelastic element constraining sister separation at a specific length, or as an elastic element that stretches proportionally to the force applied to it (Figure 3A). To distinguish between these possibilities, CIN8 and KIP1 were individually deleted from strains in which histone H3 levels were repressed. If chromatin is elastic, then centromere separation should be decreased in cin8Δ or kip1Δ cells; however, if chromatin is inelastic, centromere separation should not be affected (Figure 3A).
Figure 3. Pericentric chromatin is an elastic spindle component.
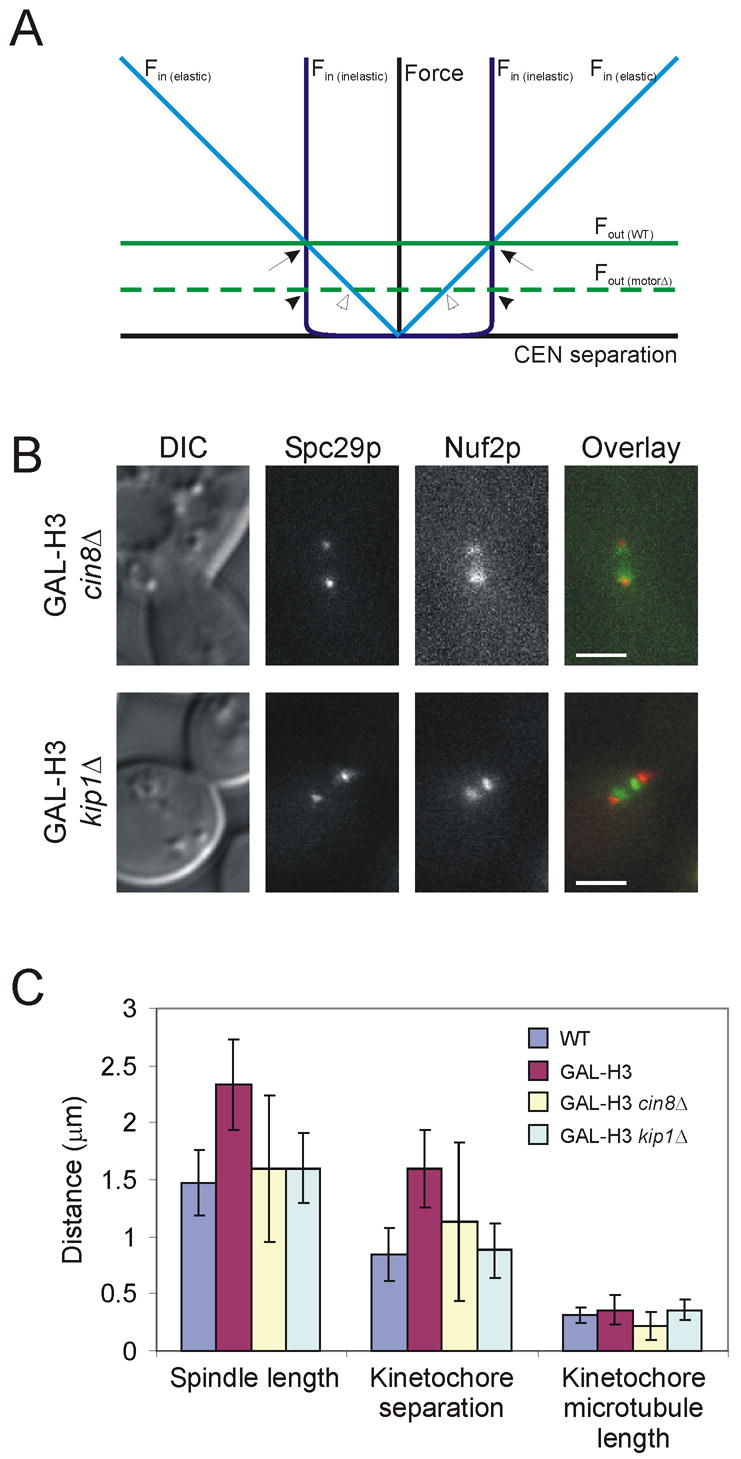
(A) Theoretical force diagram of forces acting on centromere separation. Outward forces (green lines) are assumed to be constant regardless of centromere separation distance. Deletion of CIN8 or KIP1 is predicted to lower outward forces (dashed green line). Inward force (blue lines) is assigned to chromatin. Elastic chromatin (light blue line) is modeled with increasing force as centromere separation increases. Assuming chromatin behaves as a Hookean spring, the slope of this line is the spring constant of chromatin. Inelastic chromatin (dark blue line) is modeled to contribute inward force only when approaching nearly full extension. Intersection points of outward and inward force lines predict length of sister centromere separation. Thus, if chromatin is inelastic, motor deletion would not change centromere separation (compare arrow to filled arrowhead). However, if chromatin is elastic, lowered force (by motor deletion) would result in less stretching and therefore reduced centromere separation (hollow arrowhead). (B) Spc29p-CFP and Nuf2p-GFP in H3-repressed cells with either CIN8 or KIP1 deleted. (Scale bar, 2 μm) (C) Both spindle length and kinetochore separation are decreased in cin8Δ and kip1Δ cells, demonstrating that chromatin is elastic. Error bars represent standard deviation.
