Abstract
The human MRN complex is a multisubunit nuclease that is composed of Mre11, Rad50, and Nbs1 and is involved in homologous recombination and DNA damage checkpoints. Mutations of the MRN genes cause genetic disorders such as Nijmegen breakage syndrome. Here we identified a Schizosaccharomyces pombe nbs1+ homologue by screening for mutants with mutations that caused methyl methanesulfonate (MMS) sensitivity and were synthetically lethal with the rad2Δ mutation. Nbs1 physically interacts with the C-terminal half of Rad32, the Schizosaccharomyces pombe Mre11 homologue, in a yeast two-hybrid assay. nbs1 mutants showed sensitivities to γ-rays, UV, MMS, and hydroxyurea and displayed telomere shortening similar to the characteristics of rad32 and rad50 mutants. nbs1, rad32, and rad50 mutant cells were elongated and exhibited abnormal nuclear morphology. These findings indicate that S. pombe Nbs1 forms a complex with Rad32-Rad50 and is required for homologous recombination repair, telomere length regulation, and the maintenance of chromatin structure. Amino acid sequence features and some characteristics of the DNA repair function suggest that the S. pombe Rad32-Rad50-Nbs1 complex has functional similarity to the corresponding MRN complexes of higher eukaryotes. Therefore, S. pombe Nbs1 will provide an additional model system for studying the molecular function of the MRN complex associated with genetic diseases.
Nijmegen breakage syndrome is an autosomal recessive genetic disease characterized by developmental defects, microcephaly, immune deficiency, and a high incidence of cancer (8, 38, 64). Cells from patients with Nijmegen breakage syndrome show genetic instability and hypersensitivity to γ-ray irradiation and are impaired in cellular responses to γ-ray irradiation, including radio-resistant DNA synthesis (29, 52, 57). These cytogenetic features are indistinguishable from those of another genetic disease, ataxia telangiectasia (50). The cellular defects in Nijmegen breakage syndrome and ataxia telangiectasia cells are suggested to be caused by defective responses to DNA double-strand breaks (DSBs) due to a mutation of the responsible genes, NBS1 and ATM, respectively.
DSBs are not only generated by exogenous DNA-damaging agents such as γ-ray irradiation; they also occur during normal DNA replication. There are two main pathways for the repair of such DSBs: nonhomologous end joining and homologous recombination. The Mre11-Rad50-Xrs2 protein complex (MRX complex) from Saccharomyces cerevisiae has been suggested to be involved in the initial steps of both repair pathways (24). In nonhomologous end joining, the MRX complex is suggested to stimulate intermolecular DNA end joining by DNA ligase IV (11). In homologous recombination, the processing of DSB ends to produce 3′ single-stranded tails is a very important reaction that provides substrates for homologous pairing and strand exchange reactions. From early studies, the MRX complex was thought to function as a nuclease that is involved in such DNA end processing (24). However, recent studies suggest a structural role of the MRX complex in holding DNA ends and/or sister chromatids together (12, 17, 25, 26, 28, 30).
The Mre11 and Rad50 proteins are highly conserved from S. cerevisiae to humans. However, Xrs2 is not well conserved, and the functional counterpart of Xrs2 is considered to be Nbs1 in vertebrates. The similarity of the overall sequences between those two proteins is very weak and is limited to the N-terminal forkhead-associated domain and a small C-terminal region (56). Although Nbs1 contains a BRCA1 C-terminal (BRCT) domain in the N-terminal region, Xrs2 does not contain this domain. Nbs1 binds to the Mre11 subunit of the Rad50-Mre11 complex via the C-terminal conserved region (18, 56).
The MRX(N) complex possesses exo- and endonuclease activity in vitro (21, 49, 59, 63). Mre11 contains the phosphodiesterase motif responsible for the nuclease activity (27). Rad50 is related to the SMC proteins, which contain Walker A and B motifs, responsible for ATPase activity, separated by a long coiled-coil region (26, 27). Rad50 stimulates the nuclease activity of yeast and human Mre11 (48, 60). The biochemical functions of Xrs2 and Nbs1 during the recombination process remain unclear. To date, the only biochemical activity assigned to Nbs1 is stimulation of the unwinding and hairpin cleavage activities of the human Mre11-Rad50 complex (49). The yeast Mre11-Rad50 complex cleaves hairpin structures in the absence of Xrs2 (59). Surprisingly, some mutations that eliminate the nuclease activity of yeast Mre11 do not confer telomere defects or strong DNA repair deficiency (33, 41). Therefore, it seems likely that end processing is not the major role of the complex.
Mutations in the genes that encode the components of the MRX(N) complex result in DNA damage sensitivity, genome instability, telomere shortening, and aberrant meiosis (14). These phenotypes are considered to be related to deficiencies in DNA repair abilities involving the DNA-processing activity of the MRX(N) complex. In addition, recent studies have shown other functions for the Mre11 complex in checkpoint signaling and DNA replication (6, 31, 37, 39, 68). Hypomorphic mutations in the human MRE11 gene cause ataxia telangiectasia-like disorder, whose symptoms are very similar to those of Nijmegen breakage syndrome and ataxia telangiectasia (51). In the cells from these patients, DNA synthesis is not arrested in response to ionizing radiation. Yeast mutants with mutations of mre11, rad50, or xrs2 are hypersensitive to hydroxyurea and show no delay in DNA synthesis in the presence of hydroxyurea or bleomycin, indicating a defect in the intra-S checkpoint (15, 23). Mre11 and Xrs2/Nbs1 have been shown to be phosphorylated in response to ionizing radiation, and this phosphorylation is dependent on checkpoint kinase function (22, 34, 70).
In the fission yeast Schizosaccharomyces pombe, which is distantly related to S. cerevisiae, the rad32+ gene was first identified as a gene corresponding to a mutant that showed hypersensitivity to DNA damage and was later found to be the structural and functional homologue of MRE11 (58, 65, 66). The rad50+ gene in S. pombe was identified through its sequence similarity with the S. cerevisiae, Caenorhabditis elegans, and human RAD50 genes. Both rad32+ and rad50+ are required for DNA repair and telomere maintenance (25). However, no Nbs1/Xrs2 homologue in S. pombe has been reported; one of the reasons for this is that no structural homologue can be found in the database even though the whole genome sequence of S. pombe has been determined (67).
To identify novel genes involved in homologous recombination and recombination repair in S. pombe, we previously isolated seven slr mutants, whose growth was dependent on the rad2+ function (slr stands for synthetic lethality with rad2) (62). The rad2+ gene encodes a structure-specific endonuclease homologous to mammalian Fen-1 that is required for Okazaki fragment maturation (46). Yeast cells defective in Fen-1 nuclease activity are nonviable in combination with mutations that inactivate homologous recombination (16, 54). Thus, slr mutants are often considered to be defective in homologous recombination and recombination repair. Indeed, S. pombe rhp51 (RAD51 homologue), rhp54 (RAD54 homologue), rhp57 (RAD57 homologue), rad50, and rad32 mutations are lethal when combined with the rad2 mutation (25, 45, 58, 62). In an earlier study, we characterized two genes, rhp57+ and rad60+, identified among the seven slr mutants. The former is a structural and functional homologue of S. cerevisiae RAD57, and the latter is a novel recombination gene for which no structural homologue has been found in the database (43, 62).