Histone H3 repression alone results in longer spindles (2.33μm) and greater kinetochore separation (1.60μm; Figure 3C). Deletion of CIN8 in H3 repressed cells resulted in spindles of approximately wild-type length (1.59μm), and a reduction in kinetochore separation (1.13μm; Figure 3B,C). Likewise, deletion of KIP1 in H3 repressed cells caused spindles to return to wild-type length (1.60μm) and kinetochores were separated by approximately the same distance as wild-type cells (0.88μm; Figure 3B,C). While both motor deletions resulted in approximately the same spindle length, they did not have an equal effect on kinetochore separation (Table 1). This difference is likely due to different contributions of these two motor proteins. The decrease in kinetochore separation seen in both motor deletions supports the model that chromatin is an elastic element of the spindle that is stretched proportionally to the force applied to it. Pericentric chromatin stretching contributes to the force balance that defines both centromere positioning and spindle length in metaphase.
Table 1.
Spindle organization in wild-type and histone repressed cells.
| Strain | n | Spindle length | Kinetochore cluster separation | Kinetochore microtubule length | |||
|---|---|---|---|---|---|---|---|
| Average | Std. Dev. | Average | Std. Dev. | Average | Std. Dev. | ||
| Wild-type | 71 | 1.47a | 0.28 | 0.84a | 0.23 | 0.31a | 0.07 |
| GAL-H3 | 77 | 2.33b | 0.40 | 1.60b | 0.34 | 0.36b | 0.13 |
| GAL-H3 cin8Δ* | 41 | 1.59a | 0.64 | 1.13c | 0.70 | 0.22c | 0.12 |
| GAL-H3 kip1Δ | 75 | 1.60a | 0.31 | 0.88a | 0.24 | 0.36b | 0.09 |
| GAL-H4* | 60 | 2.69c | 0.36 | 1.38c | 0.24 | 0.66d | 0.15 |
Superscripts denote statistical significance. Data sets with identical superscripts are not significantly different (p > 0.01).
Only spindles with two kinetochore clusters were included in the analysis. Spindles with multiple (>2) Nuf2p-GFP foci or declustered Nuf2p-GFP were not considered.
Histone repression increases chromatin rest length
Individual sister chromatids can be visualized by integrating lac operator arrays in cells expressing LacI-GFP [24]. We used lacO arrays positioned 1.8 kb from CEN15 in wild-type and histone repressed cells to determine the effects of lowered nucleosome concentration on the position and movements of individual chromosomes (Figure 4A). On average, lacO foci were separated by 0.6 μm in wild-type metaphase cells, and 0.9 μm following histone H3 repression. This 50% increase in separation is consistent with the increased separation of kinetochore clusters seen following histone repression (Figure 1).
Figure 4. Single centromere dynamics following histone repression.
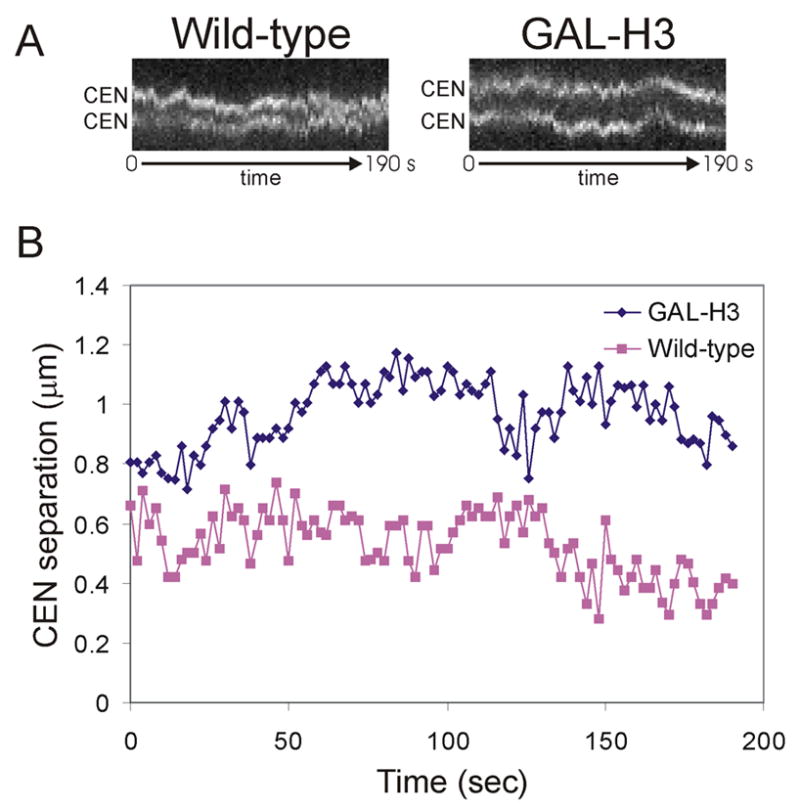
(A) Kymographs of lacO arrays positioned 1.8kb from CEN15 in wild-type and H3-repressed cells shows dyanamics of centromeres. Images were acquired in one plane every 2 sec; approximately 3 min are shown. (B) Quantitation of centromere separation and movement shows increased centromere separation following H3 repression, but similar dynamics.
Based on the model that chromatin acts as a linearly elastic spring, lowering nucleosome concentration could alter either chromatin’s rest length or spring constant [5]. For Hookean springs, the spring constant (k) relates the distance stretched (x) to the force (F) applied, as described by the equation F = −kx. If histone repression changes the spring constant (stiffness) of chromatin, we predicted that the amplitude of centromere proximal lacO array oscillations would be affected. To test this hypothesis, the distance between lacO foci was recorded every 2 seconds in wild-type metaphase cells and histone repressed cells (Figure 4B). The average distance traveled during an oscillation was calculated (see materials and methods). The average oscillation amplitude was 0.13 μm in wild-type cells, and 0.12 μm following histone H3 repression. Assuming there is no difference in the forces applied at kinetochores in these strains, the similarity in oscillation amplitude suggests that spring constant is not severely altered by histone repression. Our data suggest that the primary effect of histone repression is an increase in chromatin rest length.
Discussion
A new role for chromatin packaging: structural spindle element
To test whether chromatin’s biophysical properties would change by changing DNA-nucleosome packaging, nucleosome concentration within chromatin was lowered by allowing cells to replicate their DNA in the absence of histone gene transcription. Following histone repression, we observed an approximately 2-fold increase in the distance by which sister kinetochores were separated from each other due to the pulling forces of kinetochore microtubules. This change confirms that the mechanical properties of chromatin were altered after histone repression and that the force generated by the spindle is sufficient to stretch pericentric chromatin to greater lengths. The spindles in histone repressed cells also reached a longer steady state length in metaphase. Presumably, the spindle could have responded to the change in kinetochore cluster separation by either shortening kinetochore microtubules, or by extending the entire spindle length. The observed increase in spindle length suggests that, during metaphase, spindle length is being governed by the stretching of pericentric chromatin. Lowered chromatin packaging allowed for greater spindle extension in metaphase.
Outward spindle force has been attributed to the kinesin motor proteins Cin8p and Kip1p. While deletions of these motors lead to differences in anaphase spindle elongation rates, both have also been reported to form short spindles during metaphase [13, 25]. To validate the model that spindle length is governed by the equilibrium reached between the outward forces (generated by motors associated with overlapping interpolar microtubules) and inward forces (including the stretching of pericentric chromatin), we found that a new steady-state metaphase spindle length was achieved in cells lacking CIN8 or KIP1 in histone H3 repressed cells. In both motor deletions, spindles shortened by ∼30% to approximately wild-type length, demonstrating that chromatin stretching was reduced. Thus, chromatin is an elastic molecule that is stretched in direct opposition to the outward force generators Cin8p and Kip1p.