Since slr screening is an effective mechanism for isolating recombination-related genes, we performed a second large-scale screening and obtained further slr candidates. In this study, we characterized the eighth slr mutation, slr8. DNA sequencing of complementing plasmids revealed that slr8+ encodes a protein with a forkhead-associated domain in the N-terminal region and an Mre11-binding consensus sequence in the C-terminal region. Indeed, Slr8 protein was found to interact physically with Rad32 via the C-terminal region of Rad32 in a yeast two-hybrid assay. slr8Δ mutants showed DNA repair defect phenotypes very similar to those of rad32Δ and rad50Δ mutants and showed epistatic interactions with these mutations. Although the overall sequence similarities of the three proteins, S. cerevisiae Xrs2 and Slr8 and vertebrate Nbs1, are limited, our data allow us to conclude that slr8+ is a functional homologue of XRS2/NBS1, and thus we have designated it nbs1+ in S. pombe.
MATERIALS AND METHODS
S. pombe strains, media, and genetic methods.
The S. pombe strains used in this study are listed in Table 1. All strains are derivatives of 972 h−, 975 h+, or JY741 h−. Standard procedures and media were used for propagation and genetic manipulation (42). To measure the sensitivity of cells to γ-rays or UV light, exponentially growing cells were irradiated with γ-rays from a 60Co source at a dose rate of 100 to 200 Gy/h or with UV light from a germicidal lamp (UVP UV-Crosslinker, CL-1000) at a dose rate of 50 to 100 J/m2/min. Duplicates of irradiated cells and unirradiated cells were plated on YPAD medium (1% yeast extract, 2% polypeptone, 2% glucose, 20 μg of adenine per ml) and incubated at 30°C for 4 days, and the colonies were counted. For semiquantitative analysis of DNA repair activity, the spot assay was employed as described previously (62). Briefly, 3 μl of 10-fold dilutions of log-phase cells (0.5 × 107 cells/ml) were spotted onto a YPAD (2% agar) plate or YPAD plate containing the indicated concentration of methylmethane sulfonate (MMS) or hydroxyurea. All experiments were repeated at least twice and gave similar results.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Sourcea |
|---|---|---|
| 142 | h+ leu1-32 ura4-D18 ade6-M210 rad32::ura4+ | A. Matsuura |
| 968 | h90 | NCYC |
| 972 | h− | NCYC |
| 975 | h+ | NCYC |
| B54 | h− smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 rhp51::his3+ | This study |
| JY741 | h− leu1-32 ura4-D18 ade6-M216 | M. Yamamoto |
| KT120 | h+ leu1-32 ura4-D18 ade6-M210 rad50::LEU2 | Our lab stock |
| KW015 | h− smt-0 leu1-32 ura4-D18 his3-D1 ade6-M210 nbs1::ura4+ rad50::LEU2 | This study |
| SP154 | h+ ura4-D18 | NCYC |
| SP184 | h− smt-0 ade6-704 leu1-32 ura4-D18 rad2::ura4/pAUR2 | This study |
| SP185 | h+ ade6-704 ura4-D18 rad2::ura4/pAUR2 | This study |
| SPN100 | h− smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 nbs1::ura4+ | This study |
| SPN103 | h− smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 nbs1::ura4+ rhp51::his3+ | This study |
| SPN114 | h+ mat1PD::LEU2 leu1-32 ura4-D18 his3-D1 arg3-D1 | Our lab stock |
| SPN124 | h− smt-0 leu1-32 ura4-D18 his3-D1 arg3-D1 | Our lab stock |
| SPN138 | h+ mat1PD::LEU2 leu1-32 ura4-D18 his3-D1 arg3-D1 nbs1::ura4+ | This study |
| SPN141 | h+ mat1PD::LEU2 leu1-32 ura4-D18 his3-D1 arg3-D1 rad32::ura4+ | This study |
| SPN148 | h+ mat1PD::LEU2 leu1-32 ura4-D18 his3-D1 arg3-D1 rad50::ura4+ | This study |
| YA137 | h+ mat1PD::LEU2 ura4-D18 leu1-32 rad2::ura4+ | This study |
| Rad3D | h− ura4-D18 leu1-32 ade6-704 rad3::ura4+ | A. Carr |
NCYC, National Collection of Yeast Cultures.
slr mutant isolation.
S. pombe strain SP184 (h− smt-0 ade6-704 ura4-D18 leu1-32 rad2::ura4) carrying pAUR2, which is a derivative of pUR18 that carries the rad2+ gene and ura4+ as a selective marker (62), was mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine as described by Moreno et al. (42). Initially, approximately 100,000 colonies were roughly examined for the plasmid dependency of their growth by replica plating on Edinburgh minimal medium (EMM) plates containing 0.1% 5-fluoroorotic acid, and 521 5-fluoroorotic acid-sensitive colonies were isolated. These isolates were examined for MMS sensitivity by replica plating on EMM plates containing 0.004% MMS, and 92 MMS-sensitive colonies were obtained. Their 5-fluoroorotic acid sensitivity was confirmed by a spot assay on EMM plates containing 5-fluoroorotic acid, and 42 of the isolates were found to be sensitive to both MMS and 5-fluoroorotic acid.
In this study, we analyzed 15 of the isolates that showed much higher sensitivity to MMS than the others. These 15 isolates were then crossed with SP185 (h+ ade6-704 ura4-D18 rad2::ura4) carrying pAUR2. Three isolates were found to produce progeny with the phenotypes of MMS sensitivity and 5-fluoroorotic acid resistance, indicating that the growth of the three strains was not dependent on pAUR2. The remaining 12 strains were backcrossed three times with the wild-type strain, and the final progeny that were obtained had the genotype slr rad2+ ade ura leu.
Cloning of slr genes.
Each of the new slr mutants was transformed with the S. pombe genomic libraries constructed in the laboratories of H. Masukata (Osaka University) and A. M. Carr (University of Sussex) and spread on EMM plates containing appropriate supplements and MMS (0.004%). The transformants were examined for plasmid-dependent MMS resistance, and the plasmids which complemented the MMS sensitivity were recovered as described previously (43).
slr8 cDNA cloning.
The cDNA corresponding to the slr8+ N-terminal region was amplified by PCR with a sense primer, IWA 252 (5′-CTCATATGAACAGACAAGCTGGGTCCAG-3′), and an antisense primer, IWA184 (5′-GAGGATCCTTAAAAGTGAAACTTGAGATCATTAAATTCATCG-3′), with the S. pombe cDNA library (Clontech) as the template. The PCR product was cloned into the NdeI and BamHI sites of the vector pBSNde, giving plasmid pNT139. pBSNde was constructed by insertion of an NdeI linker (5′-CCCATATGGG-3′) into the HincII site of pBluescript II SK (+). The cDNA corresponding to the C-terminal region was amplified by PCR with a sense primer, IWA 256 (5′-CTCCATATGTGGATAATTGAGGCTGAGGCTGAGGGTGACATTC-3′), and an antisense primer, IWA37 (5′-CAGAGTCATATACTGCGTTG-3′), and the product was cloned into the NdeI and BamHI sites of pBSNde, giving plasmid pNT136.