A key element of this model is that the two Nuf2p-GFP foci observed in metaphase represent bi-oriented sister centromeres that are pulled apart. This idea is supported by three independent lines of research. First, the fluorescence intensity measured at each kinetochore cluster corresponds to 16 kinetochores [21]. Second, fluorescence is not recovered after photobleaching of GFP fusions to kinetochore proteins in a single cluster, suggesting that once separated, sister centromeres stay separated [4, 21]. Third, centromere proximal lacO arrays remain separated following bi-orientation [3, 26, 27]. This model of chromosome organization in the spindle correlates well with the findings in this study.
Chromatin elasticity
One interpretation of these results is that the mechanical properties of chromatin are similar to those of a mechanical spring. For a simple spring, force is directly proportional to the extent that it is stretched. In the case of bi-oriented sister chromatids in the metaphase spindle, the spindle exerts force on the chromosomes via the kinetochore microtubules until centromere flanking chromatin is pulled far enough to reach a force equilibrium with the spindle (Figure 5A). While this balance of forces is demonstrated by the relatively stable spindle length seen in both wild-type and histone repressed metaphase spindles, individual chromosomes are known to oscillate along the spindle axis. The movements of individual chromosomes is likely caused by at least one of the following: (1) the stochastic binding and dissociation of microtubule motor proteins at the kinetochore, (2) the regulation or binding of other MAPs at the kinetochore, or (3) the inherent dynamic properties of microtubule plus-ends. Across the 32 kinetochores in the metaphase spindle, these imbalances are averaged out and together the sister chromatids oppose the pulling forces of the spindle.
Figure 5. Modeling chromatin as a spring.
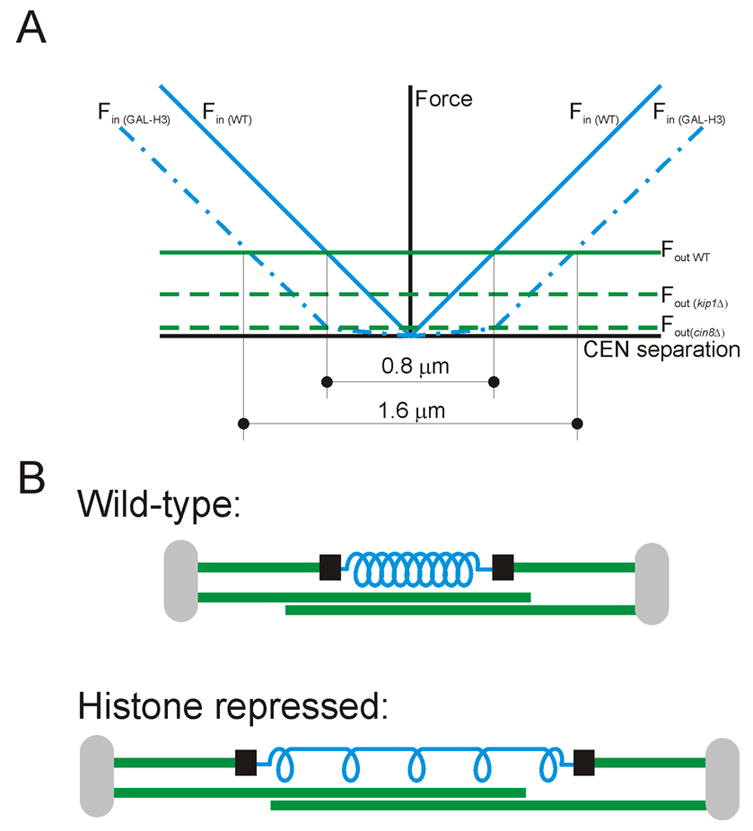
(A) Model of metaphase spindle forces based on experimental data and simplified modeling of chromatin as an elastic element. Outward spindle force is decreased in kip1Δ and cin8Δ cells and is varied by the extent that centromere separation was affected by these motor deletions (see figure 3C). Inward chromatin-dependent force is shifted outward by histone repression, representing increased rest length. (B) Schematic spindle model including chromatin as a spring. Histone repression lowers the number of incorporated nucleosomes and primarily affects the chromatin spring by increasing rest length (decreasing number of “coils” in the spring), without affecting springiness (spring constant) of the remaining nucleosomes (coils). The inward, resistive force of the stretched spring contributes to balance of forces defining centromere separation and spindle length.