The full-length open reading frame (ORF) of the cDNA was constructed by insertion of the NdeI-EcoRV fragment encoding the N-terminal portion of slr8+ from pNT139 into the NdeI and EcoRV sites of pNT136, giving pNT140. The nucleotide sequence of the full-length ORF was determined with an automated DNA sequencer (ABI Prism 3100) and shown to match SPBC6B1.09c in cosmid c6B1. The exons predicted at the Sanger Centre database (http://srs.ebi.ac.uk/srs6bin/cgi-bin/wgetz?-e+[{EMBL%20EMBLNEW}-acc:AL021838]) and the exons predicted by our cDNA sequencing are shown below. Exons predicted in the Sanger Center database were complement(26318 to 26354), complement(26205 to 26258), complement(26078 to 26166), complement(25825 to 25988), complement(24495 to 25785), and complement(24185 to 24448); exons predicted by our cDNA sequencing were complement(26318 to 26354), complement(26205 to 26258), complement(26078 to 26166), complement(25825 to 25973), complement(25703 to 25785), complement(24495 to 25660), and complement(24185 to 24448). The numbers used above correspond to the numbering used in cosmid c6B1 in the Sanger Centre database. The cDNA sequence data are available from the DDBJ/EMBL/GenBank nucleotide databases (accession no. AB099299).
3′- and 5′-RACE.
Total RNA was prepared from wild-type S. pombe strain 968 (h90) with an RNeasy mini kit (Qiagen), and then the mRNA was purified with an Oligotex-dT30 Super mRNA purification kit (TaKaRa). 3′-rapid amplification of cDNA ends (RACE) and 5′-RACE experiments were carried out with a 3′-Full RACE core set (TaKaRa) and 5′-Full RACE core set (TaKaRa), respectively, according to the manufacturer's instructions. The primer sets used for 3′-RACE and 5′-RACE were as follows: IWA328 (sense primer for 3′-RACE), GTCATCCGAGAAATCGAATGCTAACAGTA; IWA318 (reverse transcription primer for 5′-RACE), CGTATCGAGGTCCTTTAC; IWA321 (first sense primer for 5′-RACE), GCCATGCTCGTTTTACG; IWA320 (first antisense primer for 5′-RACE), CGAATCGTCAGATACATTTC; IWA322 (second sense primer for 5′-RACE), GTGAGAAAGACTACTTTACC; and IWA319 (second antisense primer for 5′-RACE), CCTACAATATAAGTTCCTGG.
Yeast two-hybrid analysis.
Gal4-based Matchmaker Two-Hybrid System 3 (Clontech) was used for the yeast two-hybrid assay according to the manufacturer's instructions. S. cerevisiae strain AH109 was used as the reporter strain, The indicated proteins were fused to the GAL4 activation domain in vector pGADT7 and the GAL4 DNA-binding domain in pGBKT7 and expressed in AH109. The reciprocal combinations of fusions with the activation domain and DNA-binding domain were examined. The interactions were judged by a spot test on three types of dropout (DO) plates: 4DO (SD medium lacking adenine, histidine, leucine, and tryptophan; high-stringency conditions), 3DO (4DO plus adenine; medium stringency), and the control 2DO (SD without leucine and tryptophan) to select plasmids.
Disruption of nbs1+ gene.
An Nbs1 knockout plasmid, pNT117, was constructed as follows. A 0.8-kb fragment containing the sequence upstream of the nbs1+ ORF, which was amplified by PCR with genomic DNA and primers IWA180 (5′-CACAAGCTTGTATACACATACTTCTCCAG-3′) and IWA181 (5′-CACCTCGAGGGTTTAGTAGATTTAGCTTC-3′) was subcloned into pBluescript II SK (+), giving plasmid pNT115. A 1.0-kb fragment containing the sequence downstream of the nbs1+ ORF, which was amplified by PCR with genomic DNA and primers IWA178 (5′-CACGGATCCGCTCTTCTTCCAAGATTTTG-3′) and IWA179 (5′-CACAAGCTTGACTACTTACCTGAGATATC-3′), was subcloned into pBluescript II SK(+), giving plasmid pNT114. Then the HindIII-BamHI fragment containing the sequence downstream of the nbs1+ ORF from pNT114 was inserted into the HindIII and BamHI sites in pNT115, giving pNT116. Next, the 1.8-kb ura4+ gene was inserted into the HindIII site in pNT116, giving knockout plasmid pNT117. An XhoI-BamHI fragment carrying the nbs1::ura4+ construct derived from knockout plasmid pNT117 was transformed into haploid strain SPN124 (smt-0 ura4-D18 leu1-32 his3-D1 arg3-D1) with the lithium acetate method (42). Stable transformants were isolated, and gene disruption was confirmed by Southern blot analysis.
Measurement of telomere length.
Telomere length was measured by Southern hybridization according to the procedure described previously (13) with the AlkPhos Direct kit module (Amersham Pharmacia Biotech). Briefly, chromosomal DNA which had been digested with ApaI and separated by electrophoresis on a agarose gel was probed with a 0.3-kb DNA fragment containing the telomeric repeat sequences derived from pNSU70 (a gift from N. Sugawara).
RESULTS AND DISCUSSION
Newly isolated slr mutants.
To identify novel genes involved in homologous recombination and recombination repair of S. pombe, we isolated slr mutants as described in Materials and Methods. From approximately 100,000 cells mutagenized with N-methyl-N′nitro-N-nitrosoguanidine, 12 colonies were identified as slr mutants (Table 2). Complementation analysis of MMS and UV sensitivities with the cloned genes rhp51+, rhp55+, and rhp57+ revealed that a mutation of mutant 11 in Table 2 was an allele of rhp57+. We subsequently attempted to isolate plasmids that complemented the MMS sensitivity of the 11 remaining mutants from two genomic libraries. Four genes have been isolated to date. Mutants 6, 7, and 23 were complemented by plasmids carrying the rad32+ gene. Sequencing of the genomic DNA revealed that the rad32+ gene was mutated in these strains. This is consistent with a previous report demonstrating that rad32 mutations are lethal in combination with a rad2 mutation (58).
TABLE 2.
Summary of slr mutant isolation
| Mutant no. | Mutant genea | S. cerevisiae homologue |
|---|---|---|
| 6 | rad32 | MRE11 |
| 7 | rad32 | MRE11 |
| 11 | rhp57 | RAD57 |
| 14 | ND | |
| 16 | ND | |
| 17 | ND | |
| 23 | rad32 | MRE11 |
| 27 | slr8 (this study) | XRS2 |
| 28 | ND | |
| 29 | ND | |
| 34 | ND | |
| 45 | ND |
ND, not determined.
One plasmid that complemented the sensitivity of mutant 27 was isolated (pNT101). pNT101 carried a 3.3-kb genomic DNA fragment of cosmid c6B1. Since there are no known genes involved in DNA repair and recombination in this region, we named it slr8+ (since we had already isolated seven slr mutants) (62) and analyzed it further in this study.
Identification of the nbs1+ gene in S. pombe.
Complementation analysis with several deletion plasmids derived from pNT101 revealed that slr8+ corresponds to the open reading frame SPBC6B1.09c. The cDNA was prepared, and its sequence revealed that slr8+ contained six introns, although only five of these were predicted in the Sanger Centre database (http://srs.ebi.ac.uk/srs6bin/cgi-bin/wgetz?-e+[{EMBL%20EMBLNEW}-acc:AL021838]) (see Materials and Methods). RACE experiments revealed that mRNA for nbs1+ was transcribed from 82 bp upstream from the putative initiation codon to 110 bp downstream from the predicted stop codon. Since there was a stop codon and no ATG codons in-frame upstream of the putative ATG, this should be the actual initiation codon. These cDNA analyses indicate that the slr8+ gene product consists of 613 amino acids. Genomic DNA sequencing of the slr8 locus revealed that the codon for amino acid Gln-88 (CAA) in Slr8 protein is mutated to a nonsense sequence (TAA) in the slr8 mutant, suggesting that the gene is responsible for the slr8 mutation. The features of the amino acid sequence of Slr8 (see below) suggest that this protein could be an Nbs1 homologue, and therefore we designated Slr8 Nbs1 in S. pombe.
Structure of S. pombe Nbs1/Slr8 protein and Nbs1 homologues.
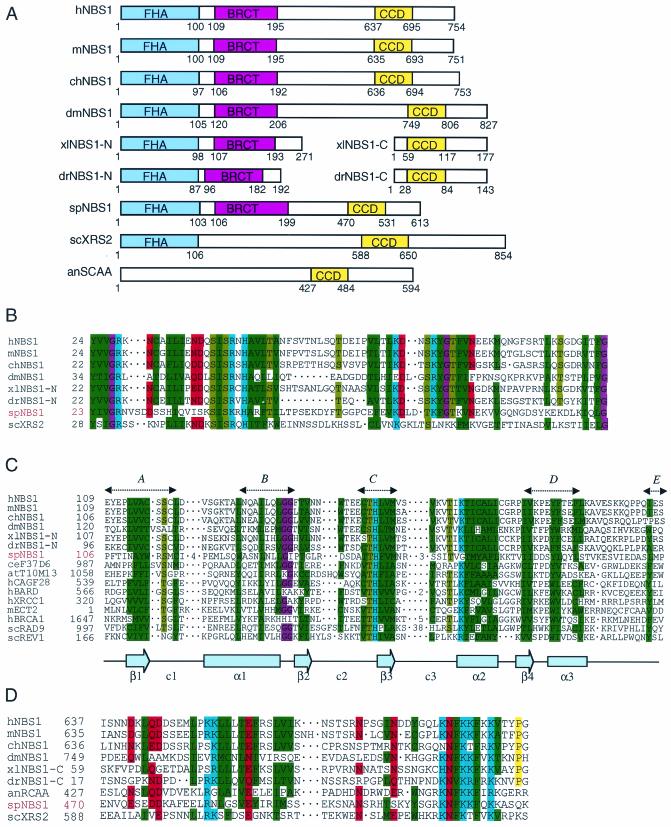
Vertebrate Nbs1 proteins contain a forkhead-associated domain in the N-terminal region, followed by a BRCT domain and a C-terminal conserved domain (CCD) (Fig. 1A). The S. pombe Slr8 also contains a forkhead-associated domain in the N-terminal region (amino acids 1 to 103) and a CCD in the C-terminal region (amino acids 470 to 531), which show 26% and 23% identity to the forkhead-associated domain and CCD in human Nbs1, respectively (Fig. 1A, B and D).
FIG.1.
Amino acid sequence analysis of S. pombe Nbs1. Alignment was performed as described previously (64). (A) Overall structural comparison among several Nbs1 and related proteins. (B) Forkhead-associated (FHA) domain. (C) BRCA1 C-terminal (BRCT) domain. The five conserved regions, designated A to E (7), of the BRCT domain based on the hydrophobic cluster analysis (HCA) are shown by a dotted arrow. The secondary structure of the XRCC1 BRCT domain (69) is shown at the bottom of the alignment. (D) C-terminal conserved domain (CCD). Protein sequences were obtained from humans (h), mice (m), chickens (ch), Aspergillus nidulans (an), Drosophila melanogaster (dm), Danio rerio (dr), Xenopus laevis (xl), S. pombe (sp), and S. cerevisiae (sc). Color of conserved positions (at most, two deviating amino acids for the forkhead-associated domain, four deviating amino acids for the BRCT domain, and three deviating amino acids for the CCD): green, hydrophobic (FYWILMVTAC); red, acidic (DEQN); blue, basic (HKR); magenta, glycine (G); brown, serine or threonine (ST); yellow, proline (P). Sequences are denoted by species identification prefixes for Caenorhabditis elegans (ce) and Arabidopsis thaliana (at) and by protein designations for hNBS1 (GenBank BAA28616); spNBS1 (GenBank AB099299); drNBS1-N (GenBank BI984731); drNBS1-N (GenBank BM775439); xlNBS1-N (GenBank CA988284); xlNBS1-C (GenBank BG022948); scXRS2 (GenBank AAB64805); mNBS1 (GenBank AB016988); chNBS1 (GenBank AAG47947); dmNBS1, also named CG6754-PB protein (GenBank NM_143716); predicted translation products of the cosmids F37D6.1 (GenBank CAA99847) and T10M13.12 (GenBank T01512); the CAG trinucleotide repeat containing cDNA CAGF28 (GenBank AAB91434); the BRCA-associated RING finger domain protein BBARD (GenBank Q99728); the DNA repair proteins XRCC1 (GenBank A36353) and REV1 (GenBank S67255); the oncoprotein ECT2 (GenBank S32372); the breast cancer susceptibility type 1 protein BRCA1 (GenBank U14680); the radiation-sensitive checkpoint protein Rad9 (GenBank M26049); and campothecin resistance-conferring protein SCAA (GenBank AAF81094), which has moderate structural similarity to hNBS1. Numbers in the alignment denote the number of amino acids omitted.
The domain search program at the NCBI server (http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi?cmd =rps) did not detect a BRCT domain in Slr8. However, hydrophobic cluster analysis (7) revealed that Slr8 contains all five of the motifs (designated A to E) characteristic of the BRCT domain (Fig. 1C). In addition to these five motifs, the sequence corresponding to the alpha helix (α2) in the BRCT domain is highly conserved in Slr8 (Fig. 1C). This region (amino acids 106 to 199) shows 18% identity to the BRCT domain of human Nbs1 (Fig. 1C). These sequence features suggest that Slr8 contains a BRCT domain in this region. Therefore, we conclude that Slr8 is a structural homologue of Nbs1, although the overall similarity is very limited. S. cerevisiae Xrs2, which is considered an Nbs1 counterpart, does not seem to possess a BRCT domain (data not shown) (14). Therefore, Nbs1 is a more appropriate name than Xrs2 for the Slr8 protein.
The forkhead-associated domains in human Chk2 and S. cerevisiae Rad53 have been shown to bind to phosphorylated proteins (1, 53). The BRCT domain is also predicted to be a protein-protein interaction domain (69). Indeed, the recombinant fragment containing the forkhead-associated/BRCT domain of human Nbs1 bind to phosphorylated histone H2AX (γ-H2AX) (32). Therefore, S. pombe Nbs1 most probably binds to phosphorylated histone H2A. The physiological significance of this interaction remains unclear and will be the subject of future studies.
A database search revealed that CG6754-PB protein from Drosophila melanogaster contains the three domains (forkhead-associated, BRCT, and CCD), suggesting that it could be the Nbs1 homologue in the fruit fly (Fig. 1A). Moreover, two protein fragments (we designated them drNBS1-N and drNBS1-C) predicted from cDNA clones in the zebra fish (Danio rerio), BI984731 and BM775439, and two protein fragments (designated xlNBS1-N and xlNBS1-C) predicted from cDNA clones in the frog (Xenopus laevis), CA988284 and BG022948, showed high similarity to the human NBS1 N-terminal fragment and C-terminal fragment, respectively (Fig. 1A). These fragments contain either the forkhead-associated and BRCT domains or CCD which show high similarity to those of human Nbs1 (Fig. 1B to D). These amino acid sequence features strongly suggest that frog and zebrafish contain NBS1 proteins, although the full-length cDNAs that encode frog or zebrafish NBS1 have not been identified. These cDNA fragments containing the forkhead-associated/BRCT domains and CCD might be truncated products of the same genes.