Chromatin elasticity can be attributed to stretching of internucleosomal linker DNA, or intermolecular interactions at the DNA-protein and protein-protein levels. In vitro studies have shown that B-form DNA resembles a worm-like chain that takes little force (few pN) to achieve almost full extension. At full extension, increasing force must be applied until finally the molecule is permanently deformed [28, 29]. In contrast, chromatin pulling experiments in vitro have demonstrated that it behaves like an elastic polymer that requires 20 pN to dissociate a nucleosome [30]. This is within the range of force estimated for single kinetochore microtubules [31, 32]. Thus, chromatin elasticity can be assigned to protein-protein interactions of higher order chromatin packaging or DNA-protein interactions at the nucleosome. By repressing histones, the fraction of DNA associated with nucleosomes is decreased and the amount of linker DNA is increased (Figure 5B). Because of this shift from compacted to partially decompacted chromatin, chromatin is stretched to greater distances. No more than 50% of nucleosomes are predicted to be absent after histone repression, so the chromatin retains its elastic properties due to the remaining nucleosomes.
Spindle differences in H3 and H4 repressed cells
While spindles were longer in both histone H3- and histone H4-repressed strains, we observed differences between these strains after histone repression. First, kinetochores (Nuf2p-GFP) were declustered in approximately half of H4-repressed cells, but rarely declustered following H3 repression. We attribute this difference to the difference in nucleosome composition at the centromere (Supplementary Note 1). Second, we found that spindles were slightly longer following H4 repression than H3 repression. This finding correlates well with our model that sister chromatid pairs are responsible for the primary inward force during metaphase. If each sister pair generates 1/16 of the total force in a wild-type cell, then the loss of kinetochore attachments would lead to less total force. The disorganization and declustering of kinetochores following H4 repression suggests there are kinetochore function defects and is consistent with previous work showing decreased kinetochore-centromere binding following histone H4 repression [33]. Poor kinetochore attachments result in less total inward force of stretched sister chromatids, and thus longer spindles.
Conclusion
In conclusion, this study has demonstrated a new role for the nucleosomal packaging of DNA, and presents a more complete model of the forces in the mitotic spindle. At the centromere, a specialized nucleosome is required to form the kinetochore that links the kinetochore microtubules to the chromosomes. Flanking the centromere, the wrapping of DNA around nucleosomes functions to both package the DNA, as well as define the biophysical properties required to resist the tension placed on it. Ultimately, chromatin stretching reaches a force balance with the spindle that defines centromere separation and spindle length.
Material and Methods
Yeast Strains and growth
Unless otherwise noted, all strains used in this study were constructed in the YEF473A background [34]. Relevant genotypic information can be found in Supplemental Table 1. Spc29p fluorescent fusion proteins were created by fusing CFP (or RFP) to the C-terminus of the protein using PCR-generated integration cassettes [35, 36]. Genes of fusion proteins remained under control of their endogenous promoter. Nuf2p-GFP was created by integration of the BstEII digestion product of pJK67.
GAL-H3 and GAL-H4 strains were constructed by first deleting HHT1 and HHF1, respectively, by integration of a PCR-generated deletion cassette [35]. Next, the endogenous promoter of the second copy of each gene (HHT2 or HHF2) was replaced with a PCR-generated cassette containing the GAL1 promoter and selectable marker [35]. Cells were plated on galactose-containing selective plates. Strains were verified by death on glucose-containing plates, large-budded arrest in liquid YPD, and PCR using oligonucleotides flanking the expected sites of integration.
CIN8 deletion was carried out by integration of a deletion cassette generated by digesting pMA1186 with PstI and SalI. KIP1 was deleted using a PCR-generated deletion cassette as previously described [35]. MCD1-6HA was created using pVG270 (digested with AgeI), kindly provided by P. Megee.
GAL-H3 and GAL-H4 strains were maintained at 32°C in galactose-containing medium. Unless otherwise noted, histone repression was carried out as follows: cells were arrested for 3 hours in G1 with 10 μg/ml alpha factor. Next, cells were washed into glucose-containing medium (YPD) and incubated for 3 hours prior to imaging. For early/mid-S phase arrest, cells were incubated for 3 hours in media containing 200 mM hydroxyurea. For microtubule depolymerization, nocodazole was used at 15 μg/ml (dissolved in DMSO). An equal volume of DMSO alone was used as a negative control.