We also found that the amino acid sequence (427 to 484) of ScaA protein in the filamentous fungus Aspergillus nidulans, which is suggested to be related to human NBS1 (5), shows high similarity to CCD (amino acids 637 to 695) in human NBS1 (Fig. 1D). Strains carrying the scaA mutation are hypersensitive to camptothecin, MMS, UV light, and γ-rays (5). The sequence of amino acids 230 to 513 of ScaA shows 20% identity and 38% similarity with that of amino acids 280 to 572 of S. pombe Nbs1. These facts further suggest that ScaA is most probably a functional Nbs1 homologue in A. nidulans, although ScaA possesses neither a forkhead-associated nor a BRCT domain.
C-terminal half of Nbs1 binds to Rad32 in a two-hybrid assay.
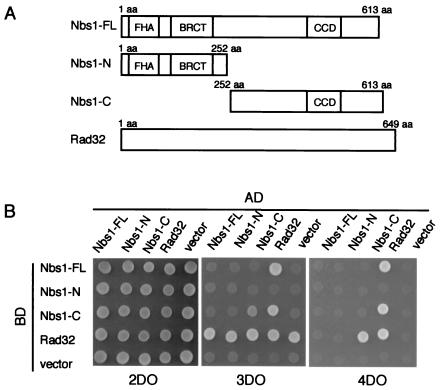
Since human Nbs1 and S. cerevisiae Xrs2 physically interact with Mre11 (10, 21, 63), we tested whether S. pombe Nbs1 also interacts with Rad32, an S. pombe homologue of Mre11. A yeast two-hybrid assay was performed in which reciprocal combinations of fusions of Nbs1 and Rad32 with the GAL4 activation domain (AD) and the GAL4 DNA binding domain (BD) were examined (Fig. 2). A BD-Nbs1-FL fusion protein interacted with the AD-Rad32 fusion at high stringency (4DO). Although we did not detect interaction between the AD-Nbs1-FL and BD-Rad32 under high-stringency conditions, we did observe this at lower stringency (3DO). The inability to detect interactions with particular combinations of activation domain and DNA-binding domain fusion proteins may have been due to a very low level of expression of the fusion protein. Indeed, Western blotting analysis revealed that the protein band corresponding to AD-Nbs1-FL was barely detectable in the extract from the S. cerevisiae tester strain (data not shown). Notably, the two-hybrid interactions were observed between the C-terminal half of Nbs1 and Rad32 under high-stringency conditions, while the N-terminal half of Nbs1 did not interact with Rad32 (Fig. 2B). This is consistent with the proposal that Nbs1 interacts with Mre11 via its small C-terminal conserved region (56). These two-hybrid interactions between Nbs1 and Rad32 suggest that S. pombe Nbs1 functions together with Rad32 in vivo.
FIG. 2.
Yeast two-hybrid interaction between Nbs1 and Rad32. (A) A simple scheme of the protein structure and constructs of Nbs1 designed for yeast two-hybrid analysis. Nbs1-FL, full-length Nbs1 (amino acids [aa] 1 to 613); Nbs1-N, N-terminal half of Nbs1 (amino acids 1 to 252); Nbs1-C, C-terminal half of Nbs1 (amino acids 252 to 613). Forkhead-associated domain (amino acids 1 to 103), BRCT domain (amino acids 106 to 199), and C-terminal conserved domain (CCD) (amino acids 470 to 531) are indicated. (B) Two-hybrid interaction of Nbs1 and Rad32. Pairwise interaction of pGBKT7 and pGADT7 was judged by a spot test on three types of dropout plates: 4DO (high stringency), 3DO (medium stringency), and 2DO (control) (see Materials and Methods).
Since the CCDs are highly conserved among Nbs1/Xrs2 proteins and the conservation is much higher than that of the forkhead-associated or BRCA domains (Fig. 1D), it is suggested that the molecular mechanism of the interaction between Nbs1 and Mre11 is conserved in eukaryotes. We also suggest that the CCD could be used for searching for Nbs1 homologues in other organisms as described above.
Construction of nbs1 disruptants.
To study the in vivo function of nbs1+ of S. pombe, we made a heterozygous strain (nbs1+/−) with a one-step gene replacement procedure, in which one of the chromosomal nbs1+ genes was replaced with a ura4+ cassette. Spores derived from the heterozygote were viable regardless of auxotrophy for uracil, indicating that nbs1+ is not essential (data not shown). However, the growth of the nbs1 disruptant was poor compared with that of the wild-type strain. The synthetic lethality of the nbs1 disruption with rad2Δ was confirmed by analyzing the segregants of nbs1+/− rad2+/− heterozygotes (data not shown). We also constructed an nbs1 null mutant from a haploid strain as described in Materials and Methods and used it for further characterization.
DNA repair activity of nbs1Δ mutants.
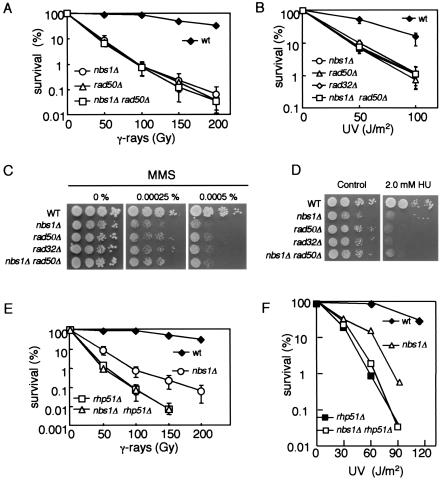
We first characterized the DNA repair activity of nbs1 null mutants. An nbs1 single mutant showed high sensitivity to γ-ray irradiation, indicating that Nbs1 is involved in DSB repair (Fig. 3A). In addition, the mutant was very sensitive to UV, MMS, and hydroxyurea, suggesting that Nbs1 is also involved in the repair of many types of DNA damage in addition to DSBs (Fig. 3B to D). The hydroxyurea sensitivity of the nbs1 mutant might imply the involvement of S. pombe Nbs1 in the S-phase checkpoint. The sensitivities of the nbs1 single mutant to these DNA damages were very similar to those of rad32 and rad50 single mutants, and combinations of these mutations with an nbs1 mutation did not affect these sensitivities (Fig. 3A to D). These results suggest that Nbs1 functions together with Rad32 and Rad50, perhaps via a complex similar to the Mre11-Rad50-(Xrs2/Nbs1) complex.
FIG. 3.
DNA damage sensitivity of nbs1Δ cells and epistasis analysis. (A) The sensitivities to γ-ray of nbs1Δ cells (SPN100), rad50Δ cells (KT120), and rad50 nbs1 double mutants (KW015). JY741 was used as a wild-type (wt) strain. (B) The sensitivities to UV light of nbs1Δ cells, rad50Δ cells, rad32Δ cells (142) and the rad50 nbs1 double mutants. (C and D). The sensitivities of nbs1Δ cells, rad50Δ cells, rad32Δ cells, and rad50 nbs1 double mutants to MMS (C) and hydroxyurea (D) determined in a spot test. (E and F). Epistasis between nbs1Δ cells and rhp51Δ cells for γ-ray (E) and UV (F) sensitivity. Wild-type (SPN124), nbs1Δ (SPN100), rhp51Δ (B54), and nbs1 rhp51 (SPN103) cells. For genotypes, see Table 1. Standard deviations are shown by error bars.
Rhp51 protein from S. pombe is the functional homologue of S. cerevisiae Rad51, which plays central roles in the homology search and DNA strand exchange reactions during homologous recombination and recombination repair (44). The sensitivities to γ-ray and UV of an nbs1 rhp51 double mutant were very similar to those of the rhp51 single mutant (Fig. 3E and F). Taken together, our results suggest that nbs1+ belongs to the same epistatic group in DNA repair as rad50+, rad32+, and rhp51+ and that nbs1+ is involved in recombination repair.
Treatment with MMS, hydroxyurea, or UV, unlike γ-ray irradiation, does not directly produce DSBs. However, the fact that rad22, rhp51, and rhp54 mutants all have significantly increased sensitivities to these treatments indicates that recombination is very important for recovery from the damage caused by those agents in S. pombe. These treatments cause replication fork arrest. When replication forks are stalled, Holliday junctions or chicken-foot structures of DNA are thought to be produced, and those would either be reversed by Rqh1 or resolved by Mus81-Eme1 endonuclease (4, 35). In the latter pathway, it is thought that DNA DSBs are generated upon cleavage by Mus81-Eme1 (19). The Rad32-Rad50-Nbs1 complex might be required for the processing of these DSBs at the replication fork to initiate recombination-dependent replication. It is also possible that the Rad32-Rad50-Nbs1 complex, rather than Mus81-Eme1 endonuclease, is directly involved in the cleavage at collapsed replication forks, since it has been suggested that Rad32 cleaves the hairpin structure generated on the lagging DNA strand during S phase (20).
Nbs1 is involved in telomere length regulation.
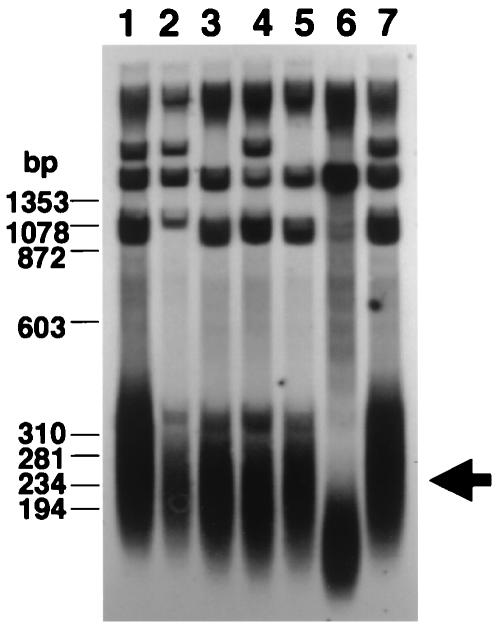
As S. pombe Rad50 and Rad32 are involved in telomere length regulation (25, 66) and Rad32 has been shown to associate with telomeres (47), we examined the telomere length of nbs1 mutants. The telomeres of the nbs1 mutant were shorter than those of the wild-type strain (Fig. 4). The telomere length of the nbs1 mutant was almost the same as those of rad32 and rad50 single mutants and the nbs1 rad50 double mutant. These results indicate that Nbs1 is required for telomere length maintenance. Recently, we found that deletion of taz1+ significantly increased the single-stranded G-rich overhang at telomere ends and that this overhang disappeared after concomitant deletion of rad32+, rad50+ (58a), or nbs1+ (data not shown). This result suggests that Nsb1 functions together with Rad32 and Rad50 at telomere ends. Human Mre11 and Rad50 bind to telomere ends in a cell cycle-independent manner, while human Nbs1 binds to telomeres only during S phase, suggesting that the MRN complex, a multisubunit nuclease composed of Mre11, Rad50, and Nbs1, plays an important role in telomere maintenance in S phase (71). Consistent with this, it has been suggested that the S. cerevisiae MRX complex may be required for recruitment of the telomerase components to telomeres during S phase (55, 61).
FIG. 4.
Nbs1 is involved in telomere length maintenance. The telomere length of wild-type, nbs1Δ, rad50Δ, rad32Δ, and rad50 nbs1 double mutant cells was evaluated by Southern hybridization. Lane 1, wild-type cells (JY741); lane 2, nbs1Δ (SPN100); lane 3, rad50Δ (KT120); lane 4, rad32Δ (142); lane 5, rad50 nbs1 double mutant (KW015); lane 6, rad3Δ (rad3D); lane 7, wild-type cells (JY741). Genomic DNA was digested with ApaI and separated by electrophoresis on a 2% agarose gel. Telomeres are indicated by an arrow. For genotypes, see Table 1.
nbs1Δ cells are elongated and exhibit abnormal nuclear morphology.
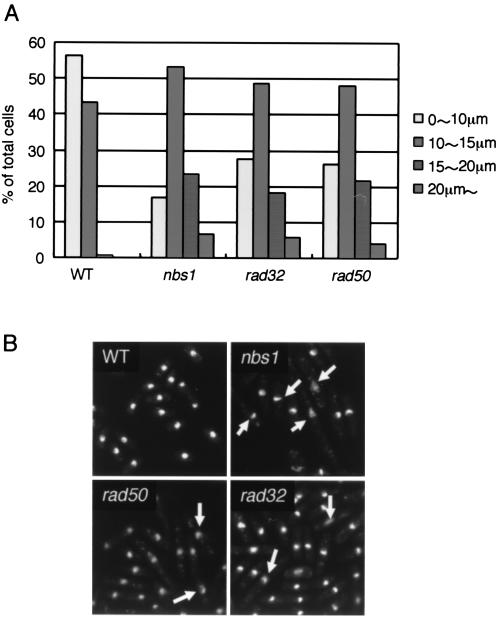
Since the growth of the nbs1Δ cells was fairly poor, we attempted to observe the cell morphology by microscopy. nbs1Δ cells as well as rad32Δ cells and rad50Δ cells were significantly elongated (Fig. 5A). rhp51Δ cells and rhp54Δ cells are also elongated (45). As the Rad32-Rad50-Nbs1 complex, Rhp51 and Rhp54 are all required for recombination repair, elongation of the cells may be due to the accumulation of DNA damage during normal mitotic growth. Such DNA damage would activate the DNA damage checkpoint, which would retard the cell cycle and cause elongation of the cells (9). Consistent with this, we observed a slight activation of the checkpoint kinase Cds1 in rad50 mutants in the absence of any DNA-damaging agents (data not shown) (3).
FIG. 5.
nbs1Δ, rad50Δ, and rad32Δ cells are all elongated and exhibit abnormal nuclear morphology. (A) Cell length of wild-type (WT, SPN114), nbs1Δ (SPN138), rad50Δ (SPN148), and rad32Δ (SPN141) cells was determined by microscopy. (B) Nuclear morphology of wild-type, nbs1Δ, rad50Δ, and rad32Δ cells. The cells were fixed with 2.5% glutaraldehyde and stained with 1 μg of DAPI per ml. Fluorescence images obtained with an epifluorescence microscope are shown. Arrows indicate nuclei with abnormal morphology. For genotypes, see Table 1.
In addition, most of the nbs1Δ cells had aberrant nuclear morphology, as visualized by 4′,6′-diamidino-2-phenylindole (DAPI) staining, whereas the nuclear chromosomal domain of wild-type cells was hemispherical or round (Fig. 5B). The nuclei of most nbs1 mutant cells appeared diffuse, and a compressed and crescent-like structure was observed within them. The rad32Δ cells and rad50Δ cells also had similar aberrant nuclear morphology (Fig. 5B). These results indicate that Nbs1, probably in a protein complex with Rad32-Rad50, is required for maintaining chromosomal structure.
Perspectives.