Microscopy
Unless otherwise noted, images were acquired at room temperature with a Nikon E600-FN microscope using a 1.4 NA 100x objective and cooled Hamamatsu Orca II camera. Cells were mounted on nutrient-containing gelatin slabs prior to imaging. Image acquisition and quantitation were performed using Metamorph 6.1 sofware (Universal Imaging). All distances were measured in triplicate as pixel distances using Metamorph 6.1 software (Universal Imaging) and converted to actual distance (μm). Data were exported to Microsoft Excel for analysis and presentation. While images presented in figures are maximum intensity projections of 5 plane z-series stacks, distance measurements were made using uncompiled images.
lacO-LacI-GFP images were acquired in one plane every 2 seconds. Total observation time was approximately 10 min, yielding over 300 data points for each cell type. Oscillation amplitude was defined as the distance traveled before a change in direction.
Fluorescence recovery after photobleaching (FRAP) experiments were carried out with a Nikon TE2000-U microscope using a 1.4 NA 100x objective and cooled Hamamatsu Orcar ER Camera, as previously described [4].
DraI accessibility assay
DraI accessibility at the centromere was performed as carried out previously [33]. Briefly, nuclei were isolated from spheroplasted yeast under native conditions and digested with increasing amounts of DraI enzyme. DNA was extracted, resolved on a 1% agarose gel, and transferred to nitrocellulose membrane. Hybridization of radiolabelled probe was detected using a phosphorimager screen, and quantitated using ImageQuant (Molecular Devices).
Chromatin immunoprecipitation
Chromatin immunoprecipation experiments were carried out on histone repressed and non-repressed cells as previously described [23, 37, 38]. Sequences for loci amplified by PCR (at CEN3 and the arm of ChrX) are available upon request.
Supplementary Material
Acknowledgments
We thank T. Jernigan and J. Haase for technical assistance; M. Gardner, and Drs. E. Yeh, A. Joglekar, and J. Molk for critical reading of the manuscript; and Dr. P. Megee for technical advice. Plasmids and strains were kindly provided by Drs. P. Megee, J. Pringle, M.A. Hoyt, P. Silver, T. Davis, and M. Yanagida. This work was supported by NIH grant GM-32238 (to K.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
David C. Bouck, Department of Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-3280.
Kerry Bloom, Department of Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-3280.
References
- 1.Scholey JM, Brust-Mascher I, Mogilner A. Cell division. Nature. 2003;422:746–752. doi: 10.1038/nature01599. [DOI] [PubMed] [Google Scholar]
- 2.Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 3.Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- 4.Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 5.Gardner MK, Pearson CG, Sprague BL, Zarzar TR, Bloom K, Salmon ED, Odde DJ. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol Biol Cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. J Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorbsky GJ, Simerly C, Schatten G, Borisy GG. Microtubules in the metaphase-arrested mouse oocyte turn over rapidly. Proc Natl Acad Sci U S A. 1990;87:6049–6053. doi: 10.1073/pnas.87.16.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- 14.Severin F, Hyman AA, Piatti S. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J Cell Biol. 2001;155:711–718. doi: 10.1083/jcb.200104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng X, Kahana JA, Silver PA, Morphew MK, McIntosh JR, Fitch IT, Carbon J, Saunders WS. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page BD, Snyder M. CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. Genes Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- 18.Antoniacci LM, Skibbens RV. Sister-chromatid telomere cohesion is nonredundant and resists both spindle forces and telomere motility. Curr Biol. 2006;16:902–906. doi: 10.1016/j.cub.2006.03.060. [DOI] [PubMed] [Google Scholar]
- 19.Kim UJ, Han M, Kayne P, Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. Embo J. 1988;7:2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 21.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 23.Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- 25.Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goshima G, Yanagida M. Time course analysis of precocious separation of sister centromeres in budding yeast: continuously separated or frequently reassociated? Genes Cells. 2001;6:765–773. doi: 10.1046/j.1365-2443.2001.00464.x. [DOI] [PubMed] [Google Scholar]
- 28.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 29.Gore J, Bryant Z, Nollmann M, Le MU, Cozzarelli NR, Bustamante C. DNA overwinds when stretched. Nature. 2006;442:836–839. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- 30.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 32.Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders MJ, Yeh E, Grunstein M, Bloom K. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol Cell Biol. 1990;10:5721–5727. doi: 10.1128/mcb.10.11.5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bi E, Pringle JR. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Bouck DC, Bloom KS. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc Natl Acad Sci U S A. 2005;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem. 1991;197:83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- 38.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