The abnormal nuclear morphology of the rad32, rad50, and nbs1 mutants might be related to a defect in chromatin cohesion. The depletion of a component of the cohesin complex, Rad21, from cells has been shown to result in disturbance of the nuclear organization (2), and a functional link between Rad21 and Rad50 has also been suggested (25). Such a link between the cohesin complex and the MRN complex has also been suggested in mammals. For example, human SMC1 is phosphorylated in response to ionizing radiation in an Nbs1-dependent manner (31, 68). More recently, a direct interaction between the Mre11-Rad50 complex and cohesin at DNA damage sites has been reported (30). However, the exact roles of the cohesin complex in DNA repair and the maintenance of nuclear organization remain unclear. More extensive investigations of the roles of S. pombe Nbs1 in the DNA damage response would reveal the molecular details of the DNA damage checkpoint pathway and of the possible functional link between the Nbs1 complex and cohesin in DNA repair and the maintenance of nuclear organization.
Although both the S. cerevisiae MRX complex and vertebrate MRN complexes have been suggested to be involved in both DNA repair and checkpoint signaling (14), there are several differences in the roles of these complexes in DNA repair and checkpoint signaling. For example, the S. cerevisiae MRX complex has been reported to be partially required for phosphorylation of the Rad53 checkpoint kinase in response to DSBs (23). On the other hand, activation of human CHK2, a functional homologue of S. cerevisiae Rad53, in response to ionizing radiation is intact in NBS1 cells (68). In addition, the S. cerevisiae MRX complex is required for nonhomologous end joining repair, while chicken Nbs1 is not (40, 57). Like vertebrate MRN complexes, S. pombe Rad50 is not required for nonhomologous end joining repair (36). Moreover, the sequence features of S. pombe Nbs1 are closer to those of homologues in higher eukaryotes than to those of S. cerevisiae Xrs2 (Fig. 1), suggesting that the S. pombe Rad32-Rad50-Nbs1 complex may have functional similarity to the MRN complexes of higher eukaryotes rather than to the MRX complex of S. cerevisiae.
Conclusions.
We succeeded in identifying the Nbs1 homologue in S. pombe. S. pombe nbs1+ is required for DNA repair, telomere length regulation, and maintenance of chromatin structure. The amino acid sequence features and the DNA repair function suggest that the S. pombe Rad32-Rad50-Nbs1 complex has functional similarities to the MRN complexes of higher eukaryotes. Accordingly, S. pombe Nbs1 will become a convenient model system for studying the molecular function of the Mre11 (Rad32)-Rad50-Nbs1 complex, which plays very important roles in DNA repair, the DNA damage checkpoint, and the maintenance of telomere and chromatin integrity. In addition, it will also provide clues to understanding the molecular mechanism of cancer development observed in ataxia telangiectasia and Nijmegen breakage syndrome patients.
Acknowledgments
We thank Akira Matsuura, Takashi Ushimaru, and Masahiro Uritani for discussion and sharing strains; Takeshi Saito, Shinji Yasuhira, and Hiroshi Utsumi for help with the γ-ray irradiation; and Nicholas Rhind and Paul Russell for sharing unpublished results. We also thank Masahiko Okuno and Yoshifumi Nishimura for valuable comments on the prediction of the BRCT domain. We thank Hisao Masukata for providing the S. pombe genomic libraries, Antony Carr for providing the S. pombe genomic libraries and critical reading of the manuscript, and Kenshi Komatsu for critical reading of the manuscript.
This work was supported in a part by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan to H.I., H.S., and M.U., by grants from the Human Frontier Science Program Organization (HFSPO) to H.I. and H.S., and by a grant from the Yokohama City Collaboration of Regional Entities for the Advancement of Technological Excellence, JST, to M.U. Part of this work was performed with the facilities of the Research Reactor Institute, Kyoto University.
M. Ueno and H. Iwasaki contributed equally to this work.
REFERENCES
- 1.Ahn, J. Y., X. Li, H. L. Davis, and C. E. Canman. 2002. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J. Biol. Chem. 277:19389-19395. [DOI] [PubMed] [Google Scholar]
- 2.Birkenbihl, R. P., and S. Subramani. 1995. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J. Biol. Chem. 270:7703-7711. [DOI] [PubMed] [Google Scholar]
- 3.Boddy, M. N., B. Furnari, O. Mondesert, and P. Russell. 1998. Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280:909-912. [DOI] [PubMed] [Google Scholar]
- 4.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates, 3rd, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 5.Bruschi, G. C., C. C. de Souza, M. R. Fagundes, M. A. Dani, M. H. Goldman, N. R. Morris, L. Liu, and G. H. Goldman. 2001. Sensitivity to camptothecin in Aspergillus nidulans identifies a novel gene, scaA+, related to the cellular DNA damage response. Mol. Genet. Genomics 265:264-275. [DOI] [PubMed] [Google Scholar]
- 6.Buscemi, G., C. Savio, L. Zannini, F. Micciche, D. Masnada, M. Nakanishi, H. Tauchi, K. Komatsu, S. Mizutani, K. Khanna, P. Chen, P. Concannon, L. Chessa, and D. Delia. 2001. Chk2 activation dependence on Nbs1 after DNA damage. Mol. Cell. Biol. 21:5214-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callebaut, I., and J. P. Mornon. 1997. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 400:25-30. [DOI] [PubMed] [Google Scholar]
- 8.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates, 3rd, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 9.Caspari, T., and A. M. Carr. 1999. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie 81:173-181. [DOI] [PubMed] [Google Scholar]
- 10.Chamankhah, M., and W. Xiao. 1999. Formation of the yeast Mre11-Rad50-Xrs2 complex is correlated with DNA repair and telomere maintenance. Nucleic Acids Res. 27:2072-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 12.Connelly, J. C., and D. R. Leach. 2002. Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27:410-418. [DOI] [PubMed] [Google Scholar]
- 13.Cooper, J. P., E. R. Nimmo, R. C. Allshire, and T. R. Cech. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385:744-747. [DOI] [PubMed] [Google Scholar]
- 14.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 15.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debrauwere, H., S. Loeillet, W. Lin, J. Lopes, and A. Nicolas. 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8:1129-1135. [DOI] [PubMed] [Google Scholar]
- 18.Desai-Mehta, A., K. M. Cerosaletti, and P. Concannon. 2001. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol. Cell. Biol. 21:2184-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753-32759. [DOI] [PubMed] [Google Scholar]
- 20.Farah, J. A., E. Hartsuiker, K. Mizuno, K. Ohta, and G. R. Smith. 2002. A 160-bp palindrome is a Rad50·Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 161:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuse, M., Y. Nagase, H. Tsubouchi, K. Murakami-Murofushi, T. Shibata, and K. Ohta. 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17:6412-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115-119. [DOI] [PubMed] [Google Scholar]
- 23.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 24.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95:583-586. [DOI] [PubMed] [Google Scholar]
- 25.Hartsuiker, E., E. Vaessen, A. M. Carr, and J. Kohli. 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20:6660-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopfner, K. P., L. Craig, G. Moncalian, R. A. Zinkel, T. Usui, B. A. Owen, A. Karcher, B. Henderson, J. L. Bodmer, C. T. McMurray, J. P. Carney, J. H. Petrini, and J. A. Tainer. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418:562-566. [DOI] [PubMed] [Google Scholar]
- 27.Hopfner, K. P., A. Karcher, L. Craig, T. T. Woo, J. P. Carney, and J. A. Tainer. 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105:473-485. [DOI] [PubMed] [Google Scholar]
- 28.Hopfner, K. P., C. D. Putnam, and J. A. Tainer. 2002. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12:115-122. [DOI] [PubMed] [Google Scholar]
- 29.Ito, A., H. Tauchi, J. Kobayashi, K. Morishima, A. Nakamura, Y. Hirokawa, S. Matsuura, K. Ito, and K. Komatsu. 1999. Expression of full-length NBS1 protein restores normal radiation responses in cells from Nijmegen breakage syndrome patients. Biochem. Biophys. Res. Commun. 265:716-721. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J. S., T. B. Krasieva, V. LaMorte, A. M. Taylor, and K. Yokomori. 2002. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 277:45149-45153. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S. T., B. Xu, and M. B. Kastan. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi, J., H. Tauchi, S. Sakamoto, A. Nakamura, K. Morishima, S. Matsuura, T. Kobayashi, K. Tamai, K. Tanimoto, and K. Komatsu. 2002. NBS1 localizes to gamma-H2AX foci through interaction with the forkhead-associated/BRCT domain. Curr. Biol. 12:1846-1851. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. E., D. A. Bressan, J. H. Petrini, and J. E. Haber. 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amsterdam) 1:27-40. [DOI] [PubMed] [Google Scholar]
- 34.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613-617. [DOI] [PubMed] [Google Scholar]
- 35.Liu, C., J. J. Pouliot, and H. A. Nash. 2002. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. USA 99:14970-14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manolis, K. G., E. R. Nimmo, E. Hartsuiker, A. M. Carr, P. A. Jeggo, and R. C. Allshire. 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20:210-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams, R. A. Zinkel, P. J. Farnham, and J. H. Petrini. 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21:6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuura, S., H. Tauchi, A. Nakamura, N. Kondo, S. Sakamoto, S. Endo, D. Smeets, B. Solder, B. H. Belohradsky, V. M. Der Kaloustian, M. Oshimura, M. Isomura, Y. Nakamura, and K. Komatsu. 1998. Positional cloning of the gene for Nijmegen breakage syndrome. Nat. Genet. 19:179-181. [DOI] [PubMed] [Google Scholar]
- 39.Mirzoeva, O. K., and J. H. Petrini. 2003. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 1:207-218. [PubMed] [Google Scholar]
- 40.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 43.Morishita, T., Y. Tsutsui, H. Iwasaki, and H. Shinagawa. 2002. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 22:3537-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muris, D. F., K. Vreeken, A. M. Carr, B. C. Broughton, A. R. Lehmann, P. H. Lohman, and A. Pastink. 1993. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 21:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muris, D. F., K. Vreeken, A. M. Carr, J. M. Murray, C. Smit, P. H. Lohman, and A. Pastink. 1996. Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J. Cell Sci. 109:73-81. [DOI] [PubMed] [Google Scholar]
- 46.Murray, J. M., M. Tavassoli, R. al-Harithy, K. S. Sheldrick, A. R. Lehmann, A. M. Carr, and F. Z. Watts. 1994. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol. Cell. Biol. 14:4878-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura, T. M., B. A. Moser, and P. Russell. 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161:1437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 49.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shiloh, Y. 1997. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu. Rev. Genet. 31:635-662. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, G. S., R. S. Maser, T. Stankovic, D. A. Bressan, M. I. Kaplan, N. G. Jaspers, A. Raams, P. J. Byrd, J. H. Petrini, and A. M. Taylor. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99:577-587. [DOI] [PubMed] [Google Scholar]
- 52.Sullivan, K. E., E. Veksler, H. Lederman, and S. P. Lees-Miller. 1997. Cell cycle checkpoints and DNA repair in Nijmegen breakage syndrome. Clin. Immunol. Immunopathol. 82:43-48. [DOI] [PubMed] [Google Scholar]
- 53.Sun, Z., J. Hsiao, D. S. Fay, and D. F. Stern. 1998. Rad53 forkhead-associated domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272-274. [DOI] [PubMed] [Google Scholar]
- 54.Symington, L. S. 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taggart, A. K., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023-1026. [DOI] [PubMed] [Google Scholar]
- 56.Tauchi, H., J. Kobayashi, K. Morishima, S. Matsuura, A. Nakamura, T. Shiraishi, E. Ito, D. Masnada, D. Delia, and K. Komatsu. 2001. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50·hMRE11·NBS1 complex DNA repair activity. J. Biol. Chem. 276:12-15. [DOI] [PubMed] [Google Scholar]
- 57.Tauchi, H., J. Kobayashi, K. Morishima, D. C. Van Gent, T. Shiraishi, N. S. Verkaik, D. VanHeems, E. Ito, A. Nakamura, E. Sonoda, M. Takata, S. Takeda, S. Matsuura, and K. Komatsu. 2002. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 420:93-98. [DOI] [PubMed] [Google Scholar]
- 58.Tavassoli, M., M. Shayeghi, A. Nasim, and F. Z. Watts. 1995. Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 23:383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58a.Tomita, K., A. Matsuura, T. Caspari, A. M. Carr, Y Akamatsu, H. Iwasaki, K. I. Mizuno, K. Ohta, M. Uritani, T. Ushimaru, K. Yoshinaga, and M. Ueno. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exol in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 23:5186-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trujillo, K. M., and P. Sung. 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50·Mre11 complex. J. Biol. Chem. 276:35458-35464. [DOI] [PubMed] [Google Scholar]
- 60.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273:21447-21450. [DOI] [PubMed] [Google Scholar]
- 61.Tsukamoto, Y., A. K. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11:1328-1335. [DOI] [PubMed] [Google Scholar]
- 62.Tsutsui, Y., T. Morishita, H. Iwasaki, H. Toh, and H. Shinagawa. 2000. A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a functional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics. 154:1451-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 64.Varon, R., C. Vissinga, M. Platzer, K. M. Cerosaletti, K. H. Chrzanowska, K. Saar, G. Beckmann, E. Seemanova, P. R. Cooper, N. J. Nowak, M. Stumm, C. M. Weemaes, R. A. Gatti, R. K. Wilson, M. Digweed, A. Rosenthal, K. Sperling, P. Concannon, and A. Reis. 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93:467-476. [DOI] [PubMed] [Google Scholar]
- 65.Wilson, S., M. Tavassoli, and F. Z. Watts. 1998. Schizosaccharomyces pombe Rad32 protein: a phosphoprotein with an essential phosphoesterase motif required for repair of DNA double strand breaks. Nucleic Acids Res. 26:5261-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson, S., N. Warr, D. L. Taylor, and F. Z. Watts. 1999. The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 27:2655-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, V. Lelaure, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 68.Yazdi, P. T., Y. Wang, S. Zhao, N. Patel, E. Y. Lee, and J. Qin. 2002. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, X., S. Morera, P. A. Bates, P. C. Whitehead, A. I. Coffer, K. Hainbucher, R. A. Nash, M. J. Sternberg, T. Lindahl, and P. S. Freemont. 1998. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J. 17:6404-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao, S., Y. C. Weng, S. S. Yuan, Y. T. Lin, H. C. Hsu, S. C. Lin, E. Gerbino, M. H. Song, M. Z. Zdzienicka, R. A. Gatti, J. W. Shay, Y. Ziv, Y. Shiloh, and E. Y. Lee. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405:473-477. [DOI] [PubMed] [Google Scholar]
- 71.Zhu, X. D., B. Kuster, M. Mann, J. H. Petrini, and T. Lange. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25:347-352. [DOI] [PubMed] [Google Scholar]